SUMMARY
The primary cilium maintains a well-regulated complement of soluble and membrane proteins, allowing it to mediate a variety of signaling pathways essential for development and tissue homeostasis [1–3]. Entry into the cilium is regulated at the base, where a complex containing nucleoporins, referred to as the “ciliary pore complex,” (CPC) has been proposed to set a size-exclusion limit for soluble molecule diffusion into the cilium [4–6]. Here, using a fluorescence-based diffusion trap system, we demonstrate that Nup98, a component of the FG hydrogel permeability barrier at the nuclear pore complex [7, 8], limits the diffusion of soluble molecules >70kDa into the cilium in cultured mammalian cells. siRNA mediated knockdown of Nup98 increases the rate of diffusion of molecules >100kDa into the cilium. The tubulin heterodimer, the building block of the axoneme [9, 10], is approximately 100kDa in size. After knockdown of Nup98, cilia become shorter, and their length is more sensitive to changes in cytoplasmic soluble tubulin levels. These data indicate a novel function of the ciliary pore complex, limiting diffusion of soluble tubulin between the ciliary matrix and the cytosol, allowing the cilium to regulate its length independently of cytosolic microtubule dynamics.
eTOC blurb:
Endicott and Brueckner show that the FG-repeat containing nucleoporin Nup98 localizes to the ciliary base, where it functions as part of a diffusion barrier limiting diffusion of molecules >70kDa, notably including tubulin, between the ciliary matrix and cytosol.
RESULTS & DISCUSSION
Nup98 localizes to the ciliary base, where it plays a role in cilia length regulation
We previously reported that GFP-tagged Nup98 localizes to the ciliary base, and functions in regulating cilium resorption [6]. Here, we investigated the role of Nup98 at the ciliary base in interphase cells. siRNA-mediated knockdown of Nup98 in hTERT Retinal-Pigmented-Epithelium-1 (RPE) cells resulted in 75–80% reduction in total Nup98 protein levels 4 days after siRNA treatment (Figure 1B) compared to cells transfected with a non-targeting medium GC control siRNA. Quantification of Nup98 fluorescence intensity at the ciliary base revealed that cells treated with Nup98 siRNA show a significant decrease in Nup98 fluorescence intensity at the ciliary base compared to controls (Figure 1C). The transfection was heterogeneous, and fluorescence intensity of Nup98 staining at the ciliary base corresponded with fluorescence intensity at the nuclear envelope (Figure 1A, Figure S1B), confirming that Nup98 antibody staining at the ciliary base is specific.
Figure 1: Nup98 is required for cilia length regulation.

See also Figure S1. (A) siRNA-mediated knockdown of Nup98 in hTERT-RPE (RPE) cells results in a loss in the fluorescence intensity of Nup98 staining at the ciliary base, confirming antibody specificity. (B) siRNA-mediated knockdown results in a 75–80% reduction of Nup98 protein levels. The Western blot was quantified in ImageJ and repeated 2 more times with similar results (quantitation shown at upper edge of Western blot). (C) Fluorescence intensity of Nup98 was measured at the ciliary base using ImageJ, and normalized to the fluorescence intensity of the mother centriole marker, Ninein. There was a specific reduction in Nup98 signal in cells treated with a Nup98 siRNA, when compared to controls. (D) Cilia length was decreased in serum-starved RPE cells treated with a Nup98 siRNA. (E) After knockdown of Nup98, RPE cells no longer show cilium elongation in response to serum-starvation.
p values in (C, D) were calculated by unpaired t test. For (E) a one-way ANOVA was performed comparing cilia length before starvation to the 4h, 8h, and 24h time points during starvation. For control siRNA, p < 0.0001. For Nup98 siRNA, p = not significant. Cilia length was measured using ImageJ. In (C), error bars are S.D.; all other error bars are S.E.M.
Interestingly, cells treated with Nup98 siRNA showed 1μm reduction in cilia length (in confluent, serum-starved cells) compared to controls (Figure 1D). RPE cells stop dividing at confluence and generate cilia, even when grown in medium supplemented with 10% FBS. When RPE cells are grown to confluence in serum-supplemented medium and then serum-starved, cilia in control RPE cells grow ~0.8μm longer over 24hours. Re-addition of serum causes cilia to return to the baseline length within 4 hours (Figure 1E). Nup98 knockdown abrogates cilia lengthening in response to serum withdrawal (Figure 1E), suggesting that Nup98 is required for cilia length regulation.
Nup98 limits the diffusion of soluble macromolecules into the cilium
We hypothesized that altered diffusion dynamics through the ciliary base underlie the cilia length defect resulting from Nup98 is knockdown in RPE cells. There is increasing evidence that nucleoporins function to regulate trafficking into the cilium. Some ciliary proteins require the Imp-β2/RanGTP pathway to enter the cilium [11–15]; the RanGTP pathway is necessary to build a cilium [16], and methods that abrogate transport through the nuclear pore complex (NPC) also block import into the cilium [4, 5]. At the nucleus, Nup98 is a key component of the FG hydrogel permeability barrier, where it also acts as a negative regulator of NPC breakdown at mitotic onset [17, 18]. Moreover, Nup98 is sufficient to rescue both directional import and passive size-exclusion in nuclei depleted of all other FG-repeat nucleoporins [7]. Thus, we hypothesized that Nup98 is a component of the size-exclusion permeability barrier at the ciliary base. To investigate if Nup98 can restrict diffusion into the cilium, we took advantage of a chemically inducible diffusion trap, established as a method for measuring diffusion into the cilium [19, 20]. A 5HT6-CFP-FRB diffusion trap was expressed in RPE cells, where it localized to the ciliary membrane (Figures S1A, 2A). 5HT6 is a receptor expressed in neurons that localizes specifically to the ciliary membrane [21]. Cells were co-transfected with constructs expressing YFP-FKBP-tag (or YFP-tag-FKBP) prey proteins, where the tag is an inert protein of a desired size, making completed prey constructs ranging from 49kDa to >600kDa (Figure 2G). Upon addition of rapamycin, the FRB and FKBP domains dimerize, causing the YFP prey construct to become trapped in the cilium by the CFP diffusion trap, if the YFP construct is below the size-exclusion limit of the cilium (Figures S1A & 2A).
Figure 2. Nup98 sets the size exclusion limit for the diffusion of soluble molecules through the ciliary base.
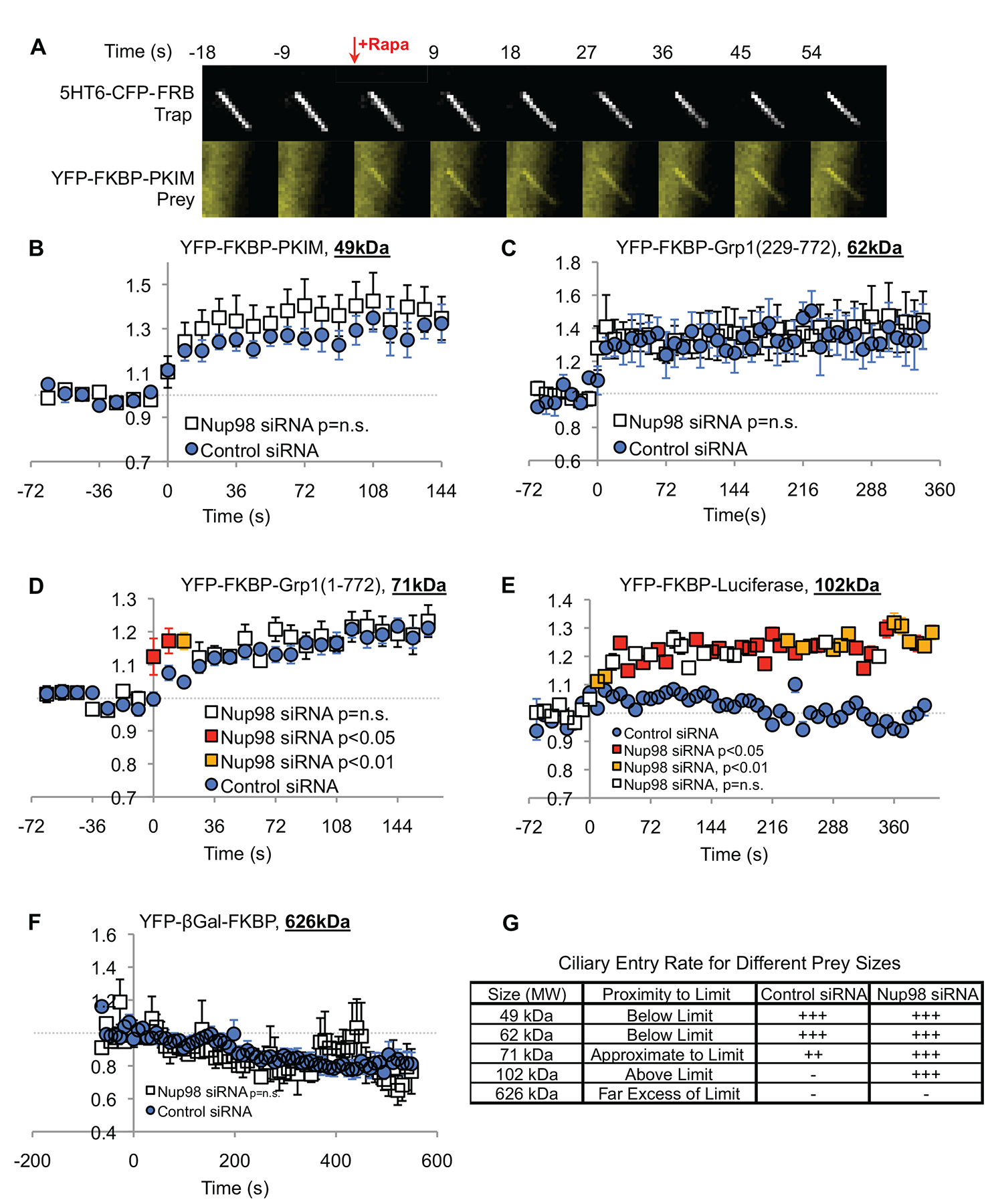
(See also Videos S1–4) (A) Montage of images at 9 second intervals, showing both the CFP and YFP channels, demonstrating that a YFP-FKBP-PKIM construct (49 kDa) is below the size exclusion limit of the ciliary base, and accumulates rapidly in the cilium after application of rapamycin. The ciliary membrane is tagged with the 5HT6-CFP-FRB diffusion trap. See also Figure S1. (B-F) CFP and YFP fluorescence intensities in the cilium were measured using a custom program (available online; see STAR Methods). The YFP fluorescence in the cilium is normalized to the CFP fluorescence intensity to prevent motion artifacts. Baseline YFP/CFP before addition of rapamycin was adjusted to 1 arbitrary unit. p-values are calculated for each time point individually, using Welch’s t test. If p = n.s., the nup98 siRNA markers are white; if p < 0.05, the markers are red; if p < 0.01 the markers are amber. (B) A 49kDa construct and (C) a 62kDa construct showed equal accumulation in cells treated with the control or nup98 siRNA. (D) A 71 kDa construct showed slightly faster accumulation in cells treated with nup98 siRNA than controls. (E) A 102kDa construct showed no accumulation in control cells, while showing significant accumulation in cells treated with nup98 siRNA. (F) A 626kDa construct showed no accumulation in any cilia measured. (G) Summary of results from diffusion trap experiments.
Previously, the size exclusion limit of RPE cilia has been determined to be ~70kDa [4]. Consistently, with the diffusion trap system, we observed that 49kDa and 62kDa prey constructs rapidly accumulated in the cilium, with no observable significant difference between cells treated with control or Nup98 siRNAs (Figure 2B, C; Video S1; Video S2). A prey construct of 71kDa, near the diffusion limit, showed slightly faster accumulation into the cilia of cells treated with Nup98 siRNA than controls (Figure 2D;). In contrast, a 102kDa construct showed no accumulation in cilia of cells treated with control siRNA, but showed robust accumulation in the cilia of cells treated with Nup98 siRNA (Figure 2E; Video S3; Video S4). A very large 626kDa prey construct, YFP-βGal-FKBP, which forms tetramers [19], showed no accumulation in any cilia measured from cells treated with either the control or Nup98 siRNA (Figure 2F). Together these data suggest that Nup98 is involved in setting the size exclusion limit at the ciliary base. Removal of Nup98 from the ciliary base increases the size-exclusion limit (Figure 2G), although very large molecules are still excluded even in the absence of Nup98.
Diffusion traps are prone to variability due to differences in expression levels, and two previous diffusion trap studies have reported higher exclusion limits for the ciliary base than what we find [19, 20]. Two alternative methods of measuring diffusion into the cilium align with our data, however: microinjection of tagged proteins and dextrans into the cytoplasm and measuring accumulation into the cilium, likely the most accurate method, finds the diffusion limit to be somewhere between 40–70kDa [4], and a diffusion-capture system for semi-permeabilized cells, which allows for specification of the prey construct concentration, finds the diffusion limit to be around 70kDa [20].
Knockdown of Nup98 does not perturb the gross architecture of the NPC
We considered that the change in soluble molecule diffusion after Nup98 knockdown could be secondary to a change in NPC architecture. Nup98 is generated from the same pre-protein (and mRNA) as Nup96 [22], which is an essential component of the Y-complexes necessary for NPC formation [8, 23, 24]. Thus, if Nup96 protein levels were dramatically reduced, we would expect a significant disruption of NPC function; however, Nup96 has a much longer half-life than Nup98 in non-dividing cells [25]. Consistently, we found that the knockdown efficiency of Nup96 was much less than that of Nup98 (Figure S1C). Nup85 also has a very long half-life when incorporated into a stable NPC, but is predicted to have a shortened half-life when not incorporated into an NPC [25]. Thus, we measured Nup85 protein in RPE cells after Nup98 knockdown, finding no decrease in Nup85 levels (Figure S1D). Finally, we examined the incorporation of nuclear basket component Nup153 into the nuclear envelope in cells treated with a Nup98 siRNA. Because the knockdown is heterogeneous, we qualitatively assessed nuclei for a correlation between Nup98 knockdown and a decrease in Nup153 incorporation; however, Nup153 staining at the nucleus does not correlate with a decrease in Nup98 staining (Figure S1E). Taken together these data suggest that the gross architecture of the NPC and the density of NPCs are intact after Nup98 knock down.
Knockdown of Nup98 does not perturb the gross architecture of the transition zone
The transition zone (TZ) is the section of the cilium that lies immediately distal to the mother centriole’s distal appendages/transition fibers [26], and nucleoporins have been shown to localize in close proximity to the TZ [27]. The TZ has a well-characterized role in gating entry of membrane proteins into the cilium; however, the role the TZ plays in gating of soluble molecule traffic into the cilium is unclear [28–30]. To confirm that there were no overt TZ abnormalities after knockdown of Nup98, we examined the effect of Nup98 knockdown on the localization of four distal appendage/TZ markers: Cep290, Tctn1, Odf2, and BBS13/MKS1 [28, 31–33], and observed no change in any of these markers (Figure S2). These data are consistent with the model of Nup98 affecting the diffusion of soluble molecules through a direct mechanism at the ciliary base.
Depletion of Nup98 does not prevent cilia biogenesis
To determine whether knockdown of Nup98 affects cilia length through an effect on cilia biogenesis, we deciliated serum-starved RPE cells treated with a control or Nup98 siRNA (timeline in Figure S3D), using an 18 hour treatment with 30mM ammonium sulfate adapted from [34]. Ammonium sulfate treatment effectively deciliates cells; and cilia regenerate to baseline quantities 24 hours after washout (Figure S3A–C). Nup98 knockdown does not affect cilia numbers before deciliation, after deciliation, or after recovery compared to control cells (Figure S3E), indicating that Nup98 knockdown does not impair the ability of cells to generate normal numbers of cilia, and that cilia biogenesis machinery remains grossly intact.
Nup98 regulates cilia tubulin dynamics
After finding that cells generate normal numbers of cilia after Nup98 knockdown, we considered the possibility that Nup98 affects cilia length through perturbation of tubulin trafficking. Evidence from Chlamydomonas, recapitulated in mammalian systems and tissues, suggests that ciliary length is maintained by a dynamic equilibrium of tubulin trafficking [9, 10, 35] (Figure 3A). Tubulin is carried into the cilium by IFT [9], and depends upon a tubulin-binding module formed from IFT74 and IFT81 [36]. Loss of IFT (even from fully formed cilia), genetically or through pharmacological inhibition, results in a loss of cilia [35, 37, 38]. In Chlamydomonas, two cilia on the same cell can maintain different concentrations of soluble tubulin in their respective matrices (depending on their state of elongation), suggesting that there is a diffusion-limiting gate through the ciliary base, allowing the ciliary matrix to maintain a distinct concentration of soluble tubulin from the cytosol [9]. The diffusion-limiting gate is not impervious to tubulin diffusion; it merely allows for a relative enrichment of soluble tubulin in the ciliary matrix during ciliary outgrowth. In cultured mammalian cells, diffusion into the cilium is limited for molecules greater than 70kDa [4, 19, 20]. We hypothesized that diffusion of molecules the size of the tubulin heterodimer (MW > 100kDa) out of the cilium is slower than the rate of tubulin import by IFT during ciliary growth, allowing enrichment of tubulin in the ciliary matrix, a condition favorable to ciliary elongation (Figure 3A). If knockdown of Nup98 raises the size-exclusion limit of the cilium to over 100kDa, we predict less enrichment of soluble tubulin in the cilium, and a shorter steady-state length (Figure 3B).
Figure 3. Nup98 regulates cilia length by gating the efflux of soluble tubulin from the ciliary matrix.
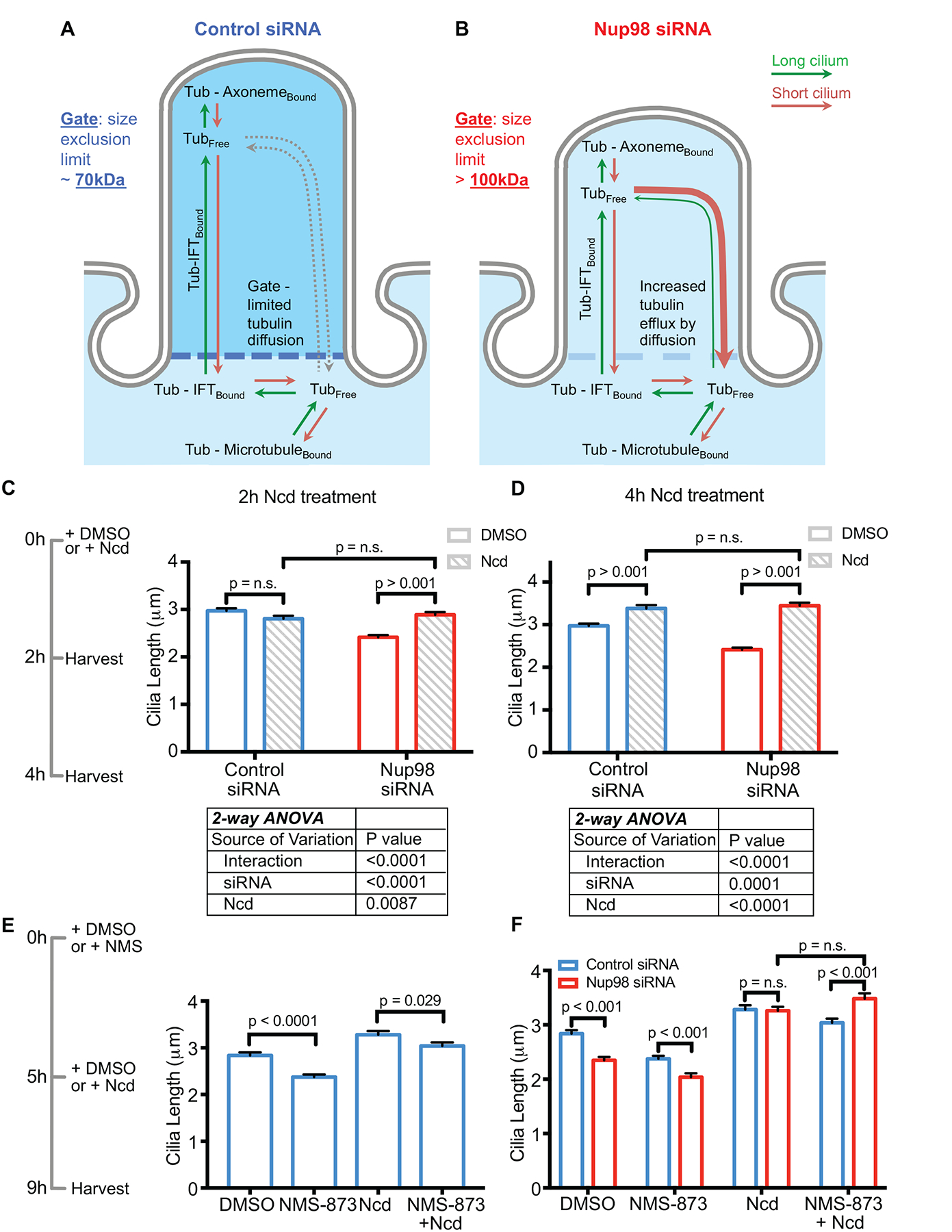
(See also Figure S4) (A) In control cells, cilia length is regulated by the availability of soluble tubulin in the ciliary matrix. (B) In Nup98 knockdown cells, there is increased efflux of tubulin from the ciliary matrix, resulting in a decrease in cilium length. (C) At 2 hours of treatment with 100nM Ncd, cilia have no change in length in control cells; however, cilia show a significant increase in length in cells treated with the Nup98 siRNA. (D) At 4 hours of treatment with 100nM Ncd, cilia in both control and Nup98 siRNA treated cell show an elongation; however, that elongation is greater in cells treated with the Nup98 siRNA. (E) Treatment of cells with NMS-873, which has been shown to destabilize anterograde IFT particles, reduces cilia length in control RPE cells, and lessens the cilia lengthening effects of Ncd. (F) When Nup98 is knocked down by siRNA and cells are treated with Ncd, partial inhibition of IFT with NMS-873 is significantly less effective at reducing cilia length when compared to control cells, also consistent with diffusion, rather than IFT, being the primary mode of tubulin import into the ciliary matrix, when Nup98 is reduced.
Error bars are S.E.M. p values were calculated by unpaired t-test, unless otherwise specified.
Recent reports describe pharmacological methods of changing cilia length [10, 37, 39]. The most direct of these is to increase the abundance of soluble tubulin in the cytosol. Low doses of nocodazole (Ncd; blocks microtubule polymerization) in the range of 100nM, have been reported to promote cilia elongation by increasing the availability of cytoplasmic tubulin, despite the fact that doses of 1μM or greater lead to cilia loss, accompanied by much more prevalent tubulin depolymerization [10].
We hypothesized that if we knocked down Nup98, raising the size-exclusion limit of the CPC, cells would show a greater elongation of the cilium in response to Ncd treatment, because the gate would not slow the diffusion of the soluble tubulin into the cilium (Figure 3A–B). We confirmed that 100nM Ncd treatment results in an increase in soluble tubulin (Figure S4A–B), by subcellular fractionation of soluble and polymerized tubulin. We analyzed ciliary length in cells treated with control or Nup98 siRNA after 2-hour and a 4-hour Ncd treatment, as shown in the timeline (Figure 3C). The 2-hour Ncd treatment had no effect on cilia length in control cells; however, it significantly increased cilia length by ~ 0.5μm in cells treated with the Nup98 siRNA (Figure 3C). Two-way ANOVA revealed that the interaction between siRNA treatment and Ncd treatment was highly significant (Figure 3C). After 4-hours of Ncd treatment, cells treated with a control siRNA showed a 0.4μm increase in average cilia length. For Nup98 siRNA-treated cells, Ncd treatment increased average cilia length by over 1μm (Figure 3D). At this (4-hour) time point, final ciliary length was approximately the same for control and Nup98 siRNA-treated cells; however, 2-way ANOVA analysis revealed a significant interaction between the siRNA treatment and the Ncd treatment. Taken together, these findings are consistent with the hypothesis that soluble tubulin diffuses into cilia faster than normal IFT-based transport if the CPC size-exclusion limit is raised by knocking down Nup98.
VCP (p97 or CDC48) is required to localize UBXN10 to cilia, and both VCP and UBXN10 are required for ciliogenesis. Short-term treatment with the VCP inhibitor NMS-873 has been shown to destabilize anterograde IFT, and to shorten cilia [37]. Since tubulin is carried into cilia by IFT, pretreatment of control cells with NMS-873 before Ncd treatment should reduce the effects of Ncd on cilia length. In other words, increased cytosolic tubulin concentration will have a reduced effect on cilia elongation if that tubulin cannot be trafficked into the cilium by IFT. Indeed, after 4h of Ncd treatment, control cells pretreated with NMS-873 have cilia significantly shorter than cells pre-treated with DMSO (Figure 3E).
We next hypothesized that if diffusion of soluble molecules (in the size range of tubulin heterodimers) into the ciliary matrix was increased by knocking down Nup98, then the loss of IFT will be less consequential to cilia length when cells are treated with Ncd, which increases the available pool of cytosolic soluble tubulin. Without Nup98 at the CPC during Ncd treatment, diffusion of tubulin into the ciliary matrix increases, and the cells will be less reliant on IFT. Like control cells, cells treated with Nup98 siRNA show a decrease in cilia length when treated with NMS-873 (Figure 3F). When cells depleted of Nup98, pre-treated for 5 hours with NMS-873, are treated with Ncd for 4 hours, their cilia reach a length significantly longer than controls treated with the same conditions (Figure 3F). Collectively, these data suggest that cilia depleted of Nup98 are more responsive to changes in cytosolic soluble tubulin concentration than controls, consistent with a model of increased diffusion of soluble tubulin between the cytoplasm and ciliary matrix.
Nup98 is required to regulate cilia length after perturbation of the actin cytoskeleton
Cytochalasin D (CD) treatment has been reported to increase cilium length [10, 40], despite an unclear consensus on the mechanism. It has been previously reported that treatment of RPE cells with 1 μM CD leads to an increases the ratio of soluble to polymerized tubulin [10]. We isolated soluble/polymerized tubulin fractions from cells treated with CD; however, we did not observe the previously reported change in soluble tubulin (Figure S4A–B). Interestingly, after CD treatment, cilia on cells treated with a Nup98 siRNA elongated to the same final length as cilia on control CD-treated cells at 2-hour, 4-hour, and 8-hour time points (Figure S4C–D). Perturbation of the actin cytoskeleton disperses negative regulator of cilia length HDAC6 [40], which has a molecular weight above the size-exclusion limit of the cilium (134kDa). Removal of Nup98 from the ciliary base could make the cilium more susceptible to the effects of negative regulators localized at the ciliary base – an effect abrogated by dispersing the actin cytoskeleton.
We reasoned that if the primary source of tubulin influx into the cilium is IFT-mediated, then pretreatment of control cells with NMS-873 before CD treatment should reduce the effects of CD on cilia length since reducing the influx of tubulin building blocks into the cilium should dampen the positive effect of CD treatment. Consistently, after 4h of CD treatment, control cells pretreated with NMS-873 have cilia averaging 3.4μm in length, while cells pre-treated with DMSO have cilia averaging 4.3μm in length, a highly significant difference (Figure S4E).
Like control cells, cells treated with Nup98 siRNA show decreased ciliary length when treated with NMS-873 (Figure S4F). When Nup98-depleted cells, pre-treated for 5 hours with NMS-873, are treated with CD for 4 hours, their cilia reach a length ~0.5μm longer than controls treated with the same conditions, a highly significant difference (Figure S4F). Combined with the data from the cells treated with Ncd, these observations support the hypothesis that loss of Nup98 sensitizes cilia length to changes in dynamics of large molecules such as tubulin, and potentially, HDAC6.
Nup85 is required for Nup98 localization to the ciliary base and cilia length regulation
As a secondary means for validating the importance of Nup98 in regulating tubulin diffusion through the ciliary base, we sought to perturb the localization of Nup98 to the ciliary base, without diminishing total Nup98 levels.
We chose to knockdown Nup85, a component of the essential Y-structure that makes up part of the symmetric core of the NPC [21]. Nup85 has a long residence time at the NPC and an extraordinarily long half-life in rat brain neurons [25, 41], consistent with a structural role in the NPC core. Knockdown of Nup85 has been reported to result in nuclei largely devoid of FG nucleoporins [42]. Nup85 localizes to the ciliary base by antibody staining (Figure 4A). siRNA-mediated knockdown of nup85 results in a ~50% reduction in Nup85 protein expression after 7 days (Figure S3J), indicating that the staining is specific. Nup85 knockdown results in a decrease in Nup98 incorporation into both NPCs and the nucleoporin complex at the ciliary base (Figure 4B and Figure S3F–I). Nup85 knockdown also results in a ~0.6 μm decrease in average cilia length (Figure 4C), just as we would expect if there is a decrease in Nup98 at the ciliary base. Because of the decrease in Nup98 at the ciliary base after the knockdown of Nup85, we hypothesized that cilia would show exaggerated elongation in response to increases in soluble cytosolic tubulin after Ncd treatment. Indeed, after 4 hours of treatment with Ncd, cilia on Nup85 knockdown cells increased in length substantially more than cilia on control cells (Figure 4C). Two-way ANOVA revealed that the interaction between the siRNA treatment and the Ncd treatment was highly significant (Figure 4D). These data demonstrate that perturbing Nup98 localization to the ciliary base results in similar tubulin trafficking defects as knocking down Nup98 directly. Moreover, these data suggest that the hierarchical assembly of the nucleoporins at the NPC is at least in part conserved at the ciliary base.
Figure 4. Nup85 is required for Nup98 to localize to the ciliary base.
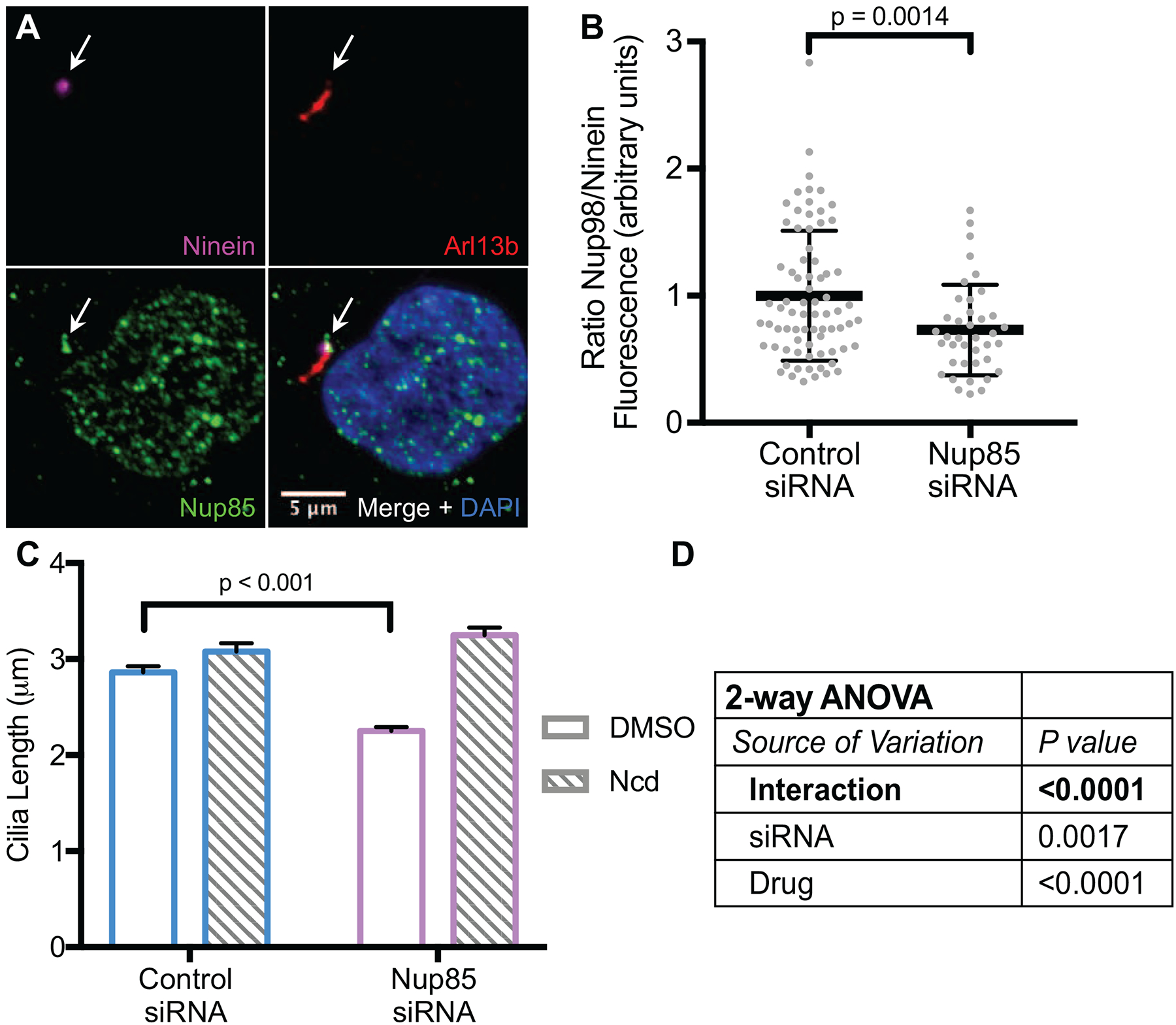
(See also Figure S3) (A) A Nup85 antibody stains both the nucleus and the ciliary base (arrows). (B) Knockdown of Nup85 causes a reduction in Nup98 fluorescence at the ciliary base. (C, D) Cilia length is reduced after the knockdown of Nup85. Cilia length increases substantially more in response to treatment with Ncd after the knockdown of Nup85, consistent with an increase in tubulin diffusion through the ciliary base.
In (B) error bars are S.D., in (C) error bars are S.E.M. p values are calculated by unpaired t test, unless otherwise specified.
Concluding remarks
Primary cilia (9+0) have the equivalent of 18 microtubule (+) ends, a number dwarfed by the number of cytosolic microtubules. We propose that in the absence of a diffusion-limiting gate (of which Nup98 is an essential component) at the ciliary base, the microtubules of the cilium must compete with the microtubules of the cytosol for soluble tubulin heterodimer building blocks, and cilia will grow shorter.
Our results indeed show that Nup98 knockdown significantly increased ciliary elongation in response to increased cytoplasmic soluble tubulin (NCD treatment), suggesting that the tubulin building blocks accumulate in the cilium faster when they can enter by diffusion (in the case of Nup98 depletion) than when they are predominantly carried in by IFT. Further, if tubulin can enter cilia by diffusion when Nup98 is knocked down and soluble tubulin levels are artificially increased by Ncd, then inhibition of IFT in Nup98 knockdown cells would be less consequential for cilia elongation than in cells with an intact diffusion barrier, which is what we observe in RPE cells. Taken together, these results point to a model where the FG nucleoporins at the ciliary base set a size-exclusion limit that under normal conditions slows the efflux of soluble tubulin from the cilium, a necessary step to maintain cilia length.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Martina Brueckner (martina.brueckner@yale.edu).
Experimental Model and Subject Details
Low-passage hTERT RPE-1 cells (ATCC Number: CRL-4000) were cultured in DMEM/F12 medium with 10% FBS at 37°C by standard procedures. hTERT RPE-1 cells are a non-diseased human cell line originating from a female donor. Cells tested negative for mycoplasma infection by InvivoGen PlasmoTest (Catalog: rep-pt1). The cell line was not authenticated, because the source was known, and the cells were phenotypically normal.
Method Details
siRNA
A Stealth siRNAs targeting the mRNAs for Nup98 and Nup85 were purchased from Invitrogen (Nup98 is 10620318; Nup98HSS143176. Nup85 is 1299001; HSS129105). The negative control Stealth siRNA was Invitrogen’s medium GC control (12935–300). siRNAs were transfected into cells with Lipofectamine RNAiMAX (ThermoFisher: 13778150), according to the manufacturer’s instructions, with two modifications: The amount of Lipofectamine was increased from 3μL to 5μL, for 1 reaction pair, and the amount of siRNA was increased from 10pmol to 50pmol. The most efficient knockdown for Nup98 was seen 4 days after transfection, and for all subsequent experiments, these conditions were used. For knockdown of Nup85, all experiments were performed 7 days after knockdown.
Live imaging of diffusion trap experiments
Cells were grown in glass-bottom, #1.0 glass, 24 well plates purchased from MatTek. 24 hours before live-imaging, cells were transfected with trap and prey constructs, using Lipofectamine 2000 (Invitrogen). Plasmid constructs were generous gifts of Takanari Inoue[19]. Before imaging cells were transferred into Leibovitz’s L-15 medium with no phenol red. Imaging was performed on a Zeiss LSM 710 DUO microscope, equipped with an XL-LSM710 S1 incubator. The incubator was maintained at 37°C during all imaging experiments. CFP and YFP were visualized by using the 457 and 514 lines of a LASOS Lasertecknik GmbH argon laser. Before starting the imaging series, Z-plane was set and maintained by a Zeiss Definite Focus module, with adjustments made every 10sec; nevertheless, images with excessive focal drift were not quantified. Rapamycin (Sigma: R0395) was added to the medium to 100nM to trigger trap/prey construct dimerization.
Analysis of Diffusion Trap Experiments
Data were analyzed by a custom program available at: https://github.com/wackywendell/diffusiontrap.
Images are first cropped to a field that included a single cilium. The cilium was outlined in the CFP channel, based on thresholding data that was manually determined in ImageJ. The outline of the cilium determined by thresholding in the CFP channel is applied to the YFP channel, where the fluorescence intensity is also measured. The YFP fluorescence value is divided by the CFP fluorescence value to reduce motion artifact. Following the analysis, the adjusted (YFP/CFP) fluorescence intensities are normalized to the average of the fluorescence intensities for the time points before addition of rapamycin. After all data was collected, t tests were performed to ensure that there was no difference in the total area of cilium quantified between cells treated with the control or with the nup98 siRNA. Furthermore, there was no significant difference in any of the raw fluorescence values measured between the cells treated with the control or with the nup98 siRNA. Each analysis used 6–13 biological replicates.
Deciliation Experiments
A variety of stressors have been shown to stimulate cells to shed their cilia in an exocyst-dependent manner [34]. We used a protocol described in [34], as follows: Ammonium sulfate (J. T. Baker: 0792–01) was added to the media to a final concentration of 30mM. Cells were incubated in ammonium sulfate for 18hours before being fixed and stained for cilia markers. For recovery experiments, cells were washed and placed in fresh media for 24 hours before being fixed for immunostaining. Cilia were counted manually in ImageJ, using the multiple selection tool. The reported experiments were performed 3 times.
Cilia length experiments
Cilia length experiments were conducted on the fourth day after siRNA transfection. Cells were serum-starved for 24 hours before drug administration. Nocodazole (Sigma: M1404) was used at 100nM. Cytochalasin D (Sigma: 22144-77-0) was used at 1μm. NMS-873 (SelleckChem: S7285) was used at 5μm. Drugs were administered at the time points indicated in the figure; however, NMS-873 was administered 5h before time point 0h. If NMS-873 and CD are administered at the same time, then NMS-873 does not prevent CD-promoted cilia elongation because of a difference in drug kinetics (data not shown). Cilia were co-stained with Arl13b and acetylated-tubulin markers. We observed no effect of Nup98 knockdown on the regular overlap of these stainings (acetylated-tubulin antibodies stain both the axoneme and the basal body, while Arl13b stains the cilia membrane). Only cilia with clear staining of both of these markers were used for measurement. The measurement reported is the length of the cilium where these markers overlap (i.e. basal body is not included in length measurement). Between 200–500 cilia were measured for each experimental group. Cilia length was measure with ImageJ software, using the segmented line tools.
Soluble/polymerized tubulin fractionation
Tubulin fractions were taken by a method developed by [10], as described here: cells were quickly washed with ice-cold PBS, before being treated with soluble protein extraction buffer A (137mM NaCl, 20mM Tris-HCl, 1% Triton X-100, 10% glycerol) for 3 minutes at 4°C. Buffer A was removed before cells were lysed in extraction buffer B (buffer A + 1% SDS). Plates were scraped and the B fraction was sonicated. A taxol (Sigma: T7402) treatment served as a control to ensure that the extraction procedure did not perturb polymerized tubulin. The experiment was performed 3 times, with similar results.
Fluorescence Microscopy
Immunofluorescent images were taken on a Zeiss Observer microscope equipped with a Hamamatsu C11440 camera and an ApoTome.2 optical interference set-up. Fluorescence intensities were quantified with ImageJ software.
Quantification and Statistical Analysis
Western blots were quantified in ImageJ. Cilia length measurements were made in ImageJ, as described in Method Details. The accumulation of the YFP prey construct in the cilia was quantified by a custom program. Statistical Analyses were performed in GraphPad Prism software. p-values are recorded for each comparison on the figures, with the statistical test used listed in the figure legend. For cilia length measurements, measurements from at least 3 independent experiments were pooled, totaling between 250–500 cilia for each condition.
Data and Software Availability
Wendell Smith wrote the software for analysis of the diffusion trap data. The software and instructions for use are available at: https://github.com/wackywendell/diffusiontrap.
Supplementary Material
Video S1 Related to Figure 2. Representative recording of YFP-FKBP-PKIM (49kDa) prey accumulating in the cilium of a cell treated with the non-targeting control siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Video S2 Related to Figure 2. Representative recording of YFP-FKBP-PKIM (49kDa) prey accumulating in the cilium of a cell treated with the Nup98 siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Video S3 Related to Figure 2. Representative recording of YFP-FKBP-Luciferase (102kDa) prey accumulating in the cilium of a cell treated with the non-targeting control siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Video S4 Related to Figure 2. Representative recording of YFP-FKBP-Luciferase (102kDa) prey accumulating in the cilium of a cell treated with the Nup98 siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Highlights:
The FG-repeat containing nucleoporin Nup98 localizes to the ciliary base
Nup98 limits the diffusion of soluble molecules >70kDa into the cilium
Nup98 regulates ciliary length by controlling tubulin diffusion in cilia
ACKNOWLEDGEMENTS
We thank Wendell Smith for the diffusion trap data analysis program. We thank members of the Brueckner and Sun labs for discussions of the project. We thank the laboratory of Takanari Inoue for the contribution of diffusion trap constructs. We thank the laboratory of Derek Toomre for the contribution of reagents. This work was supported by NIH RO1 HL124402 to MB
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Goetz SC, and Anderson KV (2010). The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satir P, and Christensen ST (2007). Overview of structure and function of mammalian cilia. Annu Rev Physiol 69, 377–400. [DOI] [PubMed] [Google Scholar]
- 3.Fliegauf M, Benzing T, and Omran H (2007). When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8, 880–893. [DOI] [PubMed] [Google Scholar]
- 4.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, and Verhey KJ (2012). A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nature cell biology 14, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, and Verhey KJ (2014). An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Current biology : CB 24, 2288–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endicott SJ, Basu B, Khokha M, and Brueckner M (2015). The NIMA-like kinase Nek2 is a key switch balancing cilia biogenesis and resorption in the development of left-right asymmetry. Development 142, 4068–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulsmann BB, Labokha AA, and Gorlich D (2012). The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150, 738–751. [DOI] [PubMed] [Google Scholar]
- 8.Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. (2016). Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craft JM, Harris JA, Hyman S, Kner P, and Lechtreck KF (2015). Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. The Journal of cell biology 208, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma N, Kosan ZA, Stallworth JE, Berbari NF, and Yoder BK (2011). Soluble levels of cytosolic tubulin regulate ciliary length control. Mol Biol Cell 22, 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, and Verhey KJ (2010). Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nature cell biology 12, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurd TW, Fan S, and Margolis BL (2011). Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. Journal of cell science 124, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torrado B, Grana M, Badano JL, and Irigoin F (2016). Ciliary Entry of the Hedgehog Transcriptional Activator Gli2 Is Mediated by the Nuclear Import Machinery but Differs from Nuclear Transport in Being Imp-alpha/beta1-Independent. PloS one 11, e0162033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madugula V, and Lu L (2016). A ternary complex comprising transportin1, Rab8 and the ciliary targeting signal directs proteins to ciliary membranes. Journal of cell science 129, 3922–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y, Xiong Y, Shi X, Wu J, Zhao Y, and Jiang J (2017). Regulation of Gli ciliary localization and Hedgehog signaling by the PY-NLS/karyopherin-beta2 nuclear import system. PLoS biology 15, e2002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan S, Whiteman EL, Hurd TW, McIntyre JC, Dishinger JF, Liu CJ, Martens JR, Verhey KJ, Sajjan U, and Margolis B (2011). Induction of Ran GTP drives ciliogenesis. Mol Biol Cell 22, 4539–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, and van Deursen JM (2001). Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proceedings of the National Academy of Sciences of the United States of America 98, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, and Kutay U (2011). Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144, 539–550. [DOI] [PubMed] [Google Scholar]
- 19.Lin YC, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, and Inoue T (2013). Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nature chemical biology 9, 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, and Nachury MV (2013). An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. The Journal of cell biology 203, 129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brailov I, Bancila M, Brisorgueil MJ, Miquel MC, Hamon M, and Verge D (2000). Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res 872, 271–275. [DOI] [PubMed] [Google Scholar]
- 22.Fontoura BM, Blobel G, and Matunis MJ (1999). A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. The Journal of cell biology 144, 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoelz A, Debler EW, and Blobel G (2011). The structure of the nuclear pore complex. Annu Rev Biochem 80, 613–643. [DOI] [PubMed] [Google Scholar]
- 24.von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. (2015). In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, and Hetzer MW (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiter JF, Blacque OE, and Leroux MR (2012). The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takao D, Wang L, Boss A, and Verhey KJ (2017). Protein Interaction Analysis Provides a Map of the Spatial and Temporal Organization of the Ciliary Gating Zone. Current biology : CB 27, 2296–2306 e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nature genetics 43, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, et al. (2011). MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. The Journal of cell biology 192, 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, and Peterson AS (2011). A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nature cell biology 14, 61–72. [DOI] [PubMed] [Google Scholar]
- 31.Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, and Witman GB (2010). CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. The Journal of cell biology 190, 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, and Tsukita S (2013). Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. The Journal of cell biology 203, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang TT, Su J, Wang WJ, Craige B, Witman GB, Tsou MF, and Liao JC (2015). Superresolution Pattern Recognition Reveals the Architectural Map of the Ciliary Transition Zone. Sci Rep 5, 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overgaard CE, Sanzone KM, Spiczka KS, Sheff DR, Sandra A, and Yeaman C (2009). Deciliation is associated with dramatic remodeling of epithelial cell junctions and surface domains. Mol Biol Cell 20, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall WF, and Rosenbaum JL (2001). Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. The Journal of cell biology 155, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. (2013). Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341, 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raman M, Sergeev M, Garnaas M, Lydeard JR, Huttlin EL, Goessling W, Shah JV, and Harper JW (2015). Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nature cell biology 17, 1356–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, and Hirokawa N (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837. [DOI] [PubMed] [Google Scholar]
- 39.Besschetnova TY, Kolpakova-Hart E, Guan Y, Zhou J, Olsen BR, and Shah JV (2010). Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Current biology : CB 20, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez de Diego A, Alonso Guerrero A, Martinez AC, and van Wely KH (2014). Dido3-dependent HDAC6 targeting controls cilium size. Nature communications 5, 3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabut G, Doye V, and Ellenberg J (2004). Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nature cell biology 6, 1114–1121. [DOI] [PubMed] [Google Scholar]
- 42.Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, and Forbes DJ (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Molecular cell 11, 853–864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1 Related to Figure 2. Representative recording of YFP-FKBP-PKIM (49kDa) prey accumulating in the cilium of a cell treated with the non-targeting control siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Video S2 Related to Figure 2. Representative recording of YFP-FKBP-PKIM (49kDa) prey accumulating in the cilium of a cell treated with the Nup98 siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Video S3 Related to Figure 2. Representative recording of YFP-FKBP-Luciferase (102kDa) prey accumulating in the cilium of a cell treated with the non-targeting control siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Video S4 Related to Figure 2. Representative recording of YFP-FKBP-Luciferase (102kDa) prey accumulating in the cilium of a cell treated with the Nup98 siRNA. Rapamycin was added to induce prey accumulation in the cilium at the indicated time.
Data Availability Statement
Wendell Smith wrote the software for analysis of the diffusion trap data. The software and instructions for use are available at: https://github.com/wackywendell/diffusiontrap.


