Abstract
Microbial cultures and/or microbial associated diseases often have a characteristic smell. Volatile organic compounds (VOCs) are produced by all microorganisms as part of their normal metabolism. The types and classes of VOC produced is wide, including fatty acids and their derivatives (e.g. hydrocarbons, aliphatic alcohols and ketones), aromatic compounds, nitrogen containing compounds, and volatile sulfur compounds. A diversity of ecological niches exist in the human body which can support a polymicrobial community, with the exact VOC profile of a given anatomical site being dependent on that produced by both the host component and the microbial species present. The detection of VOCs is of interest to various disciplines, hence numerous analytical approaches have been developed to accurately characterize and measure VOCs in the laboratory, often from patient derived samples. Using these technological advancements it is evident that VOCs are indicative of both health and disease states. Many of these techniques are still largely confined to the research laboratory, but it is envisaged that in future bedside ‘VOC profiling’ will enable rapid characterization of microbial associated disease, providing vital information to healthcare practitioners.
Introduction
Volatile organic compounds (VOCs) are produced by all microorganisms as part of their normal metabolism. Some species of bacteria have been noted to have a characteristic smell in vitro when isolated on recovery media as pure cultures, and odiferous VOC production by anaerobes in tissue infections in vivo has been recognized for many years; for example the characteristic odour of gas gangrene by cytotoxic clostridia (Boland 1929). The types and classes of VOC produced is wide, including fatty acids and their derivatives (e.g. hydrocarbons, aliphatic alcohols and ketones), aromatic compounds, nitrogen containing compounds, and volatile sulfur compounds (Schulz and Dickschat 2007).
The specific composition of VOCs produced from a given bacterial community will depend on the diversity and metabolic types/groups of species present, as well as the variety of principle carbon-energy sources available for the microbes to act upon. In the majority of cases, the largest amounts of carbon-energy are in the form of cell-impermeable polymers which require hydrolytic depolymerization before they can cross the cytoplasmic membrane, and enter into the cell (via specific transporter molecules). Figure 1 shows the main groups of macromolecules that are targeted within a host organism; polysaccharides (glycogen), proteins (e.g. collagen, albumin, globulin serum proteins, and haemoglobin), glycoproteins (mucins), lipids, phospholipids and nucleic acids. These compounds act as substrates for microbial growth via specific hydrolysis pathways (shown in figure 1) particularly during tissue infections, for example within wounds or periodontal disease (Harrington 1996). The breakdown of polymeric substrates is therefore dependent on the microflora having the right mix of hydrolytic (digesting) enzymes, and these are classified by the reactions they catalyze (enzyme commission number (EC number); table 1, Webb 1992). The majority of the microbial hydrolytic enzymes are thought to be extracellular, hence their activity is likely to benefit all species present within a microcosm. An important group of molecules in tissues and wounds are the glycosaminoglycans (mucopolysaccharides) including hyaluronic acid, chondroitin-4 and chondroitin-6, heparin and dermatan-sulfate (Wadström and Ljungh 1999). The microbial enzymes that attack these substrates are classified as hyaluronate lyases (EC 4.2.2.1; see table 1), since this is not a hydrolysis reaction, but instead involves breakage of carbon–oxygen bonds. A number of important microbial pathogens possess hyaluronate lyase (considered a ‘spreading factor’) including Staphylococcus aureus and Streptococcus pyogenes (Makris et al 2004). Collectively, the microbial enzymic degradation of available macromolecules is known to result in the production of a diversity of products, some of which are volatile.
Figure 1.
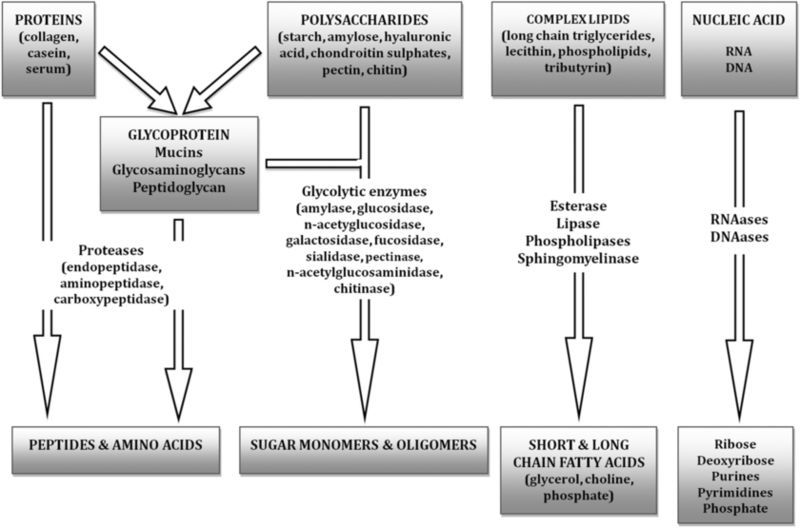
Composition and microbial hydrolysis pathways for complex feedstock. The majority of carbon-energy nutrient is in the form of soluble or insoluble polymers.
Table 1.
Common microbial hydrolytic enzymes used to breakdown polymeric substances (depolymerization).
| Microbial enzyme | EC number | Description |
|---|---|---|
| Esterase | EC 3.1: Ester bonds | Common microbial enzymes associated with breakdown and metabolism of lipids. |
| Lipase | ||
| Phosphodiesterase | ||
| Phosphatase | ||
| Nucleases (DNAase/RNAase) | ||
| DNA Glycosylases | EC 3.2: Sugars | Aid tissue breakdown. Glycoside hydrolases break off sugar chains from glycoproteins allowing them to be more easily ‘digested’ with proteases. |
| Glycoside hydrolase | ||
| Proteases (Endoprotease/Exoprotease) | EC 3.4: Peptide bonds | Cleave proteins from the termini inwards (exoproteases) or within the polypeptide chain (endoproteases). |
| Hyaluronate lyase | EC 4.2.2.1: | Cleaves hyaluronate chains (not classed as a hydrolysis reaction). |
| Carbon–oxygen bonds |
Putrefaction
Undoubtedly the earliest detection of odorous volatiles being produced by microbes would have been from the process of putrefaction. Putrefaction is known to occur when heterotrophic microorganisms (bacteria, fungi and yeast) utilize organic material from once-living tissues (plants and animals, or their products, such as milk or sewage) as a source of carbon-energy. This ‘feedstock’ has high nutritional value and the presence of easily utilizable polymeric carbohydrates, lipids, nucleic acids, fats, and proteins. Microbes breakdown these compounds using various hydrolytic enzymes (see figure 1 and table 1) and resultant products are utilized by heterotrophic species as both a source of building blocks for the synthesis of their own structural polymers (anabolism), but most frequently by the main route of catabolism, as a source of biochemical energy (NADH and ATP) obtained either from aerobic respiration (O2), anaerobic respiration (using end-terminal electron acceptors such as NO3− or SO42− in place of oxygen), or fermentation. Depending on the nature of the microbes and the local physicochemical conditions (i.e. types of organic matter, pH, salt concentration and temperature) the putrefaction process will produce VOC gases, including many that are odiferous. This includes ammonia, volatile sulfur compounds (VSC), indole and certain volatile amines, (including putrescine and cadaverine; Statheropoulos et al 2005), all products of amino acid utilization for catabolism. The amino acids and small peptides are supplied from proteins following hydrolysis and digestion by proteolytic enzymes. Therefore, microbial processes will ultimately produce a complex mixture of VOCs and/or odour depending on both intrinsic and extrinsic factors, including the composition and pH of ‘feedstock’. For example, carbohydrates do not result in major VOC odours whilst proteins and/or fats result in a variety of foul putrefactive odours (Statheropoulos et al 2005). In addition, a low pH (<6.0) is thought to allow yeasts and moulds to dominate, whilst a higher pH favours bacteria and the putrefaction processes, which will release foul-smelling VOCs (Mackie 1995), including volatile sulfide compounds (VSC), indoles and amines.
Microbial succession
In general, when complex organic compound mixtures (including those within host organisms) are exposed to complex, highly diverse mixed cultures of microorganisms, then a well described theoretical sequence of ecological shifts (succession) and corresponding metabolic ‘phases’ or stages occur. Aerobes and facultative anaerobes are the first colonizers, with high growth rate and population expansion, giving accelerated reduction of the oxygen concentration and redox. Mixed culture fermentation produces carboxylic acids that may lower the pH enabling aciduric and acidophilic species to gain a selective advantage (e.g. Lactobacillus spp., Lactococcus spp., Streptococcus spp., Propionibacterium spp.; Wilson 2005). When all small molecular weight, easy to assimilate sugars and amino acids have been fully utilized, hydrolytic mechanisms become induced or relieved of catabolite repression (Magasanik 1961) and microbial biotransformation of polymeric molecules (proteins and glycoproteins) becomes of more marked importance. The general phenomena of catabolite repression (Magasanik 1961) occurs when there is an abundance of small molecular weight (cell-transportable) nutrients (e.g. sugars or amino acids), and is a mechanism by which cells may conserve their energy (e.g. ATP) and amino-acid building blocks by repressing synthesis of enzymes involved in hydrolytic depolymerization of large molecules outside the cell. The enzymes are thus redundant under small molecule replete nutrient conditions. Even asaccharolytic species (e.g. Porphyromonas spp.) produce little proteolytic activity when grown in the presence of low molecular weight peptides and amino acids, compared to much higher amounts when grown in the presence of proteins (Minhas et al 1991).
Following proteolysis, decarboxylation reactions produce amines, and transamination and deamination reactions produce ammonium ion (Wilson 2005). The pH may then start to increase, as carbon dioxide is lost and amines and ammonia accumulate. Physicochemical conditions now favour anaerobic proteolytic species, especially strict anaerobes (e.g. Clostridium spp.). Volatile short chain fatty acids (SCFAs) will be produced through primary fermentation, producing mainly lactate, acetate, propionate and ethanol from the main carbohydrates, but is ultimately replaced by secondary fermentation, producing e.g. n-butyrate, propionate, propanol, n-butanol, and n-hexanol (Mackie 1995). The volatile SCFAs propionate and n-butyrate are produced by Propionibacterium spp. and Butyribacterium spp. respectively, as end-products of central metabolism of sugars, and additional amounts are produced by fermentation of amino acids by species such as Clostridium and Fusobacterium (MacFarlane and MacFarlane 1995). If methanogens are present (e.g. within the gastrointestinal tract) and the hydrogen partial pressure becomes sufficiently high, then acetate will be converted to methane (Jones et al 1987). Hence, the specific microbial succession (and associated microcosm metabolism) that occurs at a given anatomical location will impact on the volatiles produced.
The generalized scheme of microbial succession described above assumes that all main metabolic steps are represented within the microcosm, whereby different species can replace one another in terms of the functional metabolic groups, enabling cross feeding and full diversity of reactions and pathways. However, the human microflora is exposed to a variety of physicochemical environments at different anatomical locations, resulting in diverse but often restricted microbial ecology. For example, the diversity of species found in the large intestine of man and mammals is a strongly selected group that can withstand bile secretion and other physicochemical conditions of the gut (Wilson 2005). Skin presents a hostile environment because of its low water activity, salt content, pH, and host defence molecules, but for those strains able to tolerate these conditions easily available nutrients are present, including sweat and sebum (Roth and James 1988). In reality, the predominant microbial flora at a given anatomical site is relatively specialized, and the authors postulate that the VOC profile is therefore likely to be more characteristic of the metabolic pathways favoured by the predominant groups of organisms present (which in turn is reliant on the substrate precursors available).
VOC generating bacterial metabolic pathways
Microbial biotransformations resulting in the production of VOCs within a host can be broadly grouped into three main categories of systems. Firstly, main carbon-energy metabolic pathways (e.g. SCFAs as end products of fermentation), and whose production rate is strongly linked to growth-rate. Secondly, enzyme associated reactions whose full expression is dependent on the physicochemical environment of the cell, or presence/absence of inducer molecules. For example the generation of hydrogen sulfide by cysteine desulfhydrase in the presence of thiol-containing molecules (Starkenmann et al 2008), since even if the enzyme in the cells is fully expressed, the VOC generation rate will be rate limited depending on the concentration of enzyme substrate present (cysteine, in our example). Finally, VOCs produced from enzymatic reactions that liberate the VOCs in question from a specific type of precursor molecule produced by the mammalian host. For example, 3-hydroxy-3-methylhexanoic and other related acids are generated from amino acid, peptide or protein conjugate precursors present in axilliary secretions, via microbial aminoacylase activity (Natsch et al 2003).
Short chain fatty acid (SCFA) metabolism
Volatile SCFAs (fatty acids with <6 carbons in the aliphatic tail) are produced via numerous metabolic fermentation pathways within microbial cells, and these are summarized in table 2 along with example species which utilize each pathway. Following digestion/depolymerization, carbon-energy monomers (e.g. sugars) are metabolized into pyruvate being an intermediate in several metabolic pathways. Acetate is produced via pyruvate metabolism by many species of microorganisms, as either major or minor components of other metabolic products. Butyrate is also produced as a major end-product of pyruvate fermentation in a small number of different species of obligate anaerobe (Clostridium spp., Roseburia spp. and Eubacterium spp.; Louis and Flint 2009), which are commonly isolated from the oral cavity and gastrointestinal tract of man and animals, as well as from infected wounds. Alanine can react with α-ketoglutarate to form pyruvate and glutamate, and is then again metabolized by a whole variety of routes depending on the species of microbe, and the availability of different types of end-terminal electron acceptors. Amino acids may be deaminated, decarboxylated or transaminated depending on type. The SCFAs produced include; acetate, butyrate, propionate, butylene glycol and occasionally acetone and butanol (Mackie 1995), many of which are volatile under physiological conditions.
Table 2.
Microbial fermentation pathways. Adapted from Mackie (1995).
| Fermentation pathway | Products | Example species |
|---|---|---|
| Alcoholic | Ethanol + CO2 | Saccharomyces and other fungi |
| Zymomonas mobilis | ||
| Lactic acid | ||
| Homolactic | All pyruvate metabolized to lactate | Lactobacillus |
| Heterolactic | Pyruvate metabolized to lactic acid and other products (ethanol, CO2) | Streptococcus |
| Bacillus | ||
| Leuconostoc | ||
| Bifidobacterium bifidum | ||
| Propionic acid | Propionate (lesser amounts of acetate, lactate, succinate and CO2) | Propionibacterium pentosaceum |
| Veillonella alcalescens | ||
| Bacteroides ruminicola | ||
| Butyric acid | butyrate, acetate + H2 + CO2 (some species produce butyl alcohol, acetone, 2-proprionate ethanol or capronate) | Clostridium butyricum |
| Clostridium acetobutyricum | ||
| Clostridium kluyveri | ||
| Mixed acid | Ethanol + fatty acid mixture (acetic, lactic, formic and succinic) | Escherichia coli |
| Butanediol | 2,3-butanediol + ethanol (and smaller amounts of organic acids) | Serratia |
| Enterobacter | ||
| Erwinia | ||
| Stickland | Carboxylic acids (acetate, propionate, butyrate, isobutyrate, isovalerate) | Clostridium |
SCFA products through deamination reactions are shown in table 3, and in particular the cyclic ring amino acids phenylalanine, tyrosine and tryptophan produce interesting volatile products, including; indoleacetate, 3-methylindole (skatole), p-hydroxyphenylacetate, phenylacetate and phenylpropionate (Mackie 1995). The generalized deamination equation is
 |
1 |
Table 3.
Microbial deamination of amino acids into fatty acids. Adapted from Macfarlane and Macfarlane (1995).
| Amino acid | Deamination fatty acid product |
|---|---|
| Alanine, glycine and serine | Acetate |
| Glutamine, aspartate | Acetate, propionate and butyrate |
| Isoleucine | 2-methylbutyrate |
| Leucine | Isovalerate |
| Phenylalanine | Phenylacetate |
| Tyrosine | p-hydroxyphenylacetate, phenylacetate and phenylpropionate |
| Threonine | Propionate |
| Tryptophan | Indoleacetate and 3-methylindole |
| Valine | Isobutyrate |
SCFAs are also produced through decarboxylase reactions commonly found in bacteria, (shown in table 4), and the generalized equation is
 |
2 |
Table 4.
Microbial decarboxylation products of amino acids. Adapted from Macfarlane and Macfarlane (1995).
| Amino acid | Decarboxylase product |
|---|---|
| α-aminobutyrate | Propylamine |
| Alanine | Ethylamine |
| Arginine | Putrescine (some to pyrolidine by ring closure) and agmatine |
| Cystine | Taurine |
| Glycine | Methylamine |
| Histidine | Histamine |
| Lysine | Cadaverine (some to piperidine by ring closure) |
| Norvaline | Butylamine |
| Ornithine | Putrescine (some to pyrolidine by ring closure) |
| Phenylalanine | Phenylethylamine |
| Tyrosine | Tyramine |
| Tryptophan | Tryptamine |
Coupled oxidation and reduction reactions of amino acids to produce SCFAs is known as the Stickland reaction (see table 2) (Nisman 1954). All amino acids with exception of histidine can act as either Stickland acceptors, Stickland donors, or both donor and acceptor. The electron donor amino acid is oxidized to a volatile carboxylic acid one-carbon atom shorter than the original amino acid, whilst the electron acceptor amino acid is reduced to a volatile carboxylic acid the same length as the original amino acid (Nisman 1954).
Microbial amine production and ammonia
Many amines are bioactive (in addition to their basic properties), particularly histamine and tryptamine which are both inflammatory mediators (Allaker et al 1987), as well as cadaverine and putrescine which possess a putrid odour (Goldberg et al 1994). Trimethylamine is produced by many species of bacteria from the breakdown of choline or trimethylamine oxide (TMAO) found in meat, eggs, fish and certain vegetables (Brassica) (Groninger 1959, Yamada 1967). In fish (post mortem), TMAO is reduced by bacteria (notably Gram-negative rods) to trimethylamine (TMA), which gives rise to the characteristic off-flavour and odour of marine fish in the early stage of decay (Beatty and Gibbons 1937, Tarr 1954). In humans, TMA is produced from TMAO (in the diet) or from choline, by intestinal bacteria. From the gut, TMA is then absorbed into the bloodstream and is oxidized into odourless TMAO by the liver enzyme flavin mono-oxygenase 3 (FMO3) and excreted into urine. Trimethylaminuria (TMAU; fish odour syndrome) is a rare condition caused as a consequence of an autosomal recessive mutation in FMO3, where TMA is excreted into body fluids (including saliva and sweat) and is detected on breath giving rise to persistent or sporadic oral and body malodour (Mackay et al 2011, Whittle et al 2007). TMAO levels in the liver may vary for reasons other than TMAU mutation, including hepatic disease, anxiety and stress, diet and menstruation (Mitchell 2005, Shimizu et al 2007). A strict diet avoiding choline-rich and TMAO containing foods is advised in the management of these cases (Mitchell 2005).
Ammonium ion is a common product of many different types of reactions in microbial cells including; deamination reactions, arginine dihydrolase pathway, tryptophanase and methionine gamma lyase, and the breakdown of glutamine and glutamate. Ammonium ion production may be balanced by acid production, but if fermentable substrates are in short supply, the pH will tend to increase converting the ammonium ion present into ammonia gas. In strongly neutral buffered environments, the amount of ammonium ion production is probably not high enough to affect large pH changes. However, additional ammonium ion can be produced from the action of microbial urease enzymes on the substrate urea (found in blood, sweat, saliva and other body fluids in addition to urine), producing ammonium and bicarbonate ions (Mobley and Hausinger 1989). Bicarbonate is a weak acid and easily lost into the gas phase particularly when in the presence of carbonic anhydrase, an enzyme found in numerous microbial species, (particularly Escherichia coli; Guilloton et al 1992, Chirica et al 1997), as well as mammalian species. This allows the pH of these environments to increase and causes production of significant amounts of ammonia gas.
Volatile sulfur compounds
Although hydrogen sulfide is not organic (and so not strictly a VOC), volatile sulfur compounds (VSCs) are associated with desulfurization reactions of the sulfur containing amino acids cysteine, cystine and methionine, and plays a central role in the maintenance of a low redox potential (ideal for proteolytic anaerobes). There are two different mechanisms by which microbes produce hydrogen sulfide. One group of microbial species, collectively known as dissimilatory sulfate-reducing bacteria (SRB), reduce oxyanions of sulfer (SO4, SO3, S2O32−) as terminal electron acceptors, using cytochromes to perform ‘anaerobic respiration’ (Postgate 1984). In the mammalian oral cavity SRB (e.g. Desulfovibrio spp.) have been detected, but are generally only a small fraction of the total population (Willis et al 1995), and their presence shows a negative correlation with VSC production and oral malodour in humans (Willis et al 1999). It is much more likely that the main microbial path to hydrogen sulfide within mammalian host organisms is via desulfurization of amino acids, and also by species that can utilize glutathione or other thiol containing peptides removing the sulfur in the form of hydrogen sulfide (H2S) or methyl mercaptan (CH3SH) (Persson et al 1990). For example, Fusobacterium nucleatum is a Gram-negative, anaerobic species found in the human oral cavity with a high propensity for producing H2S (Persson et al 1990). Three or four distinct VSC producing enzymes have been isolated and characterized, including: (1) α,β-elimination of L-cysteine to produce H2S, pyruvate and ammonia; (2) β-elimination of L-cysteine to produce H2S and L-serine; (3) β-replacement reaction, where two molecules of cysteine condense to produce H2S and L-lanthione (Fukamachi et al 2002, Yoshida et al 2010, Yoshida et al 2011). A fourth mechanism has also been described involving the enzyme L-methionine γ-lyase (which normally catalyses methionine into methylmercaptan). This enzyme will produce H2S when L-methionine is replaced by L-cysteine (Kyosuke et al 2011).
Indole
Indole production from tryptophan is a property of a number of different microbial species including; E. coli (Newton and Snell 1964), Proteus vulgaris (Kamath and Yanofsky 1992), Haemophilus influenzae (Martin et al 1998), Porphyromonas gingivalis (Yoshida et al 2009) and F. nucleatum (Imamura et al 2010). The enzyme responsible is tryptophanase (more correctly named as tryptophan indole-lyase; EC4.1.99.1) and it transforms tryptophan into indole, pyruvate and ammonium ion (figure 2, Newton and Snell 1964). Indole has a low volatility coupled with a very low odour threshold. The odour has been described as foul and repellent (Tso and Adler 1974), and due to its low volatility the smell ‘lingers’ remaining for hours or days (called stench). Indole acts as a cell-to-cell signalling molecule that mediates biofilm formation in various bacteria carrying the gene for tryptophanase (Newton and Snell 1964, Martino et al 2003, Mueller et al 2009), as well as affecting the expression of several other genes, including those associated with amino acid metabolism (Wang et al 2001), multidrug exporters (Hirakawa et al 2005) and polysaccharide production (Mueller et al 2009). The enzyme consists of four identical monomers, each containing one molecule of pyridoxal 5′-phosphate (PLP). The PLP molecule forms an aldimine bond with a lysine residue (Ku et al 2006) and catalyzes the reversible hydrolytic cleavage of L-tryptophan to produce indole and ammonium pyruvate via an α,β-elimination mechanism (Snell 1975).
 |
3 |
Figure 2.
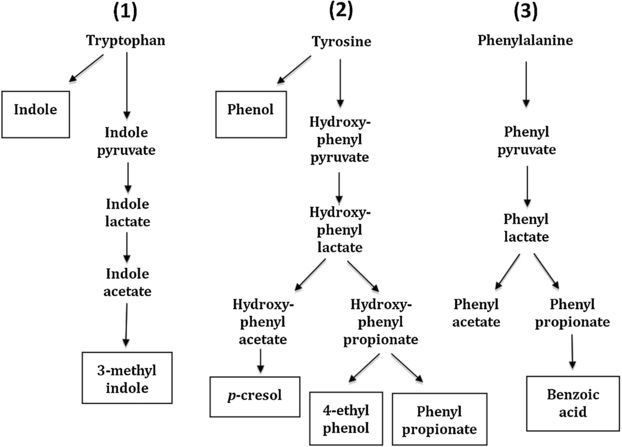
Formation of indolic (1) and phenolic (2 and 3) volatile compounds. (Adapted from Macfarlane and Macfarlane 1995.)
Methyl indole is produced from tryptophan via an indole pyruvate pathway by intestinal flora (figure 2, Yokoyama and Carlson 1974) and pathways for phenolic VOCs (phenol, p-cresol, 4-ethyl phenol, phenyl propionate, phenylacetate and benzoic acid) have also been described (figure 2, Macfarlane and Macfarlane 1995).
The effect of pH on the relative volatility of VOCs
For organic acids (including SCFAs) when the pH is above neutral they exist in the ionic dissociated (salt) form and are not volatile, whereas at decreasing pH, a greater proportion of the acid is in the associated (hydrophobic) state, hence more can partition between the liquid and gas phases. For amines, it is the other way round; when the pH is above neutral, amines exist more in their base-form, and it is only in this associated form that the compound can partition into the gas phase. Independent of the effects of pH on VOC phase transitions, pH will also variably affect the production rate of microbial VOCs, e.g. pH inducible amino decarboxylases in bacteria (Viala et al 2011). Amine production and its role in pH homeostasis in E. coli and Micrococcus lysodeikticus was described nearly 70 years ago by Gale and Epps (1942). The mechanism occurred as a consequence of low pH inducible amino acid decarboxylase enzymes, which acted on amino acids (particularly glutamate, arginine, lysine and ornithine) producing carbon dioxide and free amine base. At low pH, carbon dioxide is lost from the system (especially in the presence of microbial carbonic anhydrase activity) and the overall balance is for the system to increase the pH from acid towards neutrality. More recently it has been shown how enteric bacteria survive exposure to acidic environments, by inducing genes that provide for acid resistance and pH homeostasis (Bearson et al 1997, Hall et al 1995). E. coli has three distinct systems for acid resistance: a glucose-catabolite-repressed system and two amino acid decarboxylase-dependent systems (Castanie-Cornet et al 1999). Glutamate decarboxylase activity was thought to be the most important pathway in E. coli, the product being gamma-aminobutyric acid (GABA; which is not volatile at ambient temperatures), but interestingly lysine decarboxylase (producing the volatile cadaverine) has been shown to be central to pH homeostasis in Salmonella typhimurium (Park et al 1996). These systems overlap in ways that protect both growing and non-growing cells from acid stress across a broad range of naturally occurring acidic environments, and can result in pH dependent volatile products.
Microbial VOC detection (then and now)
The detection of VOCs is of interest to various scientific disciplines, and numerous analytical approaches have been developed to accurately characterize and measure VOCs in the laboratory. Early research studies used the Duclaux method to analyse volatile fatty acids produced by bacteria. This was based on the differential volatility of fatty acids in steam (in essence preparative chromatography), whereby sequential distillate portions could be collected and titrated with alkali to determine the concentration (Richmond 1895). The method was used very effectively to assay for formic, acetic, propionic and butyric acid (Zoller and Clark 1920), and was later improved with more refined quantitative chemical analysis of fatty acid fractions (e.g. mercuric chloride method for detection of formic acids; McNair 1933). The detection of volatile compounds has been used for many years as part of bacterial identification systems, for example the imvic (indole, methyl red, Voges-Proskauer, and citrate) system which differentiates coliforms according to their production of indole, acetoin, and mixed acid fermentation products, in addition to their ability to metabolize citrate (MacFaddin 1980). This biochemical approach to the detection of microbial volatiles continues to the present day, for example functionalized nanoporous xerogels are currently being developed for detection of the presence of acetate and indole (Crunaire et al 2011).
In most modern analytic research laboratories, the detection of microbial volatiles is carried out using gas chromatography (the separation step), and subsequent mass spectrometry, electron capture or flame ionisation detection, which coupled with analytical software enables quantification and identification of VOCs (with high sensitivity and specificity) from a given microbial species or mixed consortia. Early studies on microbial volatiles used direct gas-liquid chromatography (Gorbach et al 1976, Bogaard et al 1986), whereby compounds were identified from their retention times, using a flame ionisation detector (Ettre 1993). This requires preparation of whole microbial cultures by chemical extraction, and modified methods were developed to enable direct injection of whole microbial cultures or faeces for detection of anaerobic bacteria (Bricknell and Finegold 1973). The development of ‘head-space analysis’ of microbial cultures using gas chromatography mass spectrometry (GC-MS) provided more information, was less laborious, and was capable of automation (Larsson et al 1978a, 1978b, Zechman et al 1986). In particular, the use of mass spectrometry (via electron ionisation) has enabled qualitative and quantitative detection of microbial volatiles by measuring the mass-charge ratio of charged sample particles. Subsequent software mining of mass spectral libraries (e.g. NIST or Massfinder) then enables identification of the volatiles present based on the sample mass chromatogram produced. However, this analysis should be performed with caution since many related compounds generate similar mass spectra (Schulz and Dickschat 2007), and it has been suggested that the structure of many microbial volatiles is yet to be ‘unequivocally’ identified (Kai et al 2009). The application of head-space GC-MS has been shown capable of identifying anaerobes (Julak et al 2000, Mardh et al 1981), facultative bacteria, (Zechman et al 1986), and fungi (Larsen and Frisvad 1995). The headspace gas sampling procedure initially involved simple purge trap approaches using various adsorbent materials (e.g. charcoal, tenax, super Q), but these are thought to preferentially bind certain compounds according to their chemical composition (Kai et al 2009). Hence, as GC-MS technology has advanced, so has the sampling procedure, and pre-concentration solid phase micro-extraction (SPME; Bojko et al 2011, Julak et al 2003) and closed-loop stripping apparatus (CLSA; Hassett and Rohwer 1999) have greatly improved the sampling efficiency. GC-MS does have some disadvantages being relatively expensive and time-consuming, requiring highly skilled operators, and is reliant on the use of internal standards to identify and quantify each VC component. Therefore, although it is largely considered the ‘gold-standard’ for microbial volatile detection, other analytical approaches have been explored.
A significant development in microbial volatile analysis is soft ionisation mass spectrometry techniques, enabling direct sampling in air. For example, ion mobility mass spectrometry separates and identifies ionized molecules (including VOCs) in the gas phase based on their mobility in a carrier buffer gas, and has enabled rapid determination of microbial volatiles in humid samples (milliseconds to minutes) (Eiceman 2002, Shnayderman et al 2005). This is a promising technology with real-world applications, but suffers from being unable to differentiate all volatiles within complex samples due to insufficient sensitivity and mass resolution (Smith and Spanel 2005). To address this issue, a new approach to direct, online microbial headspace analysis has emerged in the form of proton transfer reaction mass spectrometry (PTR-MS) and selective ion flow tube mass spectrometry (SIFT-MS). These techniques are capable of determining absolute head-space concentrations of VOCs regardless of humidity, and without need for any sample preparation (Mayr et al 2003, Smith and Spanel 1996, Smith and Spanel 2005). PTR-MS uses protonated water (H3O+) as a chemical ionisation reagent enabling direct and real-time sampling and analysis of sample specimen headspace (Lindinger et al 1998). This has been used to quantify VOCs emanating from microbial cultures, enabling not only species specific volatile profile discrimination, but also temporal evolution of VOCs during bacterial growth (Bunge et al 2008, O'Hara and Mayhew 2009). In contrast, SIFT-MS uses multiple reagent ions (H3O+, O2+ and NO+) to perform soft ionisation. The use of three precursor ions enables internal verification of the VOCs detected, making identification more accurate, and enabling discrimination of isomeric and isobaric compounds (Hu and Storer 2008). The sample gas is delivered to the instrument at a constant rate where it is mixed with microwave generated reagent ions within a fast-flowing helium flow tube. The resultant reaction products are separated and counted downstream by quadrupole mass spectrometry, enabling quantitative measurements of complex volatile mixtures (Allardyce et al 2006a, Smith and Spanel 2005). Accurate identification and quantification of trace gaseous molecules is possible since the reaction time for molecular interactions is predictable within SIFT-MS instruments, being defined by predetermined rate co-efficients of the ion-molecule reactions (Smith and Spanel 1996). This technology has now been successfully applied to microbial headspace analysis (Allardyce et al 2006a, Allardyce et al 2006b), being capable of discrimination of bacterial species when coupled with multivariate analysis (Storer et al 2011, Thorn et al 2011).
Non-specific pattern recognition of microbial volatiles can be achieved through the use of electronic olfactory systems (e-nose). These comprise of non-specific, gas sensitive, chemical sensors connected to data-capture and analysis software, which generates an olfactory fingerprint of a specimen (Gibson et al 1997). Sample discrimination is then achieved through pattern recognition algorithms, principal component analysis (PCA), discriminant function analysis, cluster analysis or artificial neural networks (Spinelli et al 2010). These systems have been used successfully in detecting the presence of microbial activity (Spinelli et al 2010), and to differentiate microbial species by VOC profiling (Pavlou et al 2002a, 2002b). However, it should be noted that this is not a quantitative approach and cannot identify or provide absolute concentrations of VOCs, relying purely on comparative detection of patterns.
A new emerging approach to gas analysis is laser (absorption) and photoacoustic spectroscopy. Laser (absorption) spectroscopy, based on the attenuation of an electromagnetic signal to produce a characteristic adsorption profile, has been used for accurate detection and quantification of volatile compounds. An extensive review already exists on the use of this technology for detection of volatile sulfur compounds, whereby it was concluded that the detection limits (down to ppb) are sufficient for medical applications and could result in the future development of portable and affordable gas sensors (Ciaffoni et al 2011). Photoacoustic spectroscopy is the acoustic detection of the interaction of absorbed electromagnetic energy (light) with matter. This is based on a modulating excitation source which excites light absorbing gas molecules, subsequent transfer of kinetic energy to surrounding molecules, and generation of a sound wave which is detected by a specially designed microphone and amplifier (Chivukula et al 2007). This has been successfully used to measure complex VOCs using a spectrally tunable monochromatic radiation source, in combination with a sophisticated data analysis algorithm (Wolff et al 2011), and represents a highly sensitive and novel approach that could equally be applied to microbial volatile detection.
Relationship between VOCs and diseases or health conditions
It is widely known that microbes play an essential role in causation and prevention of disease in the human biosphere, but what is less well known is the potential role of microbial VOCs in disease processes. Many microbial VOCs have biogenic actions on mammalian cells (including cytotoxic and inflammatory effects), whereby the VOCs can be described as virulence factors or tissue aggressins. For example, experiments using Vero cells have shown carboxylic acid salts to be cytotoxic even down to concentrations of 1–2 mM, with the order of VOC cytotoxicity being: butyrate>propionate>valerate>isovalerate>isobutyrate, with human fibroblast cells being shown to be even more sensitive to the cytotoxic effects (Allaker et al 1985). Butyrate, histamine and tryptamine all produced detectible inflammatory effects on smooth muscle preparations in vitro, with 10 mM or less dosing still giving a 50% tissue response (ED50 values) particularly when added as mixtures (Greenman et al 1988). Moreover, crevicular fluid from patients with periodontal disease has been shown to contain propionate and butyrate at concentrations around 10 mM and 2.6 mM, respectively (Niederman et al 1997). Propionate and butyrate have also been shown to produce gingival inflammation in Beagle dogs when applied to the periodontium at concentrations down to 5 mM (Singer and Buckner 1981). Volatile sulfur compounds (VSCs) are a family of gases that are highly toxic to tissues even at quite low concentrations (Ng and Tonzetich 1984, Johnson et al 1992). Hydrogen sulfide (H2S) has been reported to be cytotoxic to a wide range of mammalian cells, including those found in the periodontal regions of humans (Calenic et al 2010a, 2010b, Fujimura et al 2010, Yaegaki et al 2008). It is unknown to what extent a complex mixture of microbes produces a complex mixture of cytotoxic, inflammatory or biologically active compounds (e.g. VOCs) and whether such compounds may have a synergistic effect when produced as mixtures.
The idea that complex metabolite profiles could be used to diagnose diseases in medicine was put forward by Linus Pauling and co-workers (Pauling et al 1971). They suggested the analysis of vapour and breath using gas chromatography (GC), in order to detect metabolic changes in indicative diseases, and identified over 200 different VOCs on breath condensate (Pauling et al 1971). However, the diagnosis of microbial disease through the analysis of breath samples is complicated by the diversity of VOCs present, as well as inter-individual VOC variation (Phillips et al 1999). VOC analysis for diagnosis of microbial associated disease is an emerging field in numerous areas of medical research, and will almost certainly play an important role in informing effective and appropriate treatment for individual patients.
Oral disease and malodour
Bad breath, halitosis or oral malodour, are all interchangeable terms used to describe the general condition of odiferous breath (Scully and Greenman 2012). Bad breath may rank only behind dental caries and periodontal disease as the cause of a patient's visits to the dentist, although the perception of halitosis is different in culturally diverse populations (Rayman and Almas 2008). The true prevalence of halitosis is unknown, particularly since it is not an all or nothing phenomena, and the degree to which it is severe enough to be considered socially unacceptable (also known as pathologic halitosis) will vary depending on how it is judged.
Human breath will contain VOCs that have originated from more than one source within the respiratory system. Three main VOC sources can be identified: (1) exogenous sources; VOC's inspired from background ambient gases and then expired again, (2) Extra-oral; from microbes further down the respiratory tract (oropharynx, bronchioles), or else blood-circulatory VOC's that enter the lungs and, through gaseous exchange, are removed from the body, and (3) Intra-oral; microbial VOCs from microbial biofilms that are encountered in the oral cavity. Exogenous sources depend on the ambient gases around the time and place of the sample streams, and may include industrial-synthetic sources or else VOCs emitted from plants, animals or microbes in the vicinity of the sample. The other two sources (extra-oral and intra-oral) can be differentiated by comparing lung gas samples (e.g. collected via nose) with samples obtained from the mouth, following a period of ‘holding’ the gas within the mouth (by breathing through the nose) to allow for phase equilibration with the aqueous phase (typically one or two minutes) and finally sampling the mouth gas after this period. The majority of human subjects with halitosis (80 to 90%) have oral malodour because of microbial activity from biofilms within the oral cavity (Miyazaki et al 1995). For extra-oral halitosis, (about 5–10% of all cases of halitosis), then the majority of patients have blood-borne halitosis (Tangerman and Winkel 2010). Blood-borne halitosis is frequently caused by odorous VSCs, in particular dimethyl sulfide (CH3SCH3). Extra-oral halitosis can potentially be a manifestation of a more serious disease, for which treatment is much more complicated than for intra-oral halitosis. Potential sources of (non-microbial) halitosis include liver and kidney diseases, metabolic disorders, medications and certain foods (Greenman and Saad 2009), and most of these VOCs are therefore produced by the mammalian system (van den Veld et al 2007a). It is therefore of utmost importance to differentiate between intra-oral and extra-oral halitosis. For malodour to be perceived as noticeable and unpleasant, the VOCs in question must be well above the threshold concentration for smell detection by the perceiver. The relative ‘smell’ of a pure compound can be determined by accurate dilution of the VOC in question to the point where a panel of judges can just detect it. The degree of dilution necessary to reach this point is a measure of the olfactory power (pol value) of the compound in question (Greenman et al 2004). The main unit of measurement (pol by volume) is defined as the negative log of the threshold concentration expressed in volumic or molar fractions, for example, 1 ppm = pol 6, whilst 1 ppb = pol 9 (Devos et al 1990). Approximate averaged standardized thresholds have been published for a number of compounds (Devos et al 1990).
In the population as a whole, available epidemiological data suggests that halitosis is common with estimates between 30% (Liu et al 2006, Sanz et al 2001, Ueno et al 2007) and 50% (Tessier and Kulkarni 1991), and can affect people of all ages with a tendency to increase in age (Miyazaki et al 1995). Most reports now agree that the most frequent sources of halitosis exists within the oral cavity. This includes bacterial reservoirs such as the dorsum of the tongue, saliva and periodontal pockets, where anaerobic bacteria degrade sulfur-containing amino acids to produce the foul smelling volatile sulfur compounds (VSCs); especially hydrogen sulfide (H2S), methyl mercaptan (CH3SH) and dimethyl sulfide (CH3SCH3) (Tonzetich 1971). Figure 3 illustrates the main microbial mechanisms in generation of oral malodour gases. A ‘generalized’ bacterial cell is shown since no single species can produce all the enzymes, or possesses all the pathways, for changing primary substrates into corresponding volatile compounds. However, collectively there is sufficient diversity of species within the oral cavity such that all pathways usually operate to some degree, the same being true for the diversity of hydrolytic enzymes that change secondary into primary substrates.
Figure 3.
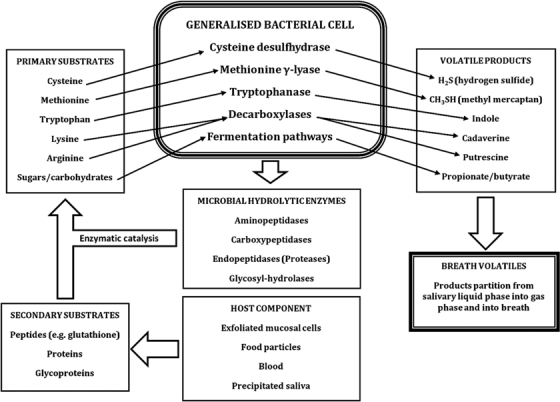
Microbial mechanisms in the generation of oral malodour gases from a ‘generalized’ bacterial cell.
The tongue biofilm coating is considered to be the most important source of VSCs, simply because it harbours the highest population of microbes (Greenman et al 2005). To date, the exact number of different species resident within the oral cavity is unknown, although several attempts have estimated numbers ranging from; 500 species (Moore et al 1985, Paster et al 2001), to 600 species (Kazor et al 2003), and up to 700 species (Aas et al 2005). Although the actual exact number will probably remain unknown due to the high diversity of resident oral flora, most estimates propose that over 50% of the microorganisms within the oral cavity are uncultivable, and their presence has only been identified by molecular rDNA testing (Aas et al 2005, Paster et al 2001). Hartley et al (1996) counted and identified microbes from tongue biofilm samples from 50 volunteers and showed a strong relationship between microbial numbers isolated (cfu cm−2) and the oral malodour status of human volunteers. For subjects grouped into low or high odour producers, significant increases in the total bacterial load and key bacterial groups, namely gram-negative anaerobes, were related to high odour, with two phenotypic characteristics (sulfide production and proteolytic activity) also strongly associated with malodour (Hartley et al 1996). Further research (Hartley et al 1999) analysed the tongue microbiota before and after use of a metronidazole mouthwash, a compound that is only effective against strict anaerobes. A reduction in oral malodour correlated closely with the reduction of strict anaerobes (predominantly Gram-negative anaerobes) whilst population levels of other species (facultative and aerobic species) remained constant throughout. Furthermore, malodour return (after 48 h) correlated with return of the strict anaerobes back to their starting numbers (Hartley et al 1999).
Although there are hundreds of different VOCs reported on breath (Van den Velde et al 2007b), the most important contributors to oral malodour are thought to be volatile sulfur compounds (VSC). The microbial degradation of cysteine residues produces H2S, and the sufhydryl anion (HS−) is a strong reducing agent that drops the redox potential (Eh) within the tongue biofilm (Kleinberg and Codipilly 2008), favouring the growth of anaerobes (McNamara et al 1972), and activating thiol-dependent enzymes including trypsin-like protease (important in the putrefaction process; Smalley et al 1994) and cysteine desulfhydrase (the very same enzymes responsible for H2S/HS− production in the first place). The detection of these compounds is complicated by the high humidity within patient breath samples, but is made possible using pre-concentration steps for GC-MS (Ochiai et al 2001), or using analytical approaches able to handle high humidity samples without need for sample preparation (e.g. SIFT-MS or PTR-MS; Smith and Spanel 2005). Most of the non-sulfur VOCs that may contribute to halitosis appear to be produced endogenously and/or in the mouth, and include acetone, 2-butanone, 2-pentanone and 1-propanol, appearing in both alveolar and mouth air, as well as indole and dimethyl selenide found mainly in alveolar air (van den Velde et al 2007a). In recent years halitosis associated sulfide detectors have been developed for this market, including; the Halimeter™ (Rosenberg et al 1991), zinc-oxide and tin-oxide semiconductor sensors (Shimura et al 1996, Maita 1996) such as the Breathtron™ (Tanda et al 2007), as well as the GC-based Oralchroma™ (Tangerman and Winkel 2008). Being compact, affordable, easy to use and capable of producing rapid results, they have been widely utilized in research and dental clinics, but lack the same level of sensitivity and specificity obtained using GC-MS or soft ionisation MS. However, the emergence of laser (absorption) spectroscopy for detection of volatile compounds may address this issue in future, being shown to have high sensitivity and specificity while potentially being scalable to bedside or chairside use (Ciaffoni et al 2011).
Periodontal disease
There are several categories of periodontal disease. The most common category (in terms of numbers of cases) is called gingivitis (inflammation of the gum tissue). This is also described as non-destructive periodontal disease since the tissue damage is limited to gum tissue without any involvement of bone or deeper tissue structures (Armitage 1999). The condition is characterized by its reversibility to normal tissue if treated appropriately, i.e. by removal or inhibition of the aetiological agents, considered to be gingival plaque as a whole (Armitage 2004). The most common form of gingivitis is termed plaque-induced gingivitis, since its magnitude relates to the amount or ‘load’ of microbes present in the gingival plaque around the gingival margin and crevice (Armitage 1999). In the absence of treatment, gingivitis may progress to periodontitis, which is a destructive form of periodontal disease. In contrast to gingivitis, there are at least six different types of termed destructive periodontal disease because they are essentially irreversible; these include; chronic periodontitis, aggressive periodontitis, periodontitis as a manifestation of systemic disease, necrotizing gingivitis/periodontitis, abscesses of the periodontium and combined periodontic-endodontic lesions (Armitage 2004). Whilst the destructive form involves specific species of periodontopathogens that have penetrated the gingival tissue, the non-destructive gingivitis might be simply the result of diffusion and penetration into gum tissue of small molecular weight biologically active molecules (e.g. VOCs butyrate, hydrogen sulfide, and/or histamine) produced by the plaque biomass ‘as a whole’ into tissue and known to be cytotoxic or inflammatory as pure compounds (Greenman et al 1988). While in some sites or individuals, gingivitis never progresses to periodontitis (Ammons et al 1972), data indicates that periodontitis is nearly always preceded by gingivitis. A group of mainly anaerobic species have been classed as ‘periodontopathogens’ and are commonly associated with chronic periodontitis. These include the proteolytic Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola, and the asaccharolytic Prevotella intermedia and Eubacterium (Loesche and Grossman 2001). Microaerophilic species associated with chronic periodontitis include Actinobacillus actinomycetemcomitans, Campylobacter rectus, and Eikenella corrodens (Loesche and Grossman 2001).
VOCs arising from saliva samples taken from periodontal disease patients (analysed immediately upon collection or upon subsequent incubation at 37 °C) have been compared for changes in VOC profile in relationship to different types of periodontal disease condition (Kostelc et al 1980, 1981). The diverse VOC profiles detected suggested that the generation rates of VOCs vary, and that increases in some VOCs related well with collagen breakdown (Kostelc et al 1980, 1981). Hydrogen sulfide is usually the most predominant VSC in chronic malodour in otherwise healthy individuals. Patients with gingivitis are thought to produce higher levels of methyl mercaptan, but not as high as those generally found in cases of periodontitis (Yaegaki and Senada 1992). Indeed, it is thought that the ratio between H2S to methyl mercaptan can be reliably used to differentiate gingivitis from periodontitis, and significant correlations have been shown between the H2S to methyl mercaptan ratio and periodontal parameters (i.e. severity) (Takeuchi et al 2010). Likely reasons for the observed differences are thought to be due to the different exposure to sulfur (S)-containing substrates. In gingivitis, the microbes are outside of the tissues and the main sources of S-containing substrates are obtained from the salivary domain. In periodontal disease, microbes are in more intimate contact with connective tissue and this is described as a serum-tissue domain, due to the presence of gingival crevicular fluid where free methionine or methionine containing peptides are at higher concentrations than in saliva.
Respiratory tract and oropharynx
The respiratory tract is the most common site of infection by pathogens and the majority are viral in nature including; adenovirus and rhinovirus (>50% of all infectious cases), influenza virus, EBV, herpes simplex, measles virus, coronavirus, respiratory syncytial virus and parainfluenza virus. Many can also cause significant infection of the throat, ear, and lungs. It is unknown to what extent virus infections would cause target mammalian cells (or migrating white blood cells) to produce a profile of VOC that would be different to normal healthy cells. In adults, respiratory infections such as tonsillitis, bronchitis, or bronchiectasis, or neoplasms, and/or the presence of tonsilloliths may be responsible for causing malodour (Castellani 1930, Rio et al 2008). VOCs from the sinuses (nasal cavity) may be detected in breath and could play a role in clinical diagnosis. In fact electronic nose technology has already been applied to this problem, and preliminary studies have shown its potential in identifying bacterial species within nose and ear swabs (Shykhon et al 2004). Interestingly, one study has shown that GC-MS headspace analysis using SPME of the VOCs from above both pure sinus-infection associated bacterial cultures and sinus mucus itself differed significantly, and that identification of bacteria-specific volatiles was possible in some cases (Preti et al 2009).
The most common bacterial infection of the throat is probably Strep. pyogenes (Group A beta-haemolytic streptococcus), but less common throat infections include Corynebacterium diphtheriae, Neisseria gonorrhoeae, Chlamydophila pneumoniae, Mycoplasma pneumoniae and Arcanobacterium haemolyticus. Strict anaerobes such as Fusobacterium may also be associated with peritonsillar abscess formation, and some cases of pharyngitis are caused by fungal infection such as Candida albicans causing oral thrush (Choby 2009). A few other causes are rare, but possibly fatal, and include parapharyngeal space infections; peritonsillar abscess (‘quinsy’) submandibular space infection (Ludwig's angina) and epiglottitis, the latter most often associated with the presence of Haemophilus influenzae type b (Johnstone and Lawy 1967). Bronchitis is characterized by inflammation of the mucous membranes of the bronchi that carry air and respiratory gases into or out from the lungs. About 90% of cases of acute bronchitis are caused by viruses, including rhinoviruses, adenoviruses, and influenza, whereas bacteria, including Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Bordetella pertussis, account for about 10% of cases (Cohen and Powderly 2004). In patients with cystic fibrosis, Staphylococcus aureus, Haemophilus influenzae, and Pseudomonas aeruginosa are the three most common organisms causing lung infections, whereas in later life 80% of patients harbour P. aeruginosa and 3.5% Burkholderia cepacia (Rowe et al 2005, Mitchell et al 2007). These respiratory disorders can result in odourous gases being expelled in air via the oral cavity and the nose (Castellani 1930, Sanz et al 2001, Yaegaki and Coil 2000). It therefore seems likely that all the main bacterial pathogens would indicate their presence in significant population numbers by emitting characteristic VOC profiles, which could then be detected in breath.
Diagnosis of respiratory tract infection through breath analysis may play a future role in identifying the various etiological agents of disease. For example, 2-aminoacetophenone has been detected (by GC-MS) in the breath of patients with cystic fibrosis, as a marker of colonisation of the lung by P. aeruginosa (Scott-Thomas et al 2010). Interestingly the volatile sample collection device significantly affected the results; when glass sampling bulbs and Tedlar® gas sampling bags were compared, this biomarker was found only to be stable within the glass sampling bulbs (Scott-Thomas et al 2010), and a more recent study has also highlighted the importance of the sampling procedure (Scott-Thomas et al 2011). Aspergillus fumigates colonisation of the respiratory tract of at-risk patients (chronic lung disease) has been associated with the presence of the volatile biomarker 2-pentylfuran in breath through GC-MS analysis (Chambers et al 2009), and this is now being developed as a novel diagnostic breath test (Chambers et al 2011). This volatile is produced by other medically important fungi including Aspergillus flavus, Aspergillus niger, Scedosporum apiospermum and Fusarium species as well as Strep. pneumoniae, and hence may have applications in other disease states. Electronic nose technology has also been applied to the diagnosis of respiratory tract infection, being shown capable of discriminating between common respiratory pathogens in vitro, including S. aureus, Strep. pneumoniae, H. influenza, and P. aeruginosa (Lai et al 2002). Moreover, it has shown good correlation with clinical pneumonia score when used to monitor breath samples from mechanically ventilated surgical intensive care patients (Hanson and Thaler 2005).
Rapid, sensitive, specific, inexpensive and non-invasive diagnostic tools are needed in order to fight tuberculosis (TB; Syhre et al 2009), and hence the use of VOC analysis technologies coupled with Mycobacterium tuberculosis specific biomarkers is being explored. This area of research has been spurred on by the observation that rats are capable of smelling the presence of TB in sputum samples, being as least as sensitive a detection method as smear microscopy (Weetjens et al 2009). Hence, non-specific e-nose studies have been shown capable of discriminating TB from non-TB cultures in vitro (Pavlou et al 2004, Fend et al 2006). Specific biomarkers of the growth of M. tuberculosis and Mycobacterium bovis cultures in vitro have also been identified (methyl phenylacetate, methyl p-anisate, methyl nicotinate and o-phenylanisole) using SPME and GC-MS (Syhre and Chambers 2008), and methyl nicotinate has shown promise as an in vivo marker of disease in the breath of patients (Syhre et al 2009). In another study the application of GC-MS consistently identified 130 volatile compounds in the headspace of Mycobacterium tuberculosis, and by using pattern recognition and fuzzy logic analysis of breath VOCs, enabled discrimination between hospitalized patients and healthy controls (Philips et al 2007). This approach has been further developed into a breath test of active pulmonary tuberculosis, by screening for the presence of both putative oxidative stress products (alkanes and alkane derivatives) and volatile metabolites of M. tuberculosis (using GC-MS) (Philips et al 2010). However, it should be noted that there is criticism of this studies’ suggestion that alkanes and alkenes are biomarkers of oxidative stress (Kwak and Preti 2011). Moreover, breath studies in general often fail to translate into diagnostic aids, largely due to the fact that many diagnostic volatiles are present in high levels in the environment, that there are high levels of inter patient variability, and often a general lack of understanding of the biosynthetic pathways involved (Kwak and Preti 2009).
Gastrointestinal system
The anaerobic microbial communities of the gut are exceedingly complex (Hold et al 2001), being characterized by high cell densities and strain diversity, and a significant degree of interspecies cross-feeding and microbial interaction (Dolfing and Gottschal 1997). The main products of microbial fermentation in the large intestine can vary significantly in their relative concentrations and production rates, depending on diet and site of production, but typically high concentrations of acetate, propionate and butyrate are produced (see figure 4), at a ratio of roughly 3:1:1 (acetate: propionate: butyrate; Cummings 1995, Cummings et al 1987). Highly-fermentable fibre residues, such as pectin, bran and highly branched starch are transformed by colonic species of bacteria into SCFAs, including significant amounts of butyrate, whereas less fermentable fibres (such as celluloses) produce fewer SCFAs (Cummings et al 2001). The production of SCFA from fibres in ruminant animals such as cattle is responsible for the high butyrate content of blood-plasma, milk and butter (Cummings et al 2001).
Figure 4.
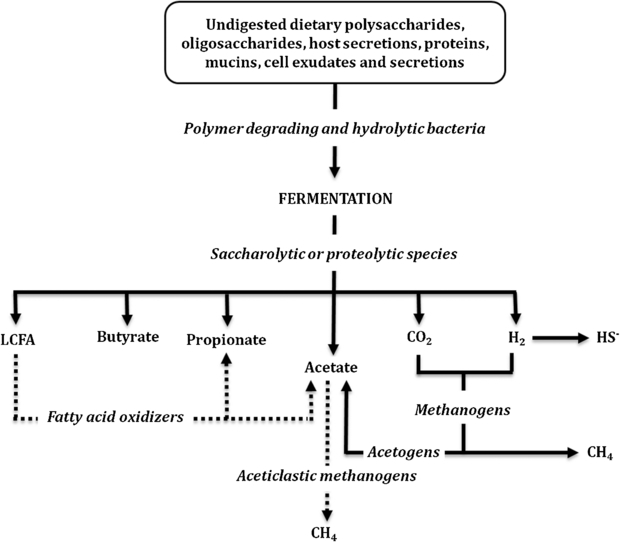
Main VOCs from fermentation in the gastrointestinal tract (large colon).
The Embden–Meyerhof–Parnas (EMP) pathway is the major human colonic pathway for the catabolism of glucose, and the formation of significant amounts of acetate by the Wood–Ljungdahl pathway (reductive acetyl co-enzymeA (CoA) Pathway). The overall fermentation in the bovine rumen also proceeds mainly via the EMP pathway, with reductive acetyl co-enzymeA (CoA) pathway accounting for up to one-third of the total acetate, probably associated with the utilization of free H2 formed as an intermediate fermentation product and the utilization of electrons by homoacetogenic, glucose-fermenting species (Miller and Wolin 1996). The other major sources of acetate are produced from the glucose carbon skeleton, probably by a combination of the homoacetate and propionate fermentations. Propionate is formed by CO2 fixation pathways, as in the Propionibacteria, and butyrate is formed from two molecules of acetyl CoA, yielding acetoacetyl-CoA, which is then converted to butyryl-CoA (Diez-Gonzalez 1999). Thereafter, butyryl-CoA may yield butyrate by two different mechanisms, either via butyrate kinase (as in some strains of ruminal Butyrivibrio fibrisolvens), or via butyryl-CoA:acetate-CoAtransferase. The overall scheme is shown in figure 4. Colonic epithelium cells use butyrate (in preference to glucose) as their main energy source (Cummings et al 1987) and it has been proposed that butyrate could provide protection from colon cancer and colitis (Archer et al 1998, Christl et al 1996). Microbial species known to produce butyrate include: Fusobacterium, Faecalibacterium, Coprococcus, Butyrivibrio and Roseburia (Barcenilla et al 2000).
Although direct VOC analysis of the GI tract would prove problematic, research into VOC analysis of the microbial populations within human stool and urine samples has been performed. Direct gas chromatography coupled with a flame ionisation detector was capable of discriminating between urinary tract infection caused by E. coli and Klebsiella spp., by analysing spent growth medium from urine samples (Manja 1983). This approach has been further developed using an e-nose system coupled to an automated headspace analyser, which is capable of screening for bacteriuria within clinical urine samples, with the same level of sensitivity as conventional culture (Aathithan et al 2001, Pavlou et al 2002b). Human faecal matter has a large microbial component and due to colonic fermentation processes gives rise to a diversity of VOCs, which are largely fatty acids and a smaller VSC component (Sato et al 2001). GC-MS analysis of ‘stool’ headspace has demonstrated that different gastrointestinal diseases produce distinct VOC profiles (Probert et al 2004, 2009). Using multivariate discriminate analysis, significant VOC profile differences have been observed between asymptomatic volunteers and patients with Campylobacter jejuni, ulcerative colitis, or Clostridium difficile infections (Garner et al 2007). Moreover, this approach has been used to characterize the VOC profile of fecal matter from premature neonates (De Lacy Costello et al 2008), and with the monitoring of certain VOC profile ‘shifts’ has potential for use as an early indicator of neonate necrotising enterocolitis (Garner et al 2009).
Vaginal infection and odour
Bacterial vaginosis is the most common cause of vaginal discharge characterized by an overgrowth of genital tract organisms and a depletion of lactobacilli (Sobel 2000). Vulvovaginal candidiasis is the next most common caused by Candida albicans (90% of cases), C. glabrata or Saccharomyces cerevisiae. Trichomoniasis is a sexually transmitted disease caused by Trichomonas vaginalis, a protozoa. On occasion, vaginal discharge may be seen in cervicitis caused by Neisseria gonorrhoeae or Chlamydia trachomatis (Buimer et al 1996).
Bacterial vaginosis is characterized by a complex change in the vaginal flora. The healthy virginal flora is dominated by Lactobacillus species that produce hydrogen peroxide and produce organic acids (mainly lactic acid) from glycogen in the vaginal epithelium; sufficient to maintain the pH to less than pH 4.7 (Vásquez et al 2002). They may also synthesize bacteriocins and other inhibitory compounds. In vaginosis, these lactobacilli are replaced by an increased concentration of other organisms, especially anaerobes such as Gardnerella vaginalis, Mycoplasma hominis, Prevotella spp., Porphyromonas spp., Bacteroides spp., anaerobic Peptostreptococcus spp., Fusobacterium spp., or Atopobium vaginae (Fredricks et al 2005). The change in the microflora at the advent of bacterial vaginosis can also be observed microscopically using Gram stain of smears from discharge samples. Gram-positive, long bacilli are predominant in a healthy microflora whilst Gram variable cocci and coccobacilli are dominant in disease. The symptoms of vaginosis can vary from asymptomatic, to one or more of the following: abnormal vaginal discharge (with an unpleasant odour), itching, burning sensation during urination and discomfort during intercourse. Gardnerella vaginalis produces proteolytic carboxylases that decompose vaginal peptides, releasing a variety of amines with fishy odour including putrescine, cadaverine and trimethylamine (Kwak and Preti 2011). These amines volatilize increasingly with rising pH, so patients often note a worsening of this symptom as the vaginal alkalinity is enhanced. Even women considered as clinically normal fall into two types based on their odorant production (Huggins and Preti (1983). These odour differences may be linked to differences in the population of bacterial flora harboured by individual females.
Skin and skin conditions
There is a large resident population of microbial species that reside and live harmlessly on the skin. The cutaneous flora consist of aerobic and facultative cocci of the micrococcaceae family (particularly Staphylococcus epidermidis), aerobic diphteroids of the genera Corynebacterium and Brevibacterium, the anaerobic diphtheroids (Propionibacterium acnes, P. granulosum, and P. avidum), and yeasts from the genus Pityrosporum (Roth and James 1988). Significant variations in both the total number of bacteria and the compositions of bacterial flora exist for different body regions, and these variations reflect differences in the amount of water and nutrients available to support bacterial growth (Roth and James 1988). For example, eccrine sweat glands, which provide water, electrolytes, and minerals; sebaceous glands, which produce a mixture of lipids; apocrine sweat glands, which along with specialized sebaceous glands secrete a substance rich in protein and lipid, all contribute to the ecology of the skin (Marples 1969). In addition, sweat contains amino acids, and minerals such as copper, iron, magnesium, zinc and calcium (Leyden et al 1987), which are important for bacterial growth and metabolism, as well as toxin production by pathogenic bacteria, for example iron dependent toxin production by Corynebacterium spp. (Wong and Groman 1984). The outermost region of the skin, the stratum corneum, is a compartment that is constantly being shed and provides a rich source of amino acids necessary for bacterial growth and metabolism. In areas of partial occlusion due to body surface-to-surface contact, such as the axilla, perineum, and toe web space, increase in CO2 tension and moisture may also be an important factor promoting growth of bacteria.
There are various skin conditions with microbial aetiological agents, including S. aureus (Feingold and Elias 1988, Hay and Addriaans 1998), Strep. pyogenes (Bisno and Stevens 1996, Hay and Addriaans 1998), S. epidermidis (Roth and James 1988, O'Brien and Collin 1992, Hay and Addriaans 1998), in addition to yeasts e.g. Candida albicans. Erythrasma is a mild, chronic, localized superficial infection of the skin caused by a group of closely related aerobic coryneform bacteria, particularly Corynebacterium minutissimum (Sarkany et al 1961, Somerville 1970). Acne vulgaris is another extremely common skin disorder that affects almost all individuals at some time during their life (Brown and Shalita 1998, Cunliffe and Simpson 1998). P. acnes is strongly associated with the condition acne vulgaris (Holland et al 1981, Brown and Shalita 1998, Holland et al 1998), and it has been hypothesized that microenvironmental changes within a pilosebaceous follicle may be important in determining the physiology of the bacterial inhabitants, with consequential effects on the follicle due to the activities of the organisms (Holland et al 1978). A number of different types of biologically active compounds are produced by P. acnes, including exocellular enzymes (Holland et al 1981), complement activating compounds (Massey et al 1978), chemotactic substances (Russell et al 1976, Gould and Cunliffe 1978), cytotoxic agents (Allaker et al 1985) and compounds that are directly inflammatory (such as the VOCs histamine and propionate) (Greenman and Holland 1985, Allaker et al 1986). In addition cell components will act as antigenic compounds. It would be expected that these differences in microbial metabolic processes would be reflected in skin VOC profiles. Some studies have now looked at volatiles emanating from skin (Riazanskaia et al 2008, Martínez-Lozano and Mora 2009), including one study which sampled, identified, and postulated on the source of a range of compounds as part of a full skin VOC profile (Gallagher et al 2008). However, few have looked directly for volatile metabolic indicators of microbial processes.
(i) Body odour from axilla
The precursors to axillary odour are produced by the apocrine glands (Shehadeh and Kligman 1963a, 1963b, Zeng et al 1992, Gower et al 1994), and the characteristic axillary odour of humans is formed by the interaction of odourless, precursor molecules found in apocrine secretions with cutaneous, axillary microorganisms (Shehadeh and Kligman 1963a, Leyden et al 1981). It is generally considered that freshly secreted sweat is sterile and that body malodour (e.g. under arm odour; UAO) is the result of biotransformation of sweat components by micro-organisms living on the surface of the skin around the apocrine glands (in the proximity of the axilla) to produce VOCs that are odoriferous. In vitro and in vivo studies of the relationship between UAO and the cutaneous microflora have shown the presence of three dominant genera (aerobic coryneforms, Micrococcaceae and propionibacteria) with no competition for habitat between these groups (Preti and Leyden 2010 and references cited therein). A relationship has also been established between severity of cases and the dominance of aerobic coryneforms within the axillary microflora populations, with Corynebacterium xerosis identified as key (Rennie et al 1990). A statistically significant association between population density of aerobic coryneform bacteria and UAO intensity was observed, whilst no such associations could be found between UAO and population densities of staphylococci, micrococci or propionibacteria (Rennie et al 1990). With regard to the production of UAO then it had been shown that Corynebacterium species are unusual in possessing ‘rare’ enzymes or metabolic pathways to biotransform 16-androstene steroids into odiferous products e.g. 5α-androstenol, 5α-androstenone, 3α(β)-androstenols, 4,16-androstadien-3-one as well as keto-substituted compounds. All these example compounds were found to be more odiferous than the 16-androstene steroids used as precursors, and found naturally in fresh apocrine sweat secretions (Rennie et al 1991). Thus the causal links to the production of high UAO were thought to be clear. However, more recent studies have shown that other pathways to odiferous VOC may be much more important regarding UAO. For example, 3-hydroxy-3-methylhexanoic acid and 3-methylhex-2-enoic acid (and other related acids) are generated via microbial aminoacylase activity from precursors naturally present in axilliary secretions (Natsch et al 2003, Natsch et al 2006). The enzyme is a zinc-dependent Nα-acyl-glutamine aminoacylase (N-AGA) produced from Corynebacterium xerosis, Corynebacterium striatum and other species of Corynebacterium, but not in Staphylococcus or Propionibacterium genus. Other precursors may include the N-terminal amino acid of apolipoprotein D that is putatively covalently bound to 3-hydroxy-3-methyl hexanoic acid, and other key odour acids in axillary sweat (Zeng et al 1996, Akiba et al 2011). However, these alternate theories regarding biogenesis still correspond with the strong correlation between a high population of corynebacteria and strong axilliary odour. These compounds are highly odiferous and their pattern of production may even be associated with MHC-allele-dependent mate selection phenomenon in rodents (Yamazaki et al 1976) and there are strong genetic influences in the patterns of VOCs in humans (Preti and Leyden 2010).
Another important class of VOC, sulfanylalkanols (3-sulfanylhexan-1-ol, 2-methyl-3-sulfanylbutan-1-ol, 3-sulfanyl-pentan-1-ol and 3-methyl-3-sulfanylhexan-1-ol), possess strong onion-sulfide smell and have been detected in axilliary odour. These VOCs are produced from C-S lyase activity of malodour-forming corynebacteria (Natsch et al 2004) and have broad substrate specificity for cysteine conjugates. The specific enzyme involved is probably the cystathionine-β-lyase enzyme (metC gene product) associated with corynebacteria but not staphylococcus (Natsch et al 2004). In addition, Staphylococcus spp. and Propionibacterium spp. are known to produce a wide range of volatile carboxylic acids, including; isovaleric, isobutyric and propionic acids, as main end-products of the primary fermentation pattern of the species, and may contribute to UAO.
(ii) Foot odour
Marshall et al (1987, 1988) examined the microflora found on normal feet and found that foot odour was associated with high population densities of staphylococci and aerobic coryneform. Feet with strong odours had significantly higher population densities of microorganisms that produced hydrolytic exoenzymes (lipase, protease, and callous degrading enzymes). Kanda et al (1990) found that isomers of SCFAs were the primary components of foot odour, and Kobayashi (1990) found that Staph. epidermidis, which is a normal resident of the skin, plays a major role in the development of foot odour. Athlete's foot (tinea pedis) is an infection of the foot caused by fungi called dermatophytes which invade the ‘dead’ outer layers of the skin (Weitzman and Summerbell 1995). The fungi thrive in warm, moist, and dark environments and the disease was thought to be fairly uncommon before human beings started wearing shoes. Pitted keratolysis occurs worldwide, seen in both tropical and temperate environments, and it can be related to occupation or sport activity. Microbial overgrowth of the normal flora occurs, with species including; Kytococcus sedentarius, Dermatophilus congolensis or species of Corynebacterium, Actinomyces or Streptomyces (Nordstrom et al 1987, Woodgyer et al 1985, Longshaw et al 2002). Under appropriate conditions such as prolonged occlusion, hyperhidrosis and increased skin surface pH, these species proliferate and produce proteinases that destroy the stratum corneum, creating pits (Holland et al 1992). Dermatophilus congolensis liberates keratinases in appropriate substrate (Hanel et al 1991), and K. sedentarius has been found to produce two keratin-degrading enzymes (Longshaw et al 2002). The malodour associated with pitted keratolysis is presumed to be the production of sulfur-compound by-products, such as thiols, sulfides, and thioesters (Nordstrom et al 1986). Strong foot odour (but without pitted skin changes) has also been described due to overgrowth of Staph. epidermidis and production of isovaleric acid from leucine (found in sweat; Ara et al 2006). Overgrowth of Bacillus subtilis and presence of Brevibacterium have also been associated with strong foot odour. The latter produces methyl mercaptan from methionine; a product of enzyme-hydrolysed skin components (Ara et al 2006).
(iii) Wound infection
When a wound is formed the integrity of the host defence mechanisms is compromised, giving rise to a new ecological niche favourable to microbial growth, which can potentially lead to an adverse effect on the process of wound healing. It is generally accepted that all acute, traumatic, surgical and chronic wounds become contaminated with microorganisms (Bowler et al 2001). This does not necessarily provide a barrier to wound healing, and it is only if a wound progresses towards infection, that there will be a failure to move through the phases of healing in a timely manner (Edwards and Harding 2004). Chronic wounds, including diabetic foot ulcers, leg ulcers and pressure ulcers, are known to be regularly colonized by a large number and diverse species of microorganisms including anaerobic organisms (Howell-Jones et al 2005), even when no clinical sign of infection is evident, possibly correlating with critical colonisation (Panuncialman and Falanga 2009). The classic sign of wound infection is inflammation (localized heat, pain, oedema and erythema), but further criteria include abscess, discharge (often purulent), discolouration (within and around wound margins), bleeding (friable) granulation tissue and wound malodour, i.e. VOCs that are odiferous (Edwards and Harding 2004).
There have been few studies of volatiles directly emanating from wounds, or wound associated clinical material, as a method of wound classification or diagnosis of infection. An early research study provided evidence for the use of direct gas-liquid chromatography of clinical wound samples (pus and serous fluid) for rapid presumptive testing of anaerobic, Gram-negative bacilli (Gorbach et al 1976), whereby isobutyric, butyric and succinic acid were determined to be signature compounds. This was corroborated by a study analysing wound pus from a variety of superficial and deep infections using the same technique, concluding that it ‘provides a rapid and reliable means for the presumptive differentiation of anaerobic from aerobic infection’ (Philips et al 1976), to the extent that this technique was integrated as a standard protocol in at least one public health laboratory. Moreover, the technique was extended to the analysis of wound swabs, and a similar diagnostic potential was observed (Reed and Sanderson 1979).
Analysis of volatiles directly emanating from fungating breast and head-and-neck cancer wounds (known to have strong odour properties), has been performed by gas chromatography-mass spectrometry-olfactometry (GC-MS-O) to separate and identify the volatile compounds present, and associate these with a corresponding human sensory perception of odour (Shirasu et al 2009). Various compounds were detected (e.g. acetic, butyric and isobutyric acid), but the main fungating wound odorant was identified as dimethyl trisulfide (C2H6S3), and although no attempt was made to identify its source, it was postulated that this may be the product of bacteria present within the wound bed (Shirasu et al 2009). Arrays of conducting polymer sensors (e-nose) have now been developed for the purpose of wound monitoring (Bailey et al 2008). One such instrument (the ‘wound-monitor’) analyses swab material, by first using a solid phase micro extraction (SPME) pre-concentration step, and then together with a pattern recognition system is hoped to assist early and rapid diagnosis of patient wounds (Persaud 2008). Early results have shown good differentiation between P. aeruginosa and S. aureus cultures (Persaud 2008). The instrument has also been shown capable of differentiating between infected and uninfected burns patients by pre-concentrating volatiles from swabs and wound dressing material, using principle component analysis (PCA) to analyse the resultant volatile spectrum (Byun et al 2010). If any such devices are to be used for direct ‘wound sniffing’ then one problem to be overcome (for all wound types), is the large diversity of human biological volatiles emanating from skin/wounds unrelated to the microbial community present. These must be further identified and characterized, so that signature microbial metabolic processes or volatile profiles can then be detected for use as a diagnostic tool. This would enable the development of a true ‘bedside’ clinical tool for rapid and non-invasive wound assessment.
In vitro microbial VOC analysis and identification: models for studying VOC production
The use of VOCs as a tool for microbial identification has been widely applied. As previously discussed, numerous existing biochemical tests are reliant on the detection of microbial VOCs in liquid phase to discriminate at the species level, for example production of indole (E. coli), hydrogen sulfide (Proteus spp. and Citrobacter spp.), and trimethylamine (E. coli and Enterobacter spp.). Modern VOC analysis tends to focus on one of two approaches (1) identifying VOC biomarkers only produced by particular species or (2) searching for a range of different VOCs, none of which are unique in themselves but together forming a unique pattern. For example, the first approach has been used in the identification of P. aeruginosa, using the grape-like odour 2-aminoacetophenone, detected by GC-MS (Cox and Parker 1979, Labows et al 1980). However, as with many other ‘species-specific’ biomarkers, there are reports of the non-specificity of this compound (Allardyce et al 2006a), and meticulous design of diagnostic approaches is required if they are to have utility in healthcare facilities.
A more recent approach has been in analysing microbial ‘volatile profiles', since this negates the need for large numbers of species-specific VOCs, and instead is reliant on commonly produced VOCs. Different species will differ in their central metabolic pathways of carbon energy sources into fermentation products (summarized in table 2). Distinct pathways include acetone-butanol fermentation (a characteristic of some Clostridium species: products include acetone, butanol and ethanol), butanediol fermentation (Aerobacter, Enterobacter, Erwinia, Klebsiella, Serratia: products include acetate, 2,3-butanediol, butylene glycol, ethanol, carbon dioxide and hydrogen), butyric-butylic fermentation (Clostridium species: products include acetate, butyrate, ethanol, isopropanol, carbon dioxide and hydrogen), homolactic fermentation (Bacillus, Lactobacillus, Streptococcus: products include lactate and small amounts of acetate), heterolactic fermentation (Bacillus, Lactobacillus, Streptococcus: products include lactate with significant amounts of acetate and ethanol), mixed acid fermentation (Escherichia, Proteus, Providencia, Salmonella, Shigella, Yersinia: products include acetate, ethanol, formate, lactate, succinate, carbon dioxide, hydrogen) and propionic fermentation (Propionibacterium: products include acetate, propionate and carbon dioxide). Therefore different species can be differentiated by reference to their principle metabolic products, some of which will be volatile, and this has enabled differentiation and identification of potentially pathogenic organisms in vitro (Pavlou et al 2002a, Thorn et al 2011).
Many different physical in vitro models have been used to study growth, metabolism and VOC production in microorganisms, and these are summarized in figure 5. All models break down into two main systems: closed (figures 5(a) and (b)) and open (figures 5(c), (d) and (e)). Open systems have nutrient flow, which brings new nutrients and removes products. Closed systems usually refer to a batch culture where the microorganisms of interest are co-incubated with media and substrates with no additional inputs (i.e. nutrients, media, substrates) or removal of outputs (except those required for analysis). A further distinction can be made for planktonic (i.e. cells in suspension; figures 5(a) and (c)) or biofilm model (figures 5(b), (d) and (e)). Analysis of VOC profiles for a wide number of bacterial species growing planktonically in batch culture mode has now been described (Bunge et al 2008, Thorn et al 2011). Although these models can reveal the presence of a wide range of characteristic VOCs for different species, the amounts produced will vary considerably over the period of incubation. In batch culture (figures 5(a) and (b)), growth of the cells goes through different phases (lag, exponential, stationary and decline) and all main physicochemical parameters (substrate concentration, product concentrations, growth rate, growth yield) are never constant. In general, this model is of limited use when trying to extrapolate to how microbes may be behaving in vivo (in response to set changes). In contrast, open systems refer to planktonic culture (figure 5(c)) or biofilm models (figures 5(d) and (e)) that have an element of continuous culture, whereby the nutrient and substrate requirements of the culture are constantly replenished and fed into the system, thus supporting continuous growth. Additionally, outputs from the biofilm (i.e. metabolites, VOCs, cells from microbial growth) are able to leave the system and are available for analysis if required, and dynamic steady states are possible (McBain 2009).
Figure 5.
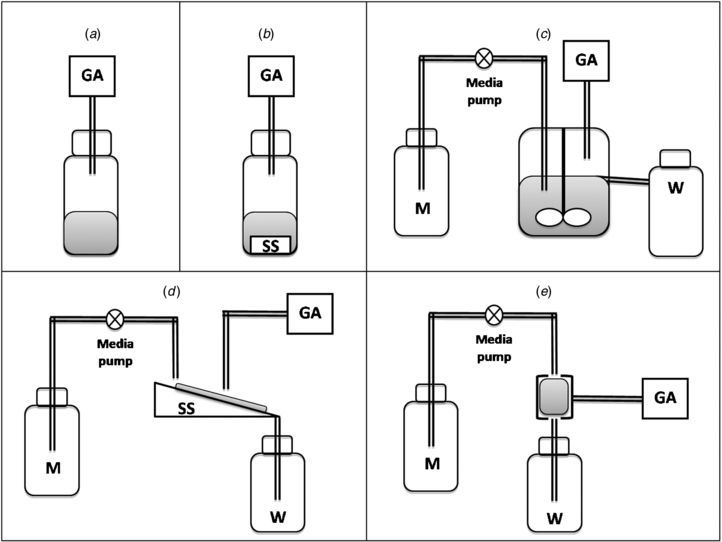
Classes of physical in vitro model for measuring VOC production rates from microbes by gas analysis (GA). (a) Planktonic batch culture; microbial headspace can be sampled from closed accumulative models. (b) Batch culture biofilm; a supporting substratum (SS) is provided to enable surface microbial growth. (c) Planktonic continuous culture; enabling steady state growth conditions, as nutrients (growth medium, M) are continuously added and waste products (W) taken away. (d) Continuous flow biofilms using impermeable substratum; nutrients (growth medium, M) are continuously added and waste products (W) taken away, enabling dynamic quasi-steady state microbial biofilms to form. (e) Continuous flow biofilms using highly permeable matrix perfusion substratum; all cells are close to nutrient micro-channels and are served with growth-limiting nutrient at approximately the same rate. More homogeneous environment with all cells growing at nutrient supply rate (i.e. µ proportional to flow rate) forming near true steady state biofilms under controlled conditions.
For the vast majority of cases of microbial infections (whether skin, oral cavity, lungs or wound) the microbes attach to a local substratum or matrix to form biofilms (Wilson 2001). Depending on the particular physicochemical conditions, the microbes utilize local nutrient substrates and multiply. The most immediate source of supply of substrates depends upon the composition of the perfusing fluids found in the nutrient domain of the biofilm. For example, supragingival dental plaque is served by nutrients derived from saliva, whilst sub-gingival plaque is served with crevicular fluid (which is a serum-derived exudate) (Marsh and Martin 1999). Typically, a small amount of growth is sustained by immediately available small molecular weight compounds (sugars, amino acids) dissolved in the fluidic domain of the biofilm. This would be blood and serum for infected wounds, bronchoalveolar lavage fluid for biofilms in the bronchus/lung, or urine and gastric fluids for urinary tract and gastrointestinal infections respectively. For modelling purposes biofilms are grown under closed (figure 5(b)) or open (figures 5(d) and (e)) conditions. For example, caries-like biofilms can be grown by inoculating dental plaque samples onto slices of hydroxyapatite, glass, or plastic surfaces and saliva periodically added or allowed to flow continuously across the biofilm. For a solid impermeable substratum (such as bone and tooth), the bacterial biofilms are only supplying nutrients from above. They form inner layers, intermediate layers and outer layers, such that inner cells are starved of nutrients by the intermediate and outer layers that are more directly exposed to salivary nutrients. A chemical gradient of growth rate limiting nutrients is formed such that the growth rate of the biofilm cells on the inside are close to zero, whilst close to the maximum dictated by local physicochemical conditions on the outside. As a result, there is a wide distribution of both growth rates and molecule residence times within the system as a whole, and hence the biofilm is heterogeneous. This is not the situation on substrata that are freely permeable. For example, biofilms formed on cellulosic or carbon-veil substrata are served with nutrients by local perfusion via micro-channels (figure 5(e)); cells are more equally supplied with nutrients and all grow at similar rates (Spencer et al 2007, Greenman et al 2008a).
In bad breath research, early experimental studies into oral malodour used a planktonic batch culture model (Tonzetich 1971, McNamara et al 1972, Tonzetich and Mcbride 1981). Kleinberg and Codipilly (1995) examined a range of gram-positive (n = 13) and gram-negative (n = 12) pure species supplemented with a range of 18 separate amino acids, and showed that gram-negative organisms produced significantly higher malodorous compounds compared to other groups. Although only batch culture, these models nevertheless helped to confirm the importance of S-metabolism in oral malodour. In dental research, a defined consortium of microorganisms is often used to investigate ecological phenomena such as species interaction, communication (quorum sensing) and competition, which would otherwise be difficult to measure for reasons of complexity. Defined consortiums make use of simplified combinations of organisms that are able to grow together when supplied with same media, and are readily identifiable by selective culture techniques. An example of a defined consortium would be the ‘Marsh consortium’ developed by Philip Marsh (CAMR, Porton down, Sailsbury, UK) for use in chemostats. This consortium comprises 10 bacterial strains (Bradshaw et al 1996), representing many of the important physiological and ecological groups that exist in plaque. Although, perhaps surprisingly VSC or microbial VOCs have been little studied using planktonic continuous culture models.
Model microcosms are stable mixed cultures of species that can grow together, being derived from real inocula from the particular ecosystem/habitat of interest. They are what remain through survival of the transfer process, from in vivo to model. The diversity of a microcosm is usually less than the original inoculum, although it is nevertheless more diverse than a defined consortium. Microcosms have the advantage of maintaining much of the complexity of the original sample, enabling the in vivo microbial community dynamics to be replicated within the biofilm model. For example, a constant depth film fermentor (CDFF) was used to develop plaque microcosm biofilms from inoculation with real plaque samples, and subsequently removed and analysed for VSC generation ex situ using a portable sulfide analyser (Pratten et al 2003). The model is an example of a continuous culture biofilm, but with biofilm formation over an impermeable substratum surface (figure 5(d)). In contrast, the ‘Sorbarod’ system is an example of a continuous flow biofilm using a highly permeable matrix (the Sorbarod; figure 5(e); Hodgson et al 1995). This system has been shown to be capable of maintaining a complex microcosm of tongue-scrape derived inoculums, and this has provided insight into the microbial ecology, physiology and VSC production of oral malodour production (Spencer et al 2007, Greenman et al 2008b, Taylor and Greenman 2010).
Future research directions
Research methods that elucidate the specific VOCs being produced by microbes or emanate from microbial associated disease processes need to be applied (e.g. GC-MS), before developing more ‘VOC focused’ or non-specific sensors (e.g. e-nose), as this information will inform and improve their development. Consequently, more research is required to further identify microbial specific VOCs, but also to ‘unequivocally’ identify the structure of these microbial associated VOCs (Kai et al 2009) so as to better enable their future detection. This situation is further complicated by the fact that the specific VOCs produced by a given microbial community in vivo will often differ from that in vitro due to varying growth substrates, presence of other (often uncultivable) competing microorganisms, and the impact of host processes. It is important to adequately define the ‘research question’ being investigated, and hence for in vitro identification systems substrates should be provided that best discriminate microbial metabolic processes as monitored through VOC analysis. However, when developing in vivo detection systems then any in vitro characterisation must attempt to best replicate the real-world environment, including provision of appropriate growth substrates, and potentially actual clinical material as has been suggested by one author investigating sinus infection (Preti et al 2009). In future it is likely that headspace gas sample VOC profiles from real conditions (in situ from a human) will be compared to an analogous model in the laboratory (see section on in vitro modelling). The differences can then be used to identify VOCs arising from the mammalian system alone, against those produced in the model (microbial alone).
Microbial infection is the root cause and/or the complicating factor of many human diseases, and traditional bacterial identification methods (e.g., culture, serological, and genetic methods) are time-consuming (from hours to days), and in many cases it may be difficult or painful to get specimens or samples (e.g. lung disease, infected wound). The ultimate goal of diagnostic VOC analysis would be online sampling of patient breath and/or body odour, enabling bedside or chairside diagnosis of their status in vivo. The rapid advancement of online VOC analysis technologies (e.g. SIFT-MS, PTR-MS) goes someway to realizing this goal, but future developments must be focused on miniaturizing technology without losing analytical or clinical sensitivity or specificity. Perhaps one of the most under-researched areas in the application of microbial VOC analysis in the clinic is time-course VOC analysis (of patient or patient samples), to assess treatment success or failure. For example, preliminary investigations have shown e-nose technology capable of following progression towards healing during wound care treatment (Edwards-Jones and Greenwood 2003). However, for this to be more widely used, VOC sampling and analysis will need to be cost effective in terms of both time and money to enable repeat patient measurements to be made.
Conclusion
It is clear that a huge array of microbial metabolic processes give rise to VOCs, and that these processes are dependent on local physicochemical conditions (e.g. anatomical location). VOC analysis as a method to monitor the progression of microbial disease (and response to treatment) has obvious advantages since it would enable timely and effective treatment decisions, reducing the need for broad spectrum antibiotics and potentially reducing the spread of microbial resistance. Moreover, being non-invasive it would have high patient acceptability. Although more research is needed to truly elucidate microbial VOC generating processes, it is envisaged that in future ‘VOC profiling’ will enable rapid characterisation of microbial associated disease, providing vital information to healthcare practitioners.
References
- Aas J A, Paster B J, Stokes L N, Olsen I, Dewhirst F E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aathithan S, Plant J C, Chaudry A N, French G L. Diagnosis of bacteriuria by detection of volatile organic compounds in urine using an automated headspace analyzer with multiple conducting polymer sensors. J. Clin. Microbiol. 2001;39:2590–3. doi: 10.1128/JCM.39.7.2590-2593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba S, Arai H, Kusuoku Y, Takagi T, Hagura K, Takeuchi K, Fuji A. The N-terminal amino acid of apolipoprotein D is putatively convalently bound to 3-hydroxy-3-methyl hexanoic acid, a key odour compound in axillary sweat. Int. J. Cosmet. Sci. 2011;33:283–6. doi: 10.1111/j.1468-2494.2010.00636.x. [DOI] [PubMed] [Google Scholar]
- Allaker R P, Greenman J, Osbourne R H, Gowers J L. Cytotoxic activity of Propionibacterium acnes and other skin organisms. Br. J. Dermatol. 1985;113:229–35. doi: 10.1111/j.1365-2133.1985.tb02069.x. [DOI] [PubMed] [Google Scholar]
- Allaker R P, Greenman J, Osborne R H. Histamine production by Propionibacterium acnes in batch and continuous culture. Microbios. 1986;48:165–72. [PubMed] [Google Scholar]
- Allaker R P, Greenman J, Osborne R H. The production of inflammatory compounds by Propionibacterium acnes and other skin organisms. Br. J. Dermatol. 1987;117:175–83. doi: 10.1111/j.1365-2133.1987.tb04114.x. [DOI] [PubMed] [Google Scholar]
- Allardyce R A, Hill A L, Murdoch D R. The rapid evaluation of bacterial growth and antibiotic susceptibility in blood cultures by selected ion flow tube mass spectrometry. Diagn. Microbiol. Infect. Dis. 2006a;55:255–61. doi: 10.1016/j.diagmicrobio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Allardyce R A, Langford V S, Hill A L, Murdoch D R. Detection of volatile metabolites produced by bacterial growth in blood culture media by selected ion flow tube mass spectrometry (SIFT-MS) J. Microbiol. Methods. 2006b;65:361–5. doi: 10.1016/j.mimet.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Ammons W F, Schectman L R, Page R C. Host tissue response in chronic periodontal disease: 1. The normal periodontium and clinical manifestations of dental and periodontal disease in the marmoset. J. Periodontol. Res. 1972;7:131–43. doi: 10.1111/j.1600-0765.1972.tb00638.x. [DOI] [PubMed] [Google Scholar]
- Ara K, Hama M, Akiba S, Koike K, Okisaka K, Hagura T, Kamiya T, Tomita F. Foot odor due to microbial metabolism and its control. Can. J. Microbiol. 2006;52:357–64. doi: 10.1139/w05-130. [DOI] [PubMed] [Google Scholar]
- Archer S, Meng S, Wu J, Johnson J, Tang R, Hodin R. Butyrate inhibits colon carcinoma cell growth through two distinctive pathways. Surgery. 1998;124:248–53. doi: 10.1016/S0039-6060(98)70127-8. [DOI] [PubMed] [Google Scholar]
- Armitage G C. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- Armitage G C. Periodontal diagnoses and classification of periodontal diseases. Periodontology. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- Bailey A L P S, Pisanelli A M, Persaud K C. Development of conducting polymer sensor arrays for wound monitoring. Sensors Actu. B. 2008;131:5–9. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [Google Scholar]
- Barcenilla A, Pryde S E, Martin J C, Duncan S H, Stewart C S, Henderson C, Flint H J. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 2000;66:1654–61. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson S, Bearson B, Foster J W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 1997;147:173–80. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- Beatty S, Gibbons N E. The measurement of spoilage in fish. J. Biol. B Can. 1937;3:77–91. [Google Scholar]
- Bisno A L, Stevens D L. Streptococcal infections of skin and soft tissues. N. Engl. J. Med. 1996;334:240–5. doi: 10.1056/NEJM199601253340407. [DOI] [PubMed] [Google Scholar]
- Bogaard A E, Hazen M J, Van Boven C P. Quantitative gas chromatographic analysis of volatile fatty acids in spent culture media and body fluids. J. Clin. Microbiol. 1986;23:523–30. doi: 10.1128/jcm.23.3.523-530.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojko B, Mirnaghi F, Pawliszyn J. Solid-phase microextraction: a multi-purpose microtechnique. Bioanalysis. 2011;3:1895–9. doi: 10.4155/bio.11.210. [DOI] [PubMed] [Google Scholar]
- Boland F K. Gas gangrene in compound fractures. Ann. Surg. 1929;90:603–13. doi: 10.1097/00000658-192910000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler P G, Duerden B I, Armstrong D G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001;14:244–69. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw D J, Marsh P D, Schilling K M, Cummins D. A modified chemostat system to study the ecology of oral biofilms. J. Appl. Bacteriol. 1996;80:124–30. doi: 10.1111/j.1365-2672.1996.tb03199.x. [DOI] [PubMed] [Google Scholar]
- Bricknell K S, Finegold S M. A simple, rapid method to process and assay fatty acids and alcohols by gas chromatography. Anal. Biochem. 1973;51:23–31. doi: 10.1016/0003-2697(73)90449-1. [DOI] [PubMed] [Google Scholar]
- Brown S K, Shalita A R. Acne vulgaris. Lancet. 1998;351:1871–6. doi: 10.1016/S0140-6736(98)01046-0. [DOI] [PubMed] [Google Scholar]
- Buimer M, van Doornum G J, Ching S, Peerbooms P G, Plier P K, Ram D, Lee H H. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by ligase chain reaction-based assays with clinical specimens from various sites: implications for diagnostic testing and screening. J. Clin. Microbiol. 1996;34:2395–400. doi: 10.1128/jcm.34.10.2395-2400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge M, Araghipour N, Mikoviny T, Dunkl J, Schnitzhofer R, Hansel A, Schinner F, Wisthaler A, Margesin R, Mark T D. On-line monitoring of microbial volatile metabolites by proton transfer reaction-mass spectrometry. Appl. Environ. Microbiol. 2008;74:2179–86. doi: 10.1128/AEM.02069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun H, Pisanelli A M, Persaud K C. Wound state monitoring for burn patients using e-nose/SPME system. ETRI J. 2010;32:440–6. doi: 10.4218/etrij.10.0109.0300. [DOI] [Google Scholar]
- Calenic B, Yaegaki K, Murata T, Imai T, Aoyama I, Sato T, Ii H. Oral malodorous compound triggers mitochondrial-dependent apoptosis and causes genomic DNA damage in human gingival epithelial cells. J. Periodontal. Res. 2010a;45:31–7. doi: 10.1111/j.1600-0765.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- Calenic B, Yaegaki K, Kozhuharova A, Imai T. Oral malodorous compound causes oxidative stress and p53-mediated programmed cell death in keratinocyte stem cells. J. Periodontol. 2010b;81:1317–23. doi: 10.1902/jop.2010.100080. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet M-P, Penfound T A, Smith D, Elliott J F, Foster J W. Control of acid resistance in Escherichia coli. J. Bacteriol. 1999;181:3525–35. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani A. Foetor oris of tonsillar origin and certain bacilli causing it. Lancet. 1930;215:623–4. doi: 10.1016/S0140-6736(00)57189-X. [DOI] [Google Scholar]
- Chambers S T, Syhre M, Murdoch D R, McCartin F, Epton M J. Detection of 2-pentylfuran in the breath of patients with Aspergillus fumigatus. Med. Mycol. 2009;47:468–76. doi: 10.1080/13693780802475212. [DOI] [PubMed] [Google Scholar]
- Chambers S T, Bhandari S, Scott-Thomas A, Syhre M. Novel diagnostics: progress toward a breath test for invasive Aspergillus fumigatus. Med. Mycol. 2011;49:S54–61. doi: 10.3109/13693786.2010.508187. [DOI] [PubMed] [Google Scholar]
- Chirica L C, Elleby B, Jonsson B H, Lindskog S. The complete sequence, expression in Escherichia coli, purification and some properties of carbonic anhydrase from Neisseria gonorrhoeae. Eur. J. Biochem. 1997;244:755–60. doi: 10.1111/j.1432-1033.1997.00755.x. [DOI] [PubMed] [Google Scholar]
- Chivukula V S, Shur M S, Čiplys D. Recent advances in application of acoustic, acousto-optic and photoacoustic methods in biology and medicine. Solid State Phys. 2007;204:3209–36. doi: 10.1002/pssa.200723313. [DOI] [Google Scholar]
- Choby B A. Diagnosis and treatment of streptococcal pharyngitis. Am. Fam. Physician. 2009;79:383–90. [PubMed] [Google Scholar]
- Christl S U, Eisner H-D, Dusel G, Kasper H, Scheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig. Dis. Sci. 1996;41:2477–81. doi: 10.1007/BF02100146. [DOI] [PubMed] [Google Scholar]
- Ciaffoni L, Peverall R, Ritchie G A. Laser spectroscopy on volatile sulfur compounds: possibilities for breath analysis. J. Breath Res. 2011;5:024002. doi: 10.1088/1752-7155/5/2/024002. [DOI] [PubMed] [Google Scholar]
- Cohen J, Powderly W. Infectious Diseases. 2nd edn. Amsterdam: Elsevier; 2004. Bronchitis, bronchiectasis, and cystic fibrosis. [Google Scholar]
- Cox C D, Parker J. Use of 2-aminoacetophenone production in identification of Pseudomonas aeruginosa. J. Clin. Microbiol. 1979;9:479–84. doi: 10.1128/jcm.9.4.479-484.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunaire S, Marcoux P R, Guillemot L H, Mallard F, Tran-Thi T H, Ngo K Q. [O02] Discriminating bacteria with functionalized nanoporous xerogels. 2nd Int. Conf. on Bio-Sensing Technology; Amsterdam. 2011. [Google Scholar]
- Cummings J H, Pomare E W, Branch W J, Naylor C P E, Macfarlane G T. Short chain fatty acids in the human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–7. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J H. Short chain fatty acids. In: Gibson G R, MacFarlane G T, editors. Human Colonic Bacteria. Michegan: CRC Press; 1995. pp. pp 101–30. [Google Scholar]
- Cummings J H, Macfarlane G T, Englyst H N. Prebiotic digestion and fermentation. Am. J. Clin. Nutr. 2001;73:415–20. doi: 10.1093/ajcn/73.2.415s. [DOI] [PubMed] [Google Scholar]
- Cunliffe W J, Simpson N B. Disorders of sebaceous glands. In: Champion R H, Burton J L, Burns D A, Brethnach S M, editors. Textbook of Dermatology. Milan: Blackwell; 1998. [Google Scholar]
- De Lacy Costello B, Ewen R, Ewer A K, Garner C E, Probert C S, Ratcliffe N M, Smith S J. An analysis of volatiles in the headspace of the faeces of neonates. Breath Res. 2008;2:037023. doi: 10.1088/1752-7155/2/3/037023. [DOI] [PubMed] [Google Scholar]
- Devos M, Patte F, Rouault J, Laffort P, Van Gemert L J. Standardized Human Olfactory Thresholds. 1st edn. New York: Oxford University Press; 1990. [Google Scholar]
- Diez-Gonzalez F, Bond D R, Jennings E, Russell J B. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch. Microbiol. 1999;171:324–30. doi: 10.1007/s002030050717. [DOI] [PubMed] [Google Scholar]
- Dolfing J, Gottschal J C. Microbe–microbe interactions. In: Mackie R I, White B A, Isaacson R E, editors. Gastrointestinal Microbiology . vol 2. New York: Chapman and Hall; 1997. pp. pp 373–433. [Google Scholar]
- Edwards-Jones V, Greenwood J E. What's new in burn microbiology? James Laing memorial prize essay 2000. Burns. 2003;29:15–24. doi: 10.1016/S0305-4179(02)00203-6. [DOI] [PubMed] [Google Scholar]
- Edwards R, Harding K G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004;17:91–6. doi: 10.1097/00001432-200404000-00004. [DOI] [PubMed] [Google Scholar]
- Eiceman G A. Ion-mobility spectrometry as a fast monitor of chemical composition. Trends Anal. Chem. 2002;21:259–75. doi: 10.1016/S0165-9936(02)00406-5. [DOI] [Google Scholar]
- Ettre L S. International union of pure and applied chemistry analytical chemistry division commission on chromatography and other analytical separations. Commission on analytical nomenclature for chromatography (IUPAC Recommendations, 1993) Pure Appl. Chem. 1993;65:81–72. doi: 10.1351/pac199365040819. [DOI] [Google Scholar]
- Feingold K R, Elias P M. Endocrine-skin interactions. Cutaneous manifestations of adrenal disease, pheochromocytomas, carcinoid syndrome, sex hormone excess and deficiency, polyglandular autoimmune syndromes, multiple endocrine neoplasia syndromes, and other miscellaneous disorders. J. Am. Acad. Dermatol. 1988;19:1–20. doi: 10.1016/S0190-9622(88)70146-2. [DOI] [PubMed] [Google Scholar]
- Fend R, Kolk A H, Bessant C, Buijtels P, Klatser P R, Woodman A C. Prospects for clinical application of electronic-nose technology to early detection of Mycobacterium tuberculosis in culture and sputum. J. Clin. Microbiol. 2006;44:2039–45. doi: 10.1128/JCM.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks D N, Fiedler T L, Marrazzo J M. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 2005;353:1899–911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Calenic B, Yaegaki K, Murata T, Ii H, Imai T, Sato T, Izumi Y. Oral malodorous compound activates mitochondrial pathway inducing apoptosis in human gingival fibroblasts. Clin. Oral Investig. 2010;14:367–73. doi: 10.1007/s00784-009-0301-5. [DOI] [PubMed] [Google Scholar]
- Fukamachi H, Nakano Y, Yoshimura M, Koga T. Cloning and characterization of the L-cysteine desulfhydrase gene of Fusobacterium nucleatum. FEMS Microbiol. Lett. 2002;215:75–80. doi: 10.1111/j.1574-6968.2002.tb11373.x. [DOI] [PubMed] [Google Scholar]
- Gale E F, Epps H M R. The effect of the pH of the medium during growth on the enzymic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem. J. 1942;36:600–19. doi: 10.1042/bj0360600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher M, Wysocki C J, Leyden J J, Spielman A I, Sun X, Preti G. Analyses of volatile organic compounds from human skin. Br. J. Dermatol. 2008;159:780–91. doi: 10.1111/j.1365-2133.2008.08748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C E, Smith S, Bardhan P K, Ratcliffe N M, Probert C S. A pilot study of faecal volatile organic compounds in faeces from cholera patients in Bangladesh to determine their utility in disease diagnosis. Trans. R. Soc. Trop. Med. Hyg. 2009;103:1171–3. doi: 10.1016/j.trstmh.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Garner C E, Smith S, Costello B dL, White P, Spencer R, Probert C S J, Ratcliffe N M. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007;21:1675–88. doi: 10.1096/fj.06-6927com. [DOI] [PubMed] [Google Scholar]
- Gibson T D, Prosser O, Hulbert J N, Marshall R W, Corcoran P, Lowery P, Ruck-Keene E A, Heron S. Detection and simultaneous identification of microorganisms from headspace samples using an electronic nose. Sensors Actuators B. 1997;44:413–22. doi: 10.1016/S0925-4005(97)00235-9. [DOI] [Google Scholar]
- Goldberg S, Kozlovsky A, Gordon D, Gelernter I, Sintov A, Rosenberg M. Cadaverine as a putative component of oral malodor. J. Dent. Res. 1994;73:1168–72. doi: 10.1177/00220345940730060701. [DOI] [PubMed] [Google Scholar]
- Gorbach S L, Mayhem J W, Bartlett J G, Thadepalli H, Onderdonk A B. Rapid diagnosis of anaerobic infections by direct gas-liquid chromatography of clinical specimens. J. Clin. Investig. 1976;57:478–84. doi: 10.1172/JCI108300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould D J, Cunliffe W J. The long-term treatment of acne vulgaris. Clin. Exp. Dermatol. 1978;3:249–52. doi: 10.1111/j.1365-2230.1978.tb01495.x. [DOI] [PubMed] [Google Scholar]
- Gower D B, Holland K T, Mallet A I, Rennie P J, Watkins W J. Comparison of 16-Androstene steroid concentrations in sterile apocrine sweat and axillary secretions: Interconversions of 16-Androstenes by the axillary microflora—a mechanism for axillary odour production in man? J. Steroid Biochem. Mol. Biol. 1994;48:409–18. doi: 10.1016/0960-0760(94)90082-5. [DOI] [PubMed] [Google Scholar]
- Greenman J, Duffield J, Spencer P, Rosenberg M, Corry D, Saad S, Lenton P, Majerus G, Nachnani S, El-Maaytah M. Study on the organoleptic intensity scale for measuring oral malodor. J. Dent. Res. 2004;83:81–5. doi: 10.1177/154405910408300116. [DOI] [PubMed] [Google Scholar]
- Greenman J, Holland K T. Effects of dilution rate on biomass and extracellular enzyme production by three species of cutaneous propionibacteria grown in continuous culture. J. Gen. Microbiol. 1985;131:1619–24. doi: 10.1099/00221287-131-7-1619. [DOI] [PubMed] [Google Scholar]
- Greenman J, Ieropoulos I, Melhuish C. Biological computing using perfusion anodophile biofilm electrodes (PABE) Int. J. Unconv. Comput. 2008a;4:23–32. [Google Scholar]
- Greenman J, Mckenzie C, Saad S, Wiegand B, Zguris J C. Effects of chlorhexidine on a tongue-flora microcosm and VSC production using an in vitro biofilm perfusion model. J. Breath Res. 2008b;2:046005. doi: 10.1088/1752-7155/2/4/046005. [DOI] [PubMed] [Google Scholar]
- Greenman J, Osborne R H, Allaker R P. The production of potentially inflammatory compounds by human dental plaque and species of periodontal bacteria. Microb. Ecol. Health D. 1988;1:245–53. doi: 10.3109/08910608809140529. [DOI] [Google Scholar]
- Greenman J, Saad B M. Relating breath malodour to food constituents and oral health. In: Wilson M, editor. In: Food Constituents and Oral Health: Current Status and Future Prospects. Cambridge: Woodhead Publishing; 2009. (Woodhead Food Series 172). [Google Scholar]
- Greenman J, Spencer P, McKenzie C, Saad S, Duffield J. Review: in vitro models for Oral Malodor. Oral Dis. 2005;11:14–23. doi: 10.1111/j.1601-0825.2005.01082.x. [DOI] [PubMed] [Google Scholar]
- Groninger S. Occurrence and Significance of Trimethylamine Oxide in Marine Animals. Special Scientific Report: Fisheries 333. 1959 (Washington, DC: United States Fish and Wildlife Service)
- Guilloton M B, Korte J J, Lamblin A F, Fuchs J A, Anderson P M. Carbonic anhydrase in Escherichia coli. A product of the cyn operon. J. Biol. Chem. 1992;267:3731–4. [PubMed] [Google Scholar]
- Hall H K, Karem K L, Foster J W. Molecular responses of microbes to environmental pH stress. Adv. Microb. Physiol. 1995;37:229–72. doi: 10.1016/S0065-2911(08)60147-2. [DOI] [PubMed] [Google Scholar]
- Hanel H, Kalisch J, Keil M, Marsch W C, Buslau M. Quantification of keratinolytic activity from Dermatophilus congolensis. Med. Microbiol. Immunol. 1991;180:45–51. doi: 10.1007/BF00191700. [DOI] [PubMed] [Google Scholar]
- Hanson C W, Thaler E R. Electronic nose prediction of a clinical pneumonia score: biosensors and microbes. Anesthesiology. 2005;102:63–8. doi: 10.1097/00000542-200501000-00013. [DOI] [PubMed] [Google Scholar]
- Harrington D J. Bacterial collagenases and collagen-degrading enzymes and their potential role in human disease. 1996;64:1885–91. doi: 10.1128/iai.64.6.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley M G, El-Maaytah M A, McKenzie C, Greenman J. The tongue microbiota of low odour and malodorous individuals. Microbiol. Ecol. Health Dis. 1996;9:215–23. doi: 10.3109/08910609609166462. [DOI] [Google Scholar]
- Hartley M G, McKenzie C, Greenman J, El Maaytah M, Scully C, Porter S R. Tongue microbiota and malodour: effects of metronidazole mouthrinse on tongue microbiota and breath odour levels. Microbiol. Ecol. Health Dis. 1999;11:226–33. doi: 10.1080/08910609908540832. [DOI] [Google Scholar]
- Hassett A J, Rohwer E R. Analysis of odorous compounds in water by isolation by closed-loop stripping with a multichannel silicone rubber trap followed by gas chromatography–mass spectrometry. J. Chromatogr. A. 1999;849:521–8. doi: 10.1016/S0021-9673(99)00621-4. [DOI] [PubMed] [Google Scholar]
- Hay R J, Addriaans B M. Bacterial infection. In: Champion R H, Burton J L, Burns D A, Breathnach S M, editors. Textbook of Dermatology. 6th edn. Oxford: Blackwell; 1998. pp. pp 1097–179. [Google Scholar]
- Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 2005;55:1113–26. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- Hodgson A E, Nelson S M, Brown M R W, Gilbert P. A simple in vitro model for growth control of bacterial biofilms. J. Appl. Bacteriol. 1995;79:87–93. doi: 10.1111/j.1365-2672.1995.tb03128.x. [DOI] [PubMed] [Google Scholar]
- Hold G L, Pryde S E, Russell V J, Furrie E, Flint H J. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 2001;39:33–9. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- Holland K T, Aldana O, Bojar R A, Cunliffe W J, Eady E A, Holland D B, Ingham E, McGeown C, Till A, Walters C. Propionibacterium acnes and acne. Dermatology. 1998;196:67–8. doi: 10.1159/000017870. [DOI] [PubMed] [Google Scholar]
- Holland K T, Cunliffe W J, Roberts C D. The role of bacteria in acne vulgaris: a new approach. Clin. Exp. Dermatol. 1978;3:253–7. doi: 10.1111/j.1365-2230.1978.tb01496.x. [DOI] [PubMed] [Google Scholar]
- Holland K T, Ingham E, Cunliffe W J. A review, the microbiology of acne. J. Appl. Bacteriol. 1981;51:195–215. doi: 10.1111/j.1365-2672.1981.tb01234.x. [DOI] [PubMed] [Google Scholar]
- Holland K T, Marshall J, Taylor D. The effect of dilution rate and pH on biomass and proteinase production by Micrococcus sedentarius grown in continuous culture. J. Appl. Bacteriol. 1992;72:429–34. doi: 10.1111/j.1365-2672.1992.tb01856.x. [DOI] [PubMed] [Google Scholar]
- Howell-Jones R S, Wilson M J, Hill K E, Howard A J, Price P E, Thomas D W. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J. Antimicrob. Chemother. 2005;55:143–9. doi: 10.1093/jac/dkh513. [DOI] [PubMed] [Google Scholar]
- Hu W P, Storer M K. Medical applications of SIFT-MS in New Zealand. Chem. NZ. 2008;72:51–4. [Google Scholar]
- Huggins G R, Preti G. Diagnostic clues from vaginal odors. Contemp. OB/GYN. 1983;22:199–220. [Google Scholar]
- Imamura T S, Yano A, Yoshida Y. Production of indole from l-Tryptophan and effects of these compounds on biofilm formation by Fusobacterium nucleatum ATCC 25586. Appl. Environ. Microbiol. 2010;76:4260–8. doi: 10.1128/AEM.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P W, Yaegaki K, Tonzetich J. Effect of volatile thiol compounds on protein metabolism by human gingival fibroblasts. J. Periodontal Res. 1992;27:553–61. doi: 10.1111/j.1600-0765.1992.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Johnstone J M, Lawy H S. Acute epiglottitis in adults due to infection with Haemophilus influenzae type b. Lancet. 1967;15:134–6. doi: 10.1016/S0140-6736(67)92968-6. [DOI] [PubMed] [Google Scholar]
- Jones W J, Nagle D P, Whitman W B. Methanogens and the diversity of archaebacteria. Microbiol. Rev. 1987;51:135–77. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julak J, Prochazkova-Francisci E, Stranska E, Rosova V. Evaluation of exudates by solid phase microextraction-gas chromatography. J. Microbiol. Methods. 2003;52:115–22. doi: 10.1016/S0167-7012(02)00148-3. [DOI] [PubMed] [Google Scholar]
- Julak J, Stranska E, Prochazkova-Francisci E, Rosova V. Blood cultures evaluation by gas chromatography of volatile fatty acids. Med. Sci. Monit. 2000;6:605–10. [PubMed] [Google Scholar]
- Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 2009;81:1001–12. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- Kamath A V, Yanofsky C. Characterization of the tryptophanase operon of Proteus vulgaris. Cloning, nucleotide sequence, amino acid homology, and in vitro synthesis of the leader peptide and regulatory analysis. J. Biol. Chem. 1992;267:19978–85. [PubMed] [Google Scholar]
- Kanda F, Yagi E, Fukuda M, Nakajima K, Ohta T, Nakata O. Elucidation of chemical compounds responsible for foot malodour. Br. J. Dermatol. 1990;122:771–6. doi: 10.1111/j.1365-2133.1990.tb06265.x. [DOI] [PubMed] [Google Scholar]
- Kazor C E, Mitchell P M, Lee A M, Stokes L N, Loesche W J, Dewhirst F E, Paster B J. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 2003;41:558–63. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberg I, Codipilly D. H2S generation and Eh reduction in cysteine challenge testing as a means of determining the potential of test products and treatments for inhibiting oral malodour. J. Breath Res. 2008;2:017018. doi: 10.1088/1752-7155/2/1/017018. [DOI] [PubMed] [Google Scholar]
- Kobayashi S. Relationship between an offensive smell given off from human foot and Staphylococcus epidermidis. Nihon Saikingaku Zasshi. 1990;45:797–800. doi: 10.3412/jsb.45.797. [DOI] [PubMed] [Google Scholar]
- Kostelc J G, Preti G, Zelson P R, Stoller N H, Tonzetich J. Salivary volatiles as indicators of periodontitis. J. Periodontal Res. 1980;15:185–92. doi: 10.1111/j.1600-0765.1980.tb00273.x. [DOI] [PubMed] [Google Scholar]
- Kostelc J G, Zelson P R, Preti G, Tonzetich J. Quantitative differences in volatiles from healthy and diseased human mouths with periodontitis. Clin. Chem. 1981;27:842–5. [PubMed] [Google Scholar]
- Ku S Y, Yip P, Howell P L. Structure of Escherichia coli tryptophanase. Acta Crystallogr. D Biol. Crystallogr. 2006;62:814–23. doi: 10.1107/S0907444906019895. [DOI] [PubMed] [Google Scholar]
- Kwak J, Preti G. Volatile disease biomarkers in breath: a critique. Curr. Pharm. Biotechnol. 2011;12:1067–74. doi: 10.2174/138920111795909050. [DOI] [PubMed] [Google Scholar]
- Kyosuke S, Yoshida Y, Nagano K, Yoshimura F. Identification of an L-methionine γ-lyase involved in the production of hydrogen sulfide from L-cysteine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology. 2011;157:2992–3000. doi: 10.1099/mic.0.051813-0. [DOI] [PubMed] [Google Scholar]
- Labows J N, McGinley K J, Webster G F, Leyden J J. Headspace analysis of volatile metabolites of Pseudomonas aeruginosa and related species by gas chromatography-mass spectrometry. J. Clin. Microbiol. 1980;12:521–6. doi: 10.1128/jcm.12.4.521-526.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S Y, Deffenderfer O F, Hanson W, Phillips M P, Thaler E R. Identification of upper respiratory bacterial pathogens with the electronic nose. Laryngoscope. 2002;112:975–9. doi: 10.1097/00005537-200206000-00007. [DOI] [PubMed] [Google Scholar]
- Larsen T O, Frisvad J C. Chemosystematics of Penicillium based on profiles of volatile metabolites. Mycol. Res. 1995;99:1167–74. doi: 10.1016/S0953-7562(09)80272-4. [DOI] [Google Scholar]
- Larsson L, Mardh P A, Odham G. Analysis of amines and other bacterial products by head-space gas chromatography. Acta Pathol. Microbiol. Scand. B. 1978a;86:207–13. doi: 10.1111/j.1699-0463.1978.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Larsson L, Mardh P A, Odham G. Detection of alcohols and volatile fatty-acids by head-space gas-chromatography in identification of anaerobic bacteria. J. Clin. Microbiol. 1978b;7:23–7. doi: 10.1128/jcm.7.1.23-27.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden J J, McGinley K J, Hoelzle K, Labows J, Kligman A M. The microbiology of the human axillae and its relation to axillary odors. J. Invest. Dermatol. 1981;77:413–6. doi: 10.1111/1523-1747.ep12494624. [DOI] [PubMed] [Google Scholar]
- Leyden J J, McGinley K J, Nordstrom K M, Webster G F. Skin microflora. J. Invest. Dermatol. 1987;88:65–72. doi: 10.1111/1523-1747.ep12468965. [DOI] [PubMed] [Google Scholar]
- Lindinger W, Hansel A, Jordan A. On-line monitoring of volatile organic compounds at pptv levels by means of proton-transfer-reaction mass spectrometry (PTR-MS) medical applications, food control and environmental research. Int. J. Mass Spectrom. 1998;173:191–241. doi: 10.1016/S0168-1176(97)00281-4. [DOI] [Google Scholar]
- Liu X N, Shinada K, Chen X C, Zhang B X, Yaegaki K, Kawaguchi Y. Oral malodor-related parameters in the Chinese general population. J. Clin. Periodontol. 2006;33:31–6. doi: 10.1111/j.1600-051X.2005.00862.x. [DOI] [PubMed] [Google Scholar]
- Loesche W J, Grossman N S. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin. Microbiol. Rev. 2001;14:727–52. doi: 10.1128/CMR.14.4.727-752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longshaw C M, Wright J D, Farrell A M, Holland K T. Kytococcus sedentarius, the organism associated with pitted keratolysis, produces two keratin-degrading enzymes. J. Appl. Microbiol. 2002;93:810–6. doi: 10.1046/j.1365-2672.2002.01742.x. [DOI] [PubMed] [Google Scholar]
- Louis P, Flint H J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- MacFaddin J F. Biochemical Tests for Identification of Medical Bacteria. 2nd edn. Baltimore, MD: Williams and Wilkins; 1980. [Google Scholar]
- Macfarlane S, Macfarlane G T. Proteolysis and amino acid fermentation. In: Gibson G R, Macfarlane G T, editors. Human Colonic Bacteria: Role in Nutrition, Physiology and Pathology. Boca Raton, FL: CRC Press; 1995. pp. pp 75–100. [Google Scholar]
- Mackay R J, McEntyre C J, Henderson C, Lever M, George P M. Trimethylaminuria: causes and diagnosis of a socially distressing condition. Clin. Biochem. Rev. 2011;32:33–43. [PMC free article] [PubMed] [Google Scholar]
- Mackie R I. Microbial production of odor components. Proc. Int. Round Table on Swine Odor Control; Iowa State University, Ames. 1995. pp. pp 18–9. [Google Scholar]
- Magasanik B. Catabolite repression. Cold Spring Harb. Symp. Quant. Biol. 1961;26:249–56. doi: 10.1101/SQB.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- Maita E. In: Bad Breath: A Multidisciplinary Approach. Steenberghe D, van Rosemberg M, editors. Belgium: Lueven University Press; 1996. chapter 19. [Google Scholar]
- Makris G, Wright J D, Ingham E, Holland K T. The hyaluronate lyase of Staphylococcus aureus—a virulence factor? Microbiology. 2004;150:2005–13. doi: 10.1099/mic.0.26942-0. [DOI] [PubMed] [Google Scholar]
- Manja K S, Rao K M. Gas-chromatographic detection of urinary tract infections caused by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 1983;17:264–6. doi: 10.1128/jcm.17.2.264-266.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardh P A, Larsson L, Odham G. Head-space gas chromatography as a tool in the identification of anaerobic bacteria and diagnosis of anaerobic infections. Scand. J. Infect. Dis. 1981;26:14–8. [PubMed] [Google Scholar]
- Marples M J. Life on the human skin. Sci. Am. 1969;220:108–115. doi: 10.1038/scientificamerican0169-108. [DOI] [PubMed] [Google Scholar]
- Marsh P D, Martin M. Oral Microbiology. Oxford: Reed Educational and Professional Publishing; 1999. pp. pp 82–103. [Google Scholar]
- Marshall J, Holland K, Gribbon E. A comparative study of the cutaneous microflora of normal feet with low and high levels of odour. J. Appl. Microbiol. 1988;65:61–8. doi: 10.1111/j.1365-2672.1988.tb04318.x. [DOI] [PubMed] [Google Scholar]
- Marshall J, Leeming J P, Holland K T. The cutaneous microbiology of normal human feet. J. Appl. Bacteriol. 1987;62:139–46. doi: 10.1111/j.1365-2672.1987.tb02391.x. [DOI] [PubMed] [Google Scholar]
- Martin K, Morlin G, Smith A, Nordyke A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J. Bacteriol. 1998;180:107–18. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Lozano P, de la Mora J F. On-line detection of human skin vapors. J. Am. Soc. Mass Spectrom. 2009;20:1060–3. doi: 10.1016/j.jasms.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Martino P D, Fursy R, Bret L, Sundararaju B, Phillips R S. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 2003;49:443–9. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- Massey A, Mowbray J F, Noble W C. Complement activation by Corynebacterium acnes. Br. J. Dermatol. 1978;98:583–4. doi: 10.1111/j.1365-2133.1978.tb01947.x. [DOI] [PubMed] [Google Scholar]
- Mayr D, Margesin R, Klingsbichel E, Hartungen E, Jenewein D, Schinner F, Mark T D. Rapid detection of meat spoilage by measuring volatile organic compounds by using proton transfer reaction mass spectrometry. Appl. Environ. Microbiol. 2003;69:4697–705. doi: 10.1128/AEM.69.8.4697-4705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain A J. In vitro biofilm models: an overview. Adv. Appl. Microbiol. 2009;69:99–132. doi: 10.1016/S0065-2164(09)69004-3. [DOI] [PubMed] [Google Scholar]
- McNair J B. A new procedure for the Duclaux method. J. Am. Chem. Soc. 1933;55:1470–4. doi: 10.1021/ja01331a024. [DOI] [Google Scholar]
- McNamara T F, Alexander J F, Lee M. The role of microorganisms in the production of oral malodour. Oral Surg. Oral Med. Oral Pathol. 1972;34:41–8. doi: 10.1016/0030-4220(72)90271-X. [DOI] [PubMed] [Google Scholar]
- Miller T L, Wolin M J. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl. Environ. Microbiol. 1996;62:1589–92. doi: 10.1128/aem.62.5.1589-1592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minhas T, Greenman J, Schaffer A G. Effects of mucin, haemoglobin and collagen on the maximum specific growth rate, biomass and hydrolytic enzyme production of Porphyromonas gingivalis in continuous culture. Microbiol. Ecol. Heal Dis. 1991;4:311–8. doi: 10.3109/08910609109140281. [DOI] [Google Scholar]
- Mitchell R S, Kumar V, Robbins S L, Abbas A K, Fausto N. Robbins Basic Pathology. Philadelphia, PA: Saunders; 2007. [Google Scholar]
- Mitchell S. Trimethylaminuria (fish-odour syndrome) and oral malodour. Oral Dis. 2005;11:10–3. doi: 10.1111/j.1601-0825.2005.01081.x. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Sakao S, Katoh Y, Takehara T. Correlation between volatile sulfur compounds and certain oral health measurements in the general population. J. Periodontol. 1995;66:679–84. doi: 10.1902/jop.1995.66.8.679. [DOI] [PubMed] [Google Scholar]
- Mobley H L, Hausinger R P. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 1989;53:85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W E, Holdeman L V, Cato E P, Smibert R M, Burmeister J A, Palcanis K G. Comparative bacteriology of juvenile periodontitis. Infect. Immun. 1985;48:507–19. doi: 10.1128/iai.48.2.507-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller R S, Beyhan S, Saini S G, Yildiz F H, Bartlett D H. Indole acts as an extracellular cue regulating gene expression in Vibrio cholera. J. Bacteriol. 2009;191:3504–16. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsch A, Derrer S, Flachsmann F, Schmid J. A broad diversity of volatile carboxylic acids, released by a bacterial aminoacylase from axilla secretions, as candidate molecules for the determination of human-body odor type. Chem. Biodiversity. 2006;3:1–20. doi: 10.1002/cbdv.200690015. [DOI] [PubMed] [Google Scholar]
- Natsch A, Gfeller H, Gygax P, Schmid J, Acuna G. A specific bacterial aminoacylase cleaves odorant precursors secreted in the human axilla. J. Biol. Chem. 2003;21:5718–27. doi: 10.1074/jbc.M210142200. [DOI] [PubMed] [Google Scholar]
- Natsch A, Schmid J, Flachsmann F. Identification of odoriferous sulfanylalkanols in human axilla secretions and their formation through cleavage of cysteine precursors by a C-S lyase isolated from axilla bacteria. Chem. Biodiversity. 2004;1:1058–72. doi: 10.1002/cbdv.200490079. [DOI] [PubMed] [Google Scholar]
- Newton W A, Snell E E. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl Acad. Sci. USA. 1964;51:382–9. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W, Tonzetich J. Effect of hydrogen sulfide and methyl mercaptan on the permeability of oral mucosa. J. Dent. Res. 1984;63:994–7. doi: 10.1177/00220345840630071701. [DOI] [PubMed] [Google Scholar]
- Niederman R, Buyle-Bodin Y, Lu B Y, Robinson P, Naleway C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 1997;76:575–9. doi: 10.1177/00220345970760010801. [DOI] [PubMed] [Google Scholar]
- Nisman B. The Stickland reaction. Bacteriol. Rev. 1954;18:16–42. doi: 10.1128/br.18.1.16-42.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom K M, McGinley K J, Capiello L, Leyden J J. The etiology of malador associated with pitted keratolysis. J. Invest. Dermatol. 1986;87:159. doi: 10.1111/1523-1747.ep12696640. [DOI] [Google Scholar]
- Nordstrom K M, McGinley K J, Cappiello L, Zechman J M, Leyden J J. Pitted keratolysis. The role of Micrococcus sedentarius. Arch. Dermatol. 1987;123:1320–5. doi: 10.1001/archderm.1987.01660340082025. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Collin J. Prosthetic vascular graft infection. Br. J. Surg. 1992;79:1262–7. doi: 10.1002/bjs.1800791205. [DOI] [PubMed] [Google Scholar]
- O'Hara M, Mayhew C A. A preliminary comparison of volatile organic compounds in the headspace of cultures of Staphylococcus aureus grown in nutrient, dextrose and brain heart bovine broths measured using a proton transfer reaction mass spectrometer. J. Breath Res. 2009;3:027001. doi: 10.1088/1752-7155/3/2/027001. [DOI] [PubMed] [Google Scholar]
- Ochiai N, Takino M, Daishima S, Cardin D B. Analysis of volatile sulfur compounds in breath by gas chromatography-mass spectrometry using a three-stage cryogenic trapping preconcentration system. J. Chromatogr. B Biomed. Sci. Appl. 2001;762:67–75. doi: 10.1016/S0378-4347(01)00343-7. [DOI] [PubMed] [Google Scholar]
- Panuncialman J, Falanga V. The science of wound bed preparation. Surg. Clin. North Am. 2009;89:611–26. doi: 10.1016/j.suc.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Paster B J, Boches S K, Galvin J L, Ericson R E, Lau C N, Levanos V A, Sahasrabudhe A, Dewhirst F E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou A K, Magan N, Jones J M, Brown J, Klatser P, Turner A P. Detection of Mycobacterium tuberculosis (TB) in vitro and in situ using an electronic nose in combination with a neural network system. Biosens. Bioelectron. 2004;20:538–44. doi: 10.1016/j.bios.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Pavlou A K, Magan N, McNulty C, Jones J, Sharp D, Brown J, Turner A P. Use of an electronic nose system for diagnoses of urinary tract infections. Biosens. Bioelectron. 2002b;17:893–9. doi: 10.1016/S0956-5663(02)00078-7. [DOI] [PubMed] [Google Scholar]
- Pavlou A, Turner A P, Magan N. Recognition of anaerobic bacterial isolates in vitro using electronic nose technology. Lett. Appl. Microbiol. 2002a;35:366–9. doi: 10.1046/j.1472-765X.2002.01197.x. [DOI] [PubMed] [Google Scholar]
- Park Y K, Bearson B, Bang S H, Bang I S, Foster J W. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 1996;20:605–11. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- Pauling L, Robinson A B, Teranishi R, Cary P. Quantitative analysis of urine vapor and breath by gas–liquid partition chromatography. Proc. Natl Acad. Sci. USA. 1971;68:2374–6. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud K C. Development of a conducting polymer sensor arrays for wound monitoring. Sensors Actuators B. 2008;131:5–9. doi: 10.1016/j.snb.2007.12.035. [DOI] [Google Scholar]
- Persson S, Edlund M B, Claesson R, Carlsson J. The formation of hydrogen sulfide and methylmercaptan by oral bacteria. Oral Microbiol. Immunol. 1990;5:195–201. doi: 10.1111/j.1399-302X.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- Phillips K D, Tearle P V, Willis A T. Rapid diagnosis of anaerobic infections by gas-liquid chromatography of clinical material. J. Clin Pathol. 1976;29:428–32. doi: 10.1136/jcp.29.5.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M, Basa-Dalay V, Bothamley G, Cataneo R N, Lam P K, Natividad M P, Schmitt P, Wai J. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb.). 2010;90:145–51. doi: 10.1016/j.tube.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Phillips M, Cataneo R N, Condos R, Ring-Erickson G A, Greenberg J, La Bombardi V, Munawar M I, Tietje O. Volatile biomarkers of pulmonary tuberculosis in the breath. Tuberculosis. 2007;87:44–52. doi: 10.1016/j.tube.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Phillips M, Herrera J, Krishnan S, Zain M, Greenberg J, Cataneo R N. Variation in volatile organic compounds in the breath of normal humans. J. Chromatogr. B Biomed. Sci. Appl. 1999;729:75–88. doi: 10.1016/S0378-4347(99)00127-9. [DOI] [PubMed] [Google Scholar]
- Postgate J R. Sulphate Reducing Bacteria. 2nd edn. Cambridge: Cambridge University Press; 1984. [Google Scholar]
- Pratten J, Pasu M, Jackson G, Flanagan A, Wilson M. Modeling oral malodor in a longitudinal study. Arch. Oral Biol. 2003;48:737–43. doi: 10.1016/S0003-9969(03)00154-7. [DOI] [PubMed] [Google Scholar]
- Preti G, Leyden J J. Genetic influences on human body odor: from genes to the axillae. Comment. J. Invest. Dermatol. 2010;130:344–6. doi: 10.1038/jid.2009.396. [DOI] [PubMed] [Google Scholar]
- Preti G, Thaler E, Hanson C W, Troy M, Eades J, Gelperin A. Volatile compounds characteristic of sinus-related bacteria and infected sinus mucus: analysis by solid-phase microextraction and gas chromatography-mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009;877:2011–8. doi: 10.1016/j.jchromb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- Probert C S, Ahmed I, Khalid T, Johnson E, Smith S, Ratcliffe N. Volatile organic compounds as diagnostic biomarkers in gastrointestinal and liver diseases. J. Gastrointest. Liver. 2009;18:337–43. [PubMed] [Google Scholar]
- Probert C S J, Jones P R H, Ratcliffe N M. A novel method for rapidly diagnosing the causes of diarrhoea. Gut. 2004;53:58–61. doi: 10.1136/gut.53.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman S, Almas K. Halitosis among racially diverse populations: an update. Int. J. Dent. Hyg. 2008;6:2–7. doi: 10.1111/j.1601-5037.2007.00274.x. [DOI] [PubMed] [Google Scholar]
- Reed P J, Sanderson P J. Detection of anaerobic wound infection by analysis of pus swabs for volatile fatty acids by gas–liquid chromatography. J. Clin. Pathol. 1979;32:1203–5. doi: 10.1136/jcp.32.12.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie P J, Gower D B, Holland K T, Mallet A I, Watkins W J. The skin microflora and the formation of human axillary odour. Int. J. Cosmet. Sci. 1990;12:197–207. doi: 10.1111/j.1467-2494.1990.tb00535.x. [DOI] [PubMed] [Google Scholar]
- Rennie P J, Gower D B, Holland K T. In-vitro and in vivo studies of human axillary odour and the cutaneous microflora. Br. J. Dermatol. 1991;124:596–602. doi: 10.1111/j.1365-2133.1991.tb04958.x. [DOI] [PubMed] [Google Scholar]
- Riazanskaia S, Blackburn G, Harker M, Taylor D, Thomas C L. The analytical utility of thermally desorbed polydimethylsilicone membranes for in vivo sampling of volatile organic compounds in and on human skin. Analyst. 2008;133:1020–7. doi: 10.1039/b802515k. [DOI] [PubMed] [Google Scholar]
- Richmond H D. Duclaux's method for the estimation of ‘volatile fatty acids,’ the laws governing ‘volatility’ deduced there-from, and their application to analysis, more especially to that of butter. Analyst. 1895;20:217–38. doi: 10.1039/an8952000217. [DOI] [Google Scholar]
- Rio A C, Franchi-Teixeira A R, Nicola E M. Relationship between the presence of tonsilloliths and halitosis in patients with chronic caseous tonsillitis. Br. Dent. J. 2008;204:E4. doi: 10.1038/bdj.2007.1106. [DOI] [PubMed] [Google Scholar]
- Rosenberg M, Septon I, Eli I, Bar-Ness R, Gelernter I, Brenner S, Gabbay J. Halitosis measurement by an industrial sulfide monitor. J. Periodontol. 1991;62:487–9. doi: 10.1902/jop.1991.62.8.487. [DOI] [PubMed] [Google Scholar]
- Roth R R, James W D. Microbial ecology of the skin. Annu. Rev. Microbiol. 1988;42:441–64. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- Rowe S M, Miller S, Sorscher E J. Cystic fibrosis. N. Engl. J. Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- Russell R J, McInroy R J, Wilkinson P C, White R G. A lipid chemotactic factor from anaerobic coryneform bacteria including Corynebacterium parvum with activity for macrophages and monocytes. Immunology. 1976;30:935–49. [PMC free article] [PubMed] [Google Scholar]
- Sanz M, Roldán S, Herrera D. Fundamentals of breath malodour. J. Contemp. Dent. Pract. 2001;2:1–17. [PubMed] [Google Scholar]
- Sarkany I, Taplin D, Blank H. The etiology and treatment of erythrasma. J. Invest. Dermatol. 1961;37:283–90. [PubMed] [Google Scholar]
- Sato H, Hirose T, Kimura T, Moriyama Y, Nakashimab Y. Analysis of malodorous volatile substances of human waste: feces and urine. J. Health Sci. 2001;47:483–90. doi: 10.1248/jhs.47.483. [DOI] [Google Scholar]
- Schulz S, Dickschat J S. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 2007;24:814–42. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- Scott-Thomas A, Pearson J, Chambers S. Potential sources of 2-aminoacetophenone to confound the Pseudomonas aeruginosa breath test, including analysis of a food challenge study. J. Breath Res. 2011;5:046002. doi: 10.1088/1752-7155/5/4/046002. [DOI] [PubMed] [Google Scholar]
- Scott-Thomas A J, Syhre M, Pattemore P K, Epton M, Laing R, Pearson J, Chambers S T. 2-aminoacetophenone as a potential breath biomarker for Pseudomonas aeruginosa in the cystic fibrosis lung. BMC Pulm. Med. 2010;10:56. doi: 10.1186/1471-2466-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully C, Greenman J. Halitology (breath odour: aetiopathogenesis and management) Oral Dis. 2012 doi: 10.1111/j.1601-0825.2011.01890.x. doi: 10.1111/j.1601-0825.2011.01890.x. [DOI] [PubMed] [Google Scholar]
- Shehadeh N, Kligman A M. The effect of topical antibacterial agents on the bacterial flora of the axillae. J. Invest. Dermatol. 1963a;40:61–71. doi: 10.1038/jid.1963.10. [DOI] [PubMed] [Google Scholar]
- Shehadeh N, Kligman A M. The bacteria responsible for apocrine odor: II. J. Invest. Dermatol. 1963b;41:1–5. [Google Scholar]
- Shimizu M, Cashman J R, Yamazaki H. Transient trimethylaminuria related to menstruation. BMC Med. Genet. 2007;8:2. doi: 10.1186/1471-2350-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura M, Yasuno Y, Iwakura M, Shimada Y, Sakai S, Suzuki K, Sakamoto S. A new monitor with a zinc-oxide thin film semiconductor sensor for the measurement of volatile sulfur compounds in mouth air. J. Periodontol. 1996;67:396–402. doi: 10.1902/jop.1996.67.4.396. [DOI] [PubMed] [Google Scholar]
- Shirasu M, Nagai S, Hayashi R, Ochiai A, Touhara K. Dimethyl trisulfide as a characteristic odor associated with fungating cancer wounds. Biosci. Biotechnol. Biochem. 2009;73:2117–20. doi: 10.1271/bbb.90229. [DOI] [PubMed] [Google Scholar]
- Shnayderman M, et al. Species-specific bacteria identification using differential mobility spectrometry and bioinformatics pattern recognition. Anal. Chem. 2005;77:5930–7. doi: 10.1021/ac050348i. [DOI] [PubMed] [Google Scholar]
- Shykhon M E, Morgan D W, Dutta R, Hines E L, Gardner J W. Clinical evaluation of the electronic nose in the diagnosis of ear, nose and throat infection: a preliminary study. J. Laryngol. Otol. 2004;118:706–9. doi: 10.1258/0022215042244660. [DOI] [PubMed] [Google Scholar]
- Singer R E, Buckner B A. Butyrate and propionate: important components of toxic dental plaque extracts. Infect. Immun. 1981;32:458–63. doi: 10.1128/iai.32.2.458-463.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley J W, Birss A J, McKee A S, Marsh P D. Changes in the affinity of haemin-binding by Porphyromonas gingivalis W50 under different environmental conditions. Microb. Ecol. Health Dis. 1994;7:9–15. doi: 10.3109/08910609409141569. [DOI] [Google Scholar]
- Smith D, Spanel P. The novel selected-ion flow tube approach to trace gas analysis of air and breath. Rapid Commun. Mass Spectrom. 1996;10:1183–98. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1183::AID-RCM641>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Smith D, Spanel P. Selected ion flow tube mass spectrometry (SIFT-MS) for online trace gas analysis. Mass Spectrom. Rev. 2005;24:661–700. doi: 10.1002/mas.20033. [DOI] [PubMed] [Google Scholar]
- Snell E E. Tryptophanase: structure, catalytic activities, and mechanism of action. Adv. Enzymol. Relat. Areas Mol. Biol. 1975;42:287–333. doi: 10.1002/9780470122877.ch6. [DOI] [PubMed] [Google Scholar]
- Sobel J D. Bacterial vaginosis. Annu. Rev. Med. 2000;51:349–56. doi: 10.1146/annurev.med.51.1.349. [DOI] [PubMed] [Google Scholar]
- Somerville D A. Erythrasma in normal young adults. J. Med. Microbiol. 1970;3:57–64. doi: 10.1099/00222615-3-1-57. [DOI] [PubMed] [Google Scholar]
- Spencer P, Greenman J, McKenzie C, Gafan G, Spratt D, Flanagan A. In vitro biofilm model for studying tongue flora and malodour. J. Appl. Microbiol. 2007;103:985–92. doi: 10.1111/j.1365-2672.2007.03344.x. [DOI] [PubMed] [Google Scholar]
- Spinelli F, Noferini M, Vanneste J L, Costa G. Potential of the electronic-nose for the diagnosis of bacterial and fungal diseases in fruit trees. EPPO Bull. 2010;40:59–67. doi: 10.1111/j.1365-2338.2009.02355.x. [DOI] [Google Scholar]
- Statheropoulos M, Spiliopoulou C, Agapiou A. A study of volatile organic compounds evolved from the decaying human body. Forensic Sci. Int. 2005;153:147–55. doi: 10.1016/j.forsciint.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Starkenmann C, Troccaz M, Howell K. The role of cysteine and cysteine-S conjugates as odour precursors in the flavour and fragrance industry. Flavour Fragr. J. 2008;23:369–81. doi: 10.1002/ffj.1907. [DOI] [Google Scholar]
- Storer M K, Hibbard-Melles K, Davis B, Scotter J. Detection of volatile compounds produced by microbial growth in urine by selected ion flow tube mass spectrometry (SIFT-MS) J. Microbiol. Methods. 2011;87:111–3. doi: 10.1016/j.mimet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Syhre M, Chambers S T. The scent of Mycobacterium tuberculosis. Tuberculosis (Edinb.). 2008;88:317–23. doi: 10.1016/j.tube.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Syhre M, Manning L, Phuanukoonnon S, Harino P, Chambers S T. The scent of Mycobacterium tuberculosis—part II breath. Tuberculosis (Edinb.). 2009;89:263–6. doi: 10.1016/j.tube.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Machigashira M, Yamashita D, Kozono S, Nakajima Y, Miyamoto M, Takeuchi N, Setoguchi T, Noguchi K. The association of periodontal disease with oral malodour in a Japanese population. Oral Dis. 2010;16:702–6. doi: 10.1111/j.1601-0825.2010.01685.x. [DOI] [PubMed] [Google Scholar]
- Tanda N, Washio J, Ikawa K, Suzuki K, Koseki T, Iwakura M. A new portable sulfide monitor with a zinc-oxide semiconductor sensor for daily use and field study. J. Dent. 2007;35:552–7. doi: 10.1016/j.jdent.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Tangerman A, Winkel E G. The portable gas chromatograph OralChroma™: a method of choice to detect oral and extra-oral halitosis. J. Breath Res. 2008;2:017010. doi: 10.1088/1752-7155/2/1/017010. [DOI] [PubMed] [Google Scholar]
- Tangerman A, Winkel E G. Extra-oral halitosis: an overview. J. Breath Res. 2010;4:017003. doi: 10.1088/1752-7155/4/1/017003. [DOI] [PubMed] [Google Scholar]
- Tarr L A. Microbiological deterioration of fish post mortem, its detection and control. Bacteriol. Rev. 1954;18:1–15. doi: 10.1128/br.18.1.1-15.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B, Greenman J. Modelling the effects of pH on tongue biofilm using a sorbarod biofilm perfusion system. J. Breath Res. 2010;4:017107. doi: 10.1088/1752-7155/4/1/017107. [DOI] [PubMed] [Google Scholar]
- Tessier J F, Kulkarni G V. Bad breath: etiology, diagnosis and treatment. Oral Health. 1991;81:19–22. [PubMed] [Google Scholar]
- Thorn R M S, Reynolds D M, Greenman J. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J. Microbiol. Methods. 2011;84:258–64. doi: 10.1016/j.mimet.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Tonzetich J. Direct gas chromatographic analysis of sulfur compounds in mouth air in man. Arch. Oral Biol. 1971;16:587–97. doi: 10.1016/0003-9969(71)90062-8. [DOI] [PubMed] [Google Scholar]
- Tonzetich J, McBride B C. Characterization of volatile sulfur production by pathogenic and non-pathogenic strains of oral Bacteroides. Arch. Oral Biol. 1981;26:963–9. doi: 10.1016/0003-9969(81)90104-7. [DOI] [PubMed] [Google Scholar]
- Tso W W, Adler J. Negative chemotaxis in Escherichia coli. J. Bacteriol. 1974;118:560–76. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Yanagisawa T, Shinada K, Ohara S, Kawaguchi Y. Prevalence of oral malodor and related factors among adults in Akita Prefecture. J. Med. Dent. Sci. 2007;54:159–65. [Google Scholar]
- Van den Velde S, Quirynen M, van Hee P, van Steenberghe D. Halitosis associated volatiles in breath of healthy subjects. J. Chromatogr. B. 2007a;853:54–61. doi: 10.1016/j.jchromb.2007.02.048. [DOI] [PubMed] [Google Scholar]
- Van den Velde S, Quirynen M, van Hee P, van Steenberghe D. Differences between alveolar air and mouth air. Anal. Chem. 2007b;79:3425–9. doi: 10.1021/ac062009a. [DOI] [PubMed] [Google Scholar]
- Vásquez A, Jakobsson T, Ahrné S, Forsum U, Molin G. Vaginal lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 2002;40:2746–9. doi: 10.1128/JCM.40.8.2746-2749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J P, Méresse S, Pocachard B, Guilhon A A, Aussel L, Barras F. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PLoS One. 2011;6:e22397. doi: 10.1371/journal.pone.0022397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadström T, Ljungh A. Glycosaminoglycan-binding microbial proteins in tissue adhesion and invasion: key events in microbial pathogenicity. J. Med. Microbiol. 1999;48:223–33. doi: 10.1099/00222615-48-3-223. [DOI] [PubMed] [Google Scholar]
- Wang D, Ding X, Rather P N. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 2001;183:4210–6. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb E C. Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. San Diego, CA: Academic; 1992. [Google Scholar]
- Weetjens B J, et al. African pouched rats for the detection of pulmonary tuberculosis in sputum samples. Int. J. Tuberc. Lung Dis. 2009;13:737–43. [PubMed] [Google Scholar]
- Weitzman I, Summerbell R C. The dermatophytes. Clin. Microbiol. Rev. 1995;8:240–59. doi: 10.1128/cmr.8.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C L, Fakharzadeh J E, Preti G. Human breath odors and their use in diagnosis. Ann. NY Acad. Sci. 2007;1098:252–66. doi: 10.1196/annals.1384.011. [DOI] [PubMed] [Google Scholar]
- Willis C L, Gibson G R, Allison C, Macfarlane S, Holt J S. Growth, incidence and activities of dissimilatory sulfate-reducing bacteria in the human oral cavity. FEMS Microbiol. Lett. 1995;15:267–71. doi: 10.1111/j.1574-6968.1995.tb07591.x. [DOI] [PubMed] [Google Scholar]
- Willis C L, Gibson G R, Holt J, Allison C. Negative correlation between oral malodour and numbers and activities of sulphate-reducing bacteria in the human mouth. Arch. Oral Biol. 1999;44:665–70. doi: 10.1016/S0003-9969(99)00056-4. [DOI] [PubMed] [Google Scholar]
- Wilson M. Bacterial biofilms and human disease. Sci. Prog. 2001;84:235–54. doi: 10.3184/003685001783238998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W. Microbial Inhabitants of Humans. 1st edn. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Wolff M, Bruhns H, Zhang W. Photoacoustic detection of volatile organic compounds. Proc. SPIE. 2011;8073 [Google Scholar]
- Wong T P, Groman N. Production of diphtheria toxin by selected isolates of Corynebacterium ulcerans and Corynebacterium pseudotuberculosis. Infect. Immun. 1984;43:1114–6. doi: 10.1128/iai.43.3.1114-1116.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgyer A J, Baxter M, Rush-Munro F M, Brown J, Kaplan W. Isolation of Dermatophilus congolensis from two New Zealand cases of pitted keratolysis. Australas. J. Dermatol. 1985;26:29–35. doi: 10.1111/j.1440-0960.1985.tb01811.x. [DOI] [PubMed] [Google Scholar]
- Yaegaki K, Coil J M. Examination, classification, and treatment of halitosis; clinical perspectives. J. Can. Dent. Assoc. 2000;66:257–61. [PubMed] [Google Scholar]
- Yaegaki K, Sanada K. Volatile sulfur compounds in mouth air from clinically healthy subjects and patients with periodontal disease. J. Periodontal Res. 1992;27:233–8. doi: 10.1111/j.1600-0765.1992.tb01673.x. [DOI] [PubMed] [Google Scholar]
- Yaegaki K, Qian W, Murata T, Imai T, Sato T, Tanaka T, Kamoda T. Oral malodorous compound causes apoptosis and genomic DNA damage in human gingival fibroblasts. J. Periodontal Res. 2008;43:391–9. doi: 10.1111/j.1600-0765.2007.01052.x. [DOI] [PubMed] [Google Scholar]
- Yamada K. Occurrence and origin of trimethylamine oxide in fishes and marine invertebrates. Bull. Jpn. Soc. Sci. Fish. 1967;33:591–603. doi: 10.2331/suisan.33.591. [DOI] [Google Scholar]
- Yamazaki K, Boyse E A, Miké V, Thaler H T, Mathieson B J, Abbott J, Boyse J, Zayas Z A, Thomas L. Control of mating preferences in mice by genes in the major histocompatibility complex. J. Exp. Med. 1976;144:1324–35. doi: 10.1084/jem.144.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama M T, Carlson J R. Dissimilation of tryptophan and related indolic compounds by ruminal microorganisms in vitro. Appl. Microbiol. 1974;27:540–8. doi: 10.1128/am.27.3.540-548.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshda Y, Sasaki T, Ito S, Tamura H, Kunimatsu K, Kato H. Identification and molecular characterization of tryptophanase encoded by tnaA in Porphyromonas gingivalis. Microbiology. 2009;155:968–78. doi: 10.1099/mic.0.024174-0. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Ito S, Kamo M, Kezuka Y, Tamura H, Kunimatsu K, Kato H. Production of hydrogen sulfide by two enzymes associated with biosynthesis of homocysteine and lanthionine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology. 2010;156:2260–9. doi: 10.1099/mic.0.039180-0. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Suwabe K, Nagano K, Kezuka Y, Kato H, Yoshimura F. Identification and enzymatic analysis of a novel protein associated with production of hydrogen sulfide and L-serine from L-cysteine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology. 2011;157:2164–71. doi: 10.1099/mic.0.048934-0. [DOI] [PubMed] [Google Scholar]
- Zechman J, Aldinger S, Labows J. Characterization of pathogenic bacteria by automated headspace concentration—gas chromatography. J. Chromatogr. 1986;377:49–57. doi: 10.1016/S0378-4347(00)80760-4. [DOI] [PubMed] [Google Scholar]
- Zeng C, Spielman A I, Vowels B R, Leyden J J, Biemann K, Preti G. A human axillary odorant is carried by apolipoprotein D. Proc. Natl Acad. Sci. USA. 1996;93:6626–30. doi: 10.1073/pnas.93.13.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X-N, Leyden J J, Brand J G, Spielman A I, McGinley K J, Preti G. An investigation of human apocrine gland secretion for axillary odor precursors. J. Chem. Ecol. 1992;18:1039–55. doi: 10.1007/BF00980061. [DOI] [PubMed] [Google Scholar]
- Zoller H F, Clark W M. The production of volatile fatty acids by bacteria of the dysentery group. J. Gen. Physiol. 1920;20:325–30. doi: 10.1085/jgp.3.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]


