Abstract
Candida albicans is an opportunistic fungal pathogen and a commensal organism that commonly colonizes mucosal surfaces, including those inside the human mouth. To help control C. albicans, human saliva contains the antifungal peptide histatin 5 (Hst-5), which has strong antifungal activity against C. albicans. However, the pathogen produces secreted aspartic proteases (Saps) that cleave Hst-5 at lysine residues and eliminate its antifungal properties. We designed variants of Hst-5 with its lysine residues substituted with arginine or leucine to evaluate the effect on proteolysis by Saps. We found site-, residue-, and Sap-dependent effects from single amino-acid substitutions. The K17R and K17L modifications led to dramatic results, with over 77% and 100% intact peptide remaining after incubation with Sap9 and Sap2, respectively, compared to 47% and 61% of Hst-5. This decrease in proteolysis was accompanied by a reduction in cleavage on the C-terminal side of K17, suggesting the Saps prefer lysine at K17 for cleavage. Incubation with C. albicans cells and culture supernatant corroborated the results with purified Saps and highlighted their biological relevance. The modifications to Hst-5 do not diminish the antifungal activity of Hst-5, and, in fact, the K17R, K17L, and K11R peptides retained significantly more antifungal activity after treatment with Saps than Hst-5. Our results indicate that single amino-acid modifications drastically impact both proteolysis at the modification sites and the overall level of proteolysis of the peptide, demonstrating the potential of designing peptides for resistance to proteolysis as a means for improving therapeutic efficacy.
Keywords: Secreted aspartic proteases, histatin-5, proteolysis, Candida albicans, antimicrobial peptides
INTRODUCTION
Candida albicans is a fungal species that is part of the normal human flora. It commonly colonizes mucosal surfaces, including the oral cavity in over 35% of healthy people [1]. However, C. albicans is also an opportunistic pathogen, and, under conditions of immune system disruption, such as infection with the human immunodeficiency virus (HIV), it causes an array of infections, most notably oral candidiasis, which is also called oral thrush.
To help prevent infections by C. albicans and other organisms, the human oral saliva contains an arsenal of proteins and peptides [2]. One such peptide that fights against C. albicans is the antimicrobial peptide histatin 5 (Hst-5). It is one of twelve members of the histatin family of histidine-rich peptides secreted by salivary glands in the human mouth [3], with the 24-amino-acid Hst-5 having the strongest activity [3, 4]. As part of host innate immunity, Hst-5 has been proposed to play a crucial role in maintaining C. albicans at commensal levels in the oral cavity [5]. Reduced levels of Hst-5 have been reported in HIV-positive patients [6] and may contribute to their high susceptibility to oral thrush. Unlike some other common antimicrobial peptides, Hst-5 does not act on C. albicans cells by forming pores on the cell membrane [7]; instead, the peptide acts intracellularly, ultimately leading to ion imbalance and volume loss that causes cell death [8].
Although Hst-5 has potent activity against C. albicans, the fungal pathogen produces a family of ten secreted aspartic acids (Saps), some of which can degrade and inactivate Hst-5. The Saps play a role in a number of cellular processes and attributes, including cell adhesion, cell integrity, and virulence [9]. While Sap1 to Sap8 are fully secreted to the extracellular environment, Sap9 and Sap10 remain attached to the cell wall via a glycosylphosphatidylinositol (GPI) anchor [9, 10]. Each Sap contains the two conserved aspartic acids characteristic of aspartic proteases, along with four conserved cysteine residues [11].
Although Sap9 is the most highly expressed Sap in strains isolated from patients with both oral and vaginal Candida infections [12], studies on the substrate specificity of the Saps have focused most extensively on Sap2. Sap2 has a broad specificity that includes immune host proteins, such as immunoglobulin A and lactoferrin, and antimicrobial peptides like Hst-5 [5, 10, 13, 14]. Recently, the interaction of Sap2 and other Saps with antimicrobial peptides, including Hst-5, has been more carefully studied [5, 13, 14]. Meiller et al. showed that at physiological pH Hst-5 is vulnerable to proteolysis by Sap2, Sap9, and, to a lesser extent, Sap10 [5]. Furthermore, Bochenska et al. showed Hst-5 can also be cleaved by Sap1, Sap3–4, and Sap7–8 when tested at the optimal pH condition for each Sap [15]. In both studies, a lysine residue was typically present on at least one side of the sites cleaved by the Saps. These results suggest Saps may target the lysine residues in the Hst-5 sequence, which is supported by previous studies with peptide libraries that indicate Saps prefer hydrophobic residues or basic residues [16–19]. It is important to note, however, these previous studies typically used libraries of random amino acid sequences with no biological relevance [16–18] and did not consider the effect of cleavage on the functionality of the peptides. Furthermore, the residue specificity was determined by varying the residue on only one side of the cleavage site, while fixing the residue on the other side [17, 18]. Thus, they do not account for the potential roles of residues further way from the cut site.
To improve the understanding of the interaction between Hst-5 and Saps, we designed Hst-5 variants with substitutions made at the lysine residues. While earlier investigations have elucidated how modifications to the sequence of Hst-5 modulate antifungal activity [20–25], the effect of modifications on degradation specifically by aspartic proteases has not yet been explored. The designed variants were evaluated for susceptibility to proteolysis by C. albicans Saps and for antifungal activity against C. albicans. Our results demonstrate that even a single amino acid modification is sufficient to significantly modulate the degradation of Hst-5 by the proteases or C. albicans cells, while maintaining antifungal potency. Based on these findings, designing peptides for reduced proteolysis could be a viable approach for engineering antimicrobial peptides with increased resistance to proteolysis and, thus, longer-lived antimicrobial potency
RESULTS AND DISCUSSION
To study the interaction of Hst-5 with C. albicans Saps, we evaluated the proteolysis of Hst-5 and eight variants by Sap2 and Sap9, which substantially cleaved Hst-5 in previous work from Meiller et al. [5]. Our design of the Hst-5 variants focused on the four lysine residues in the peptide, as we observed that lysine residues are prominent at the reported cleavage sites of Hst-5 with Saps or C. albicans cells [5, 15]. We hypothesized that these lysine residues are important for the recognition or cleavage of Hst-5 and replaced each of the lysine residues with either an arginine or a leucine (Table 1). The arginine substitutions were selected to preserve the positive charge, as the cationic nature of antimicrobial peptides often plays a role in their function [26, 27]. The leucine substitutions were selected to remove the positive charge, which could affect the interaction of the cleavage sites with the aspartic acid residues in the active site of the Saps or the interaction of the Saps with the peptides as a whole.
Table 1.
Variants of Hst-5 with arginine or leucine substitutions at lysine residues.
| Peptide | Sequencea | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
| Hst-5 | D | S | H | A | K | R | H | H | G | Y | K | R | K | F | H | E | K | H | H | S | H | R | G | Y |
| K5R | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| K5L | - | - | - | - | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| K11R | - | - | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - |
| K11L | - | - | - | - | - | - | - | - | - | - | L | - | - | - | - | - | - | - | - | - | - | - | - | - |
| K13R | - | - | - | - | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - |
| K13L | - | - | - | - | - | - | - | - | - | - | - | - | L | - | - | - | - | - | - | - | - | - | - | - |
| K17R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - |
| K17L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | - | - | - | - | - | - | - |
Dash indicates residue was unchanged from the parent Hst-5.
Lysine substitutions modulate susceptibility to proteolysis by purified Saps
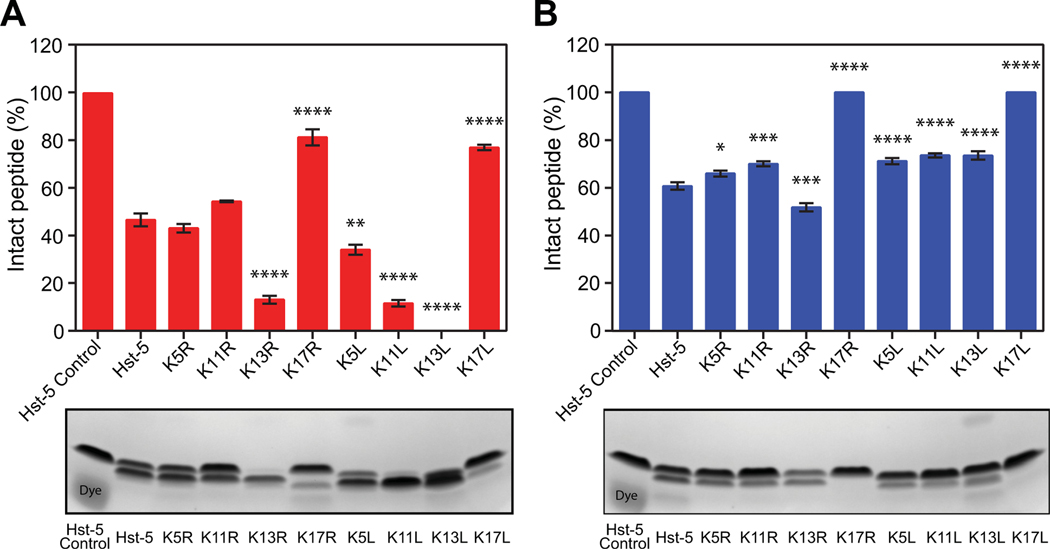
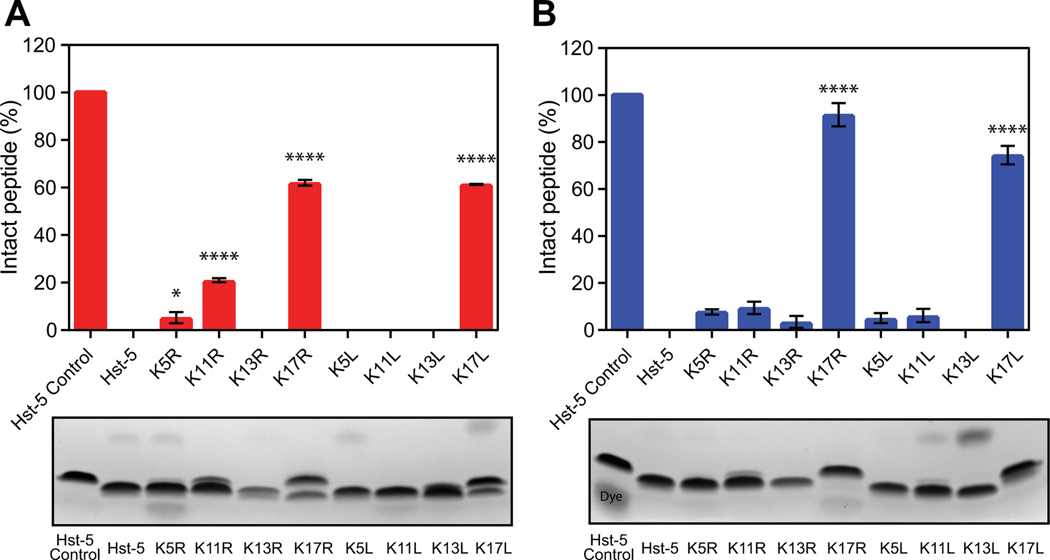
To determine whether the single-residue substitutions have an effect on the overall degradation of Hst-5, we incubated Hst-5 and each modified peptide with purified recombinant Sap9 and Sap2. Hst-5 and each variant in Table 1 were incubated with or without 3.13 μg·mL–1 Sap9 or 6.25 μg·mL–1 Sap2 for 2 h at 37 °C in 1 mM sodium phosphate buffer (NaPB). We then separated the intact peptide from the degradation products using gel electrophoresis and quantified the level of degradation using densitometric analysis of Coomassie-stained gels (Fig. 1). Both arginine and leucine substitutions at the K17 site led to a dramatic decrease in degradation by Sap9 and Sap2. Following incubation with Sap9, 82% and 77% of K17R and K17L, respectively, remained intact compared to 47% of Hst-5 (Fig. 1A). No detectable degradation of the K17R and K17L variants was visible after incubation with Sap2, while only 61% of the parent Hst-5 peptide remained intact (Fig. 1B).
Fig. 1.
Degradation of parent Hst-5 and Hst-5 variants by purified (A) Sap9 and (B) Sap2. The peptides (150 μg·mL–1) and Saps (3.13 μg·mL–1 Sap9 and 6.25 μg·mL–1 Sap2) were incubated for 2 h at 37 °C. Samples were run on a gel, and the amount of intact peptide was quantified by densitometry to compare the amount of intact peptide (upper band) to the peptide fragments. Error bars represent standard error of the mean (n = 3). The number of asterisks indicates the level of statistical significance against parent Hst-5 incubated with Sap: * for p< 0.05, ** for p< 0.01, *** for p< 0.001, and **** for p< 0.0001. The lower band in the Hst-5 control lanes is due to Coomassie dye.
With the exception of the K17 residue, modification of lysine residues to leucine made the Hst-5 variants more susceptible to degradation by Sap9. The K5L, K11L, and K13L peptides all showed greater degradation than their arginine-substituted counterparts or parent Hst-5 (Fig. 1A). In fact, the K13L peptide was degraded to the extent that no intact peptide could be detected on the gel.
While leucine substitutions resulted in more proteolysis of the modified peptides by Sap9, they led to a decrease in proteolysis by Sap2 (Fig. 1B). With the exception of K13R, all of the Hst-5 variants exhibited a decrease in degradation compared to the parent Hst-5 after incubation with Sap2.
These results demonstrate the ability to easily detect changes in a peptide’s susceptibility to proteolysis by gel electrophoresis and indicate that this approach is feasible for exploring how aspartic proteases interact with antimicrobial peptides. Additionally, they demonstrate that single-residue changes can have a major impact on the susceptibility of a peptide to degradation by Saps.
Proteolysis by purified Saps is consistent with proteolysis by C. albicans cells
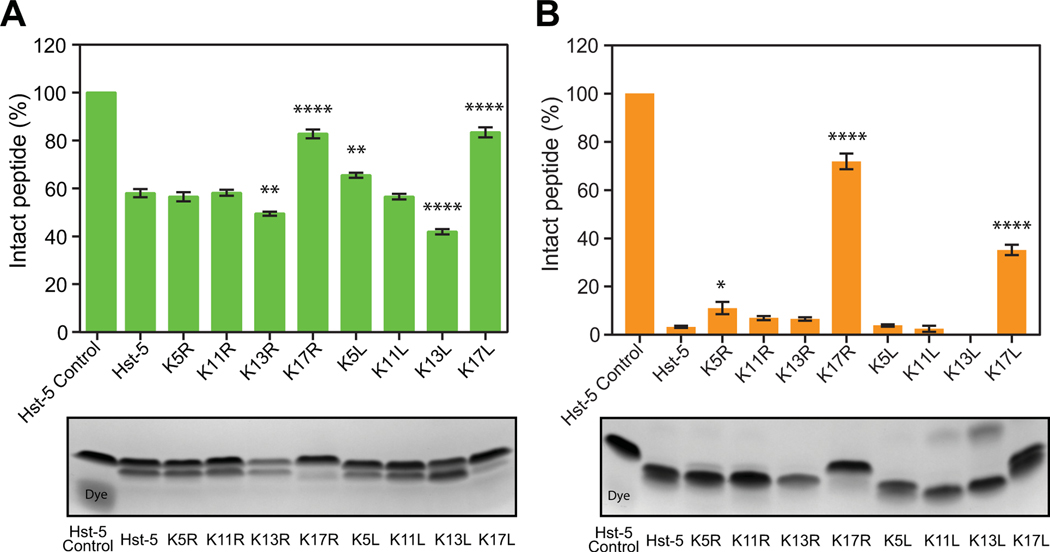
After observing the effect of residue modifications on the cleavage of Hst-5 by the purified recombinant Saps, we evaluated whether incubation of the peptides with Saps natively produced by C. albicans cells (rather than with Saps produced recombinantly) would yield similar results. Hst-5 and the variants were incubated with C. albicans cells or with the supernatant from a C. albicans culture. For incubation with C. albicans cells, the cells were washed and resuspended in 100 mM NaPB. The high ionic strength of this buffer prevents internalization of the peptides by C. albicans [28], allowing analysis of degradation of the peptides without the confounding effects of peptide internalization, which would reduce the amount of peptide available for degradation by the Saps in the buffer. For the supernatant samples, the supernatant from a subculture grown for 17.5 h was used to ensure sufficient secretion of Saps into the culture supernatant, and the buffer was exchanged to 1 mM NaPB. Following incubation with the cells or the buffer-exchanged culture supernatant, the peptide samples were run on a gel and stained, and densitometric analysis revealed a pattern that exhibits characteristics similar to the results observed with the individual purified Saps (Fig. 2). Both K17 modifications resulted in an increase in the amount of intact peptide remaining. The resistance to proteolysis was particularly apparent when the peptides were incubated with the culture supernatant. While almost no intact parent Hst-5 remained following incubation with the supernatant, 72% of K17R and 35% of K17L remained intact. The larger difference between the arginine and leucine substitutions at the K17 site is potentially due to the presence of additional proteolytic enzymes in the supernatant, including Saps other than Sap2 and Sap9.
Fig. 2.
Degradation of parent Hst-5 and Hst-5 variants by (A) C. albicans cells and (B) C. albicans culture supernatant. The peptides (150 μg·mL–1) and C. albicans cells (1×109 cells·mL–1) (A) or culture supernatant (B) were incubated for 2 h at 37 °C. Samples were run on a gel, and the amount of intact peptide was quantified by densitometry to compare the amount of intact peptide (upper band) to the peptide fragments. Error bars represent standard error of the mean [n = 6 for (A) and n = 3 for (B)]. The number of asterisks indicates the level of statistical significance against parent Hst-5 incubated with cells: * for p< 0.05, ** for p< 0.01, and **** for p< 0.0001.The lower band in the Hst-5 control lane is due to Coomassie dye.
The rest of the modified peptides followed a pattern of proteolysis consistent with whether more of the anchored Sap9 (Fig. 2A) or secreted Sap2 (Fig. 2B) was likely present in the assay. Incubation with cells (likely dominated by cell-wall anchored Saps) led to an increase in the degradation of K13L compared to the parent Hst-5, while it had no significant effect on K5R, which follows the purified Sap9 proteolysis patterns. Meanwhile, the K5R peptide showed an increase in resistance to proteolysis when incubated with the supernatant (likely dominated by secreted Saps) (Fig. 2B), as was observed with purified Sap2. These outcomes demonstrate that both the cell-wall anchored Sap9 and the fully secreted Sap2 play a role in proteolysis by C. albicans. Overall, results from incubation with the purified Saps are consistent with the results from incubation with the fungal cells and supernatant, indicating that using purified Saps is a reasonable approach for studying biologically relevant effects of peptide sequence on susceptibility to Saps produced by cells.
Mass spectrometry confirms the effects of lysine modifications
To gain a more in-depth understanding of how the modifications to the peptide sequence affect the cleavage of Hst-5 by Saps, we used mass spectrometry to determine the cleavage sites and the relative abundance of peptide fragments. After incubation of the peptides with each Sap, the four-amino-acid peptide MRFA was added as an internal standard, and samples were directly injected into the mass spectrometer.
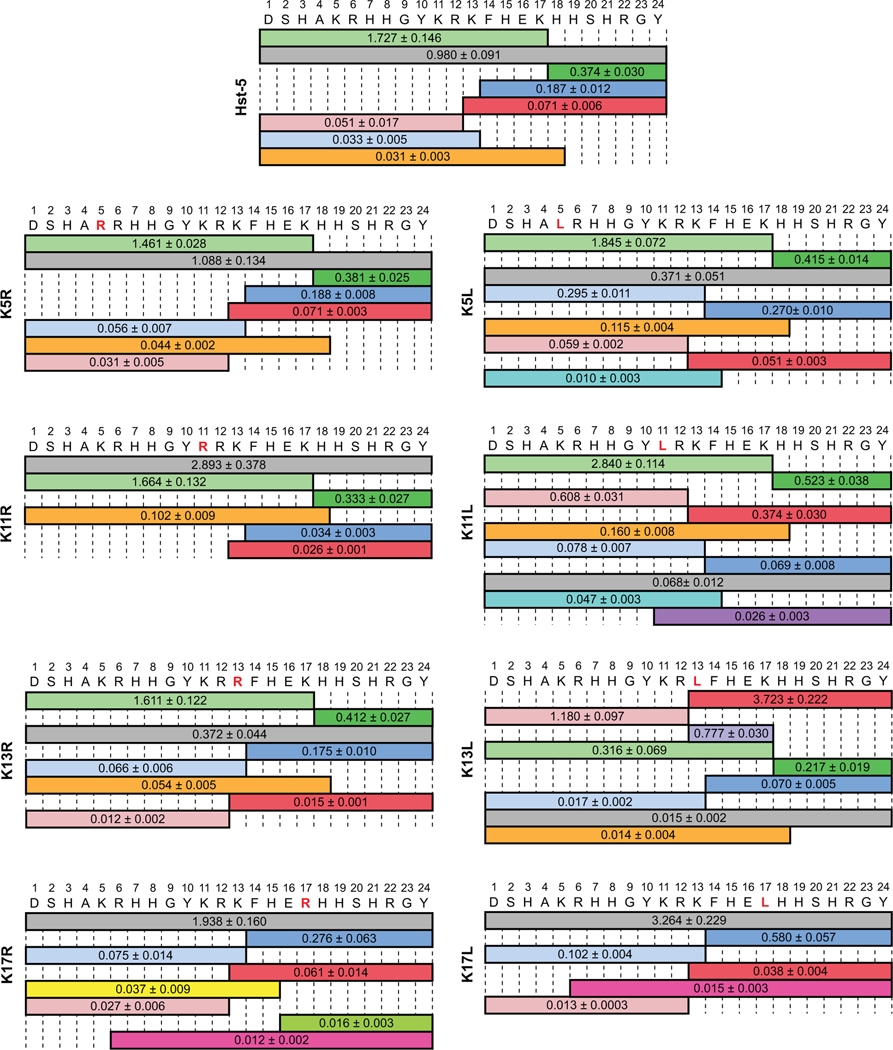
Mass spectrometry of the parent Hst-5 peptide incubated with Sap9 showed that the signal for the degradation fragment containing amino acids 1–17 of Hst-5 was higher than the intact peptide (Fig. 3). We observed cleavage on both sides of K13 and on the C-terminal side of K17, which are cleavage sites that have been previously reported for Sap9 and Sap2 [5, 15]. No cleavage was seen on either side of the K11 residue, consistent with the work of Bochenska et al. [15], though Meiller et al. did report cleavage at the N-terminal side of this residue [5]. We also detected cleavage between the H18 and H19 residues of the parent Hst-5, which has previously been observed after incubation with C. albicans cells [29] but not specifically associated with Sap9.
Fig. 3.
Relative mass spectrometry signal intensity of intact peptide (gray) and peptide fragments produced by incubation of parent Hst-5 and Hst-5 variants (modified residue in red) with 3.13 μg·mL–1 Sap9. The values on each fragment indicate the signal for the fragment relative to the signal for an internal standard. Fragments with signals greater than 0.01 relative to the standard are shown. The relative signal is the mean with standard error (n = 3).
In general, the degradation of the Hst-5 variants with Sap9 produced results in agreement with the gel electrophoresis data. For K17R and K17L, the most intense signal came from the intact peptide, as expected from the large percentage of intact peptide seen in the gel electrophoresis results (Fig. 1A). Furthermore, while the parent Hst-5 was cleaved on the C-terminal side of K17, neither K17R nor K17L showed significant cleavage at this site. K11R also shows the intact peptide to be the species with the highest signal. With the exception of the peptides with K17 substitutions, the variants with leucine substitutions showed relatively lower levels of intact peptide than the corresponding arginine-substituted peptides. Furthermore, K13L showed an apparent shift in cleavage site preference compared to the parent Hst-5. The fragments containing amino acids 1–12 and 13–24 had higher signals for K13L than the fragments containing amino acids 1–17 and 18–24, while the latter fragments had higher signals for the other modified peptides and parent Hst-5. The loss of a cleavage site for the K17-modified peptides and the shift in cleavage site preference for K13L show that the substitutions affect cleavage around the modified residues. In addition, the presence of more intact peptide for K11R indicates that substitutions can also affect the peptides as a whole.
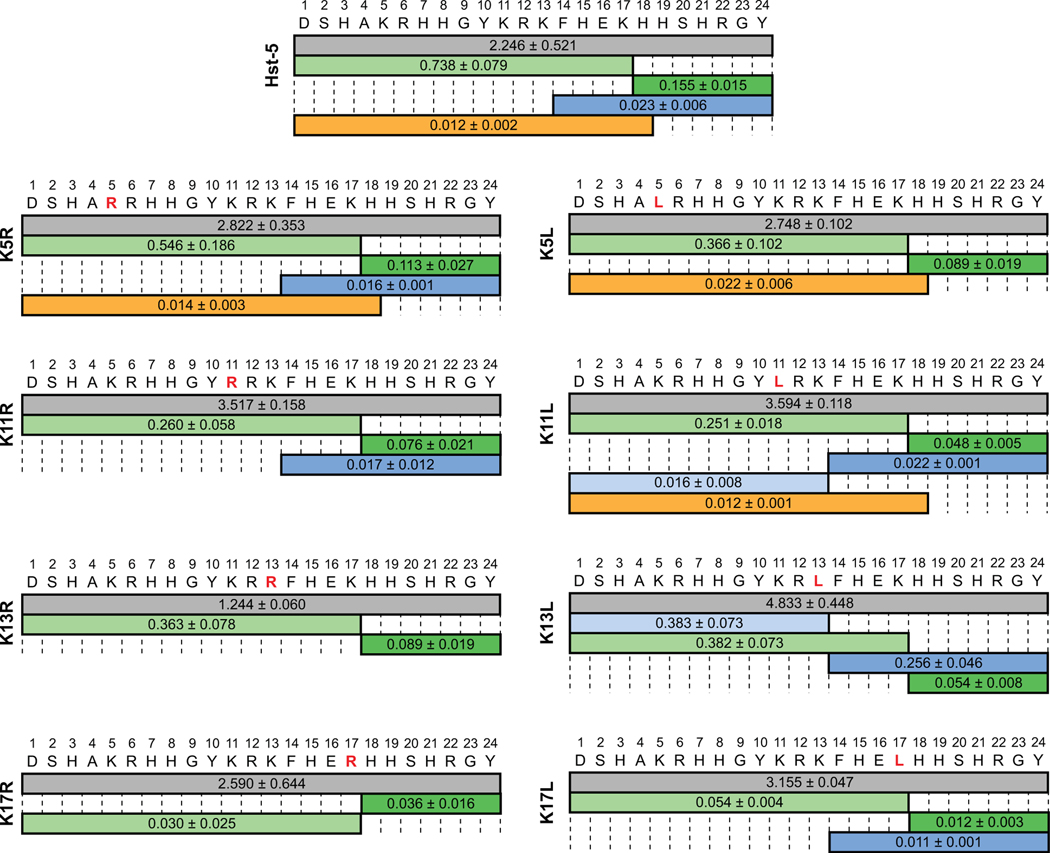
In contrast to incubation with Sap9, incubation of the parent Hst-5 peptide with Sap2 resulted in the intact peptide having the highest signal under the conditions tested (Fig. 4). A lower number of distinct fragments was detected for Hst-5 degradation by Sap2 compared to Sap9. For example, the fragments containing amino acids 1–12 and 13–24 were not significantly detected in incubation with Sap2 but were detected with Sap9. The cleavage sites that did occur with Sap2 were also observed with Sap9 and are in agreement with previously reported results [5, 15], except that cleavage between H18 and H19 was not previously specifically attributed to Sap2. Meiller et al. also reported a cleavage site at the C-terminal side of K5 [5], which neither we nor Bochenska et al. observed [15].
Fig. 4.
Relative mass spectrometry signal intensity of intact peptide (gray) and peptide fragments produced by incubation of parent Hst-5 and Hst-5 variants (modified residue in red) with 6.25 μg·mL–1 Sap2. The values on each fragment indicate the signal for the fragment relative to the signal for an internal standard. Fragments with signals greater than 0.01 relative to the standard are shown. The relative signal is the mean with standard error (n = 3).
As with incubation of the parent Hst-5, incubation of all the Hst-5 variants with Sap2 resulted in higher signals for intact peptides than for proteolytic fragments. The outcome was in line with the gel electrophoresis result, which showed that, overall, the Hst-5 variants incubated with Sap2 had more intact peptide remaining than when incubated with Sap9 (Fig. 1). Substitutions at K17 did lead to a significant decrease in signals from fragments formed by cleavage at the C-terminal side of K17; however, unlike Sap9, Sap2 still cleaved K17L and K17R at this site.
The trends seen with the gel electrophoresis (Fig. 1) and mass spectrometry results (Fig. 3 and 4) generally corroborate each other. Enhanced resistance to proteolysis was observed around the K17 residue for substitutions to both arginine and leucine with both Saps. The similar increase in resistance to degradation at K17, independent of the charge of the substituted residue, indicates that Sap9 and Sap2 have a preference for lysine at the K17 site within the Hst-5 sequence and not simply a preference for a basic residue. Unlike Sap9, Sap2 still cleaved at the C-terminal side of K17, suggesting the preference is more stringent for Sap9 than Sap2. At the other locations where lysine residues were modified, Sap9 appears to favor an uncharged leucine over a positively charged arginine or lysine. The leucine residues at these sites are N- or C-terminal to an arginine in the peptide sequence, and their preference by Sap9 agrees with previous work indicating Sap9 favors cleavage of peptides that contain leucine at the N-terminal side of an arginine [16]. Sap2, on the other hand, appears to favor lysine at most of the locations in Hst-5 that we studied. Within the Hst-5 sequence, the preference is residue-dependent rather than charge-dependent, as Sap2 preferred lysine over both arginine and leucine. The one exception was at the K13 site, where Sap2 appears to prefer an arginine. Collectively, these results confirm our gel electrophoresis results and indicate single amino-acid modifications can affect overall susceptibility to cleavage and significantly reduce cleavage at the modification site.
Most residue substitutions do not diminish antifungal activity
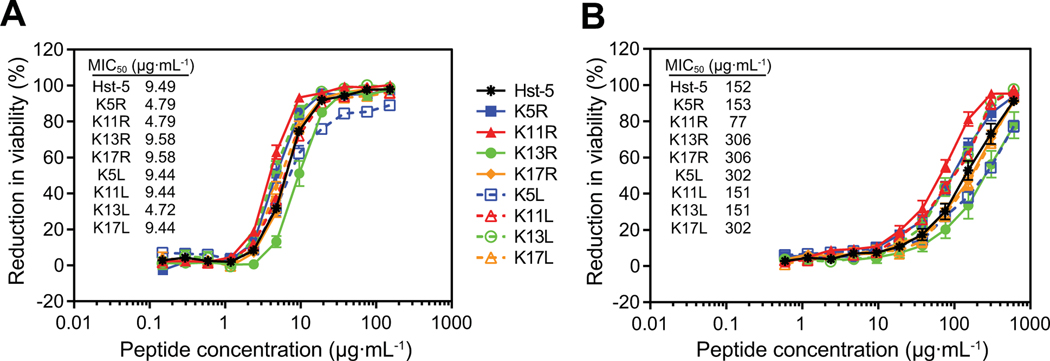
Some previously reported modifications to Hst-5 had a negative impact on the peptide’s antifungal activity [20–22]. To ensure that modification of lysine residues does not adversely affect the antifungal activity, we performed an antifungal activity assay to determine the reduction in the viability of C. albicans cells after exposure to the parent Hst-5 peptide and the Hst-5 variants (Fig. 5). Serially diluted peptides were incubated with C. albicans cells at 2.5×105 cells·mL–1 or 2.5×107 cells·mL–1 C. albicans for 30 min at 30 °C in 1 mM NaPB. The mixtures were then diluted, transferred to YPD media, and incubated overnight to determine the reduction in cell viability due to incubation with the peptides.
Fig. 5.
Antifungal activity of parent Hst-5 and the Hst-5 variants. Serially diluted peptides were incubated with (A) 2.5×105 cells·mL–1 or (B) 2.5×107 cells·mL–1 C. albicans for 30 min at 30 °C. Error bars represent standard error of the mean (n = 6). MIC50 values for the peptides are provided.
When incubated with C. albicans at 2.5×105 cells·mL–1, Hst-5 variants behaved similarly to parent Hst-5, with an increasing reduction in viability with increasing peptide concentration (Fig. 5A). Hst-5 had a minimum inhibitory concentration for 50% inhibition of growth (MIC50) of 9.49 μg·mL–1, and the MIC50 values for the Hst-5 variants were within one dilution factor of Hst-5 (Fig. 5A). The similar growth inhibition curves and MIC50 values show that the antifungal activity of Hst-5 is tolerant to substitutions of its lysine residues.
Increasing the cell concentration to 2.5×107 cells·mL–1 resulted in a shift of the MIC50 value for Hst-5 to 152 μg·mL–1 (Fig. 5B), which is expected with the hundred-fold increase in cell density. With this higher cell density, the growth inhibition curves for each peptide were more widely separated, allowing differences between the antifungal activities of the peptides to be more apparent (Fig. 5B). The K11R curve showed enhanced antifungal activity that was not apparent at the lower cell concentration. The K11 residue is present in both truncated peptides that were previously identified as active fragments for antifungal activity—fragment C14 (residues 11–24) [30] and fragment P-113 (residues 4–15) [23]. Therefore, the K11R modification is at a critical location in the peptide and altering the interaction of the peptide with the Saps at this residue could have a larger effect on the antimicrobial activity of the peptide and its degradation fragments than most other lysine residues. Interestingly, neither K17R nor K17L showed improvement in antifungal activity despite the gel and mass spectrometry results that demonstrated more intact peptide. This may be a function of the incubation time in the assay. The antifungal assay we used (based on the assay used by several other groups [22, 23, 31]) incorporates a short contact time for the peptides and C. albicans to minimize the effect of cell division on assay results. Since the kinetics of proteolysis of the in vivo Saps may be slow enough that their effect on Hst-5 is not easily observed under these conditions, improvements of the K17 modifications on antifungal activity may not be observed at the conditions tested. Overall, the antifungal activity assay demonstrates that replacement of a lysine with an arginine or a leucine does not abolish the antifungal activity of the Hst-5 and may lead to improvements. While there have been no previous studies evaluating the effect of lysine to arginine modifications on the antifungal activity of Hst-5, Helmerhorst et al. reported that a lysine to leucine substitution in a fragment of Hst-5 called dh-5 showed similar activity to the peptide containing lysine [32], further supporting the tolerance of Hst-5 to lysine-to-leucine modifications.
Several Hst-5 variants retain antifungal activity after treatment with Saps
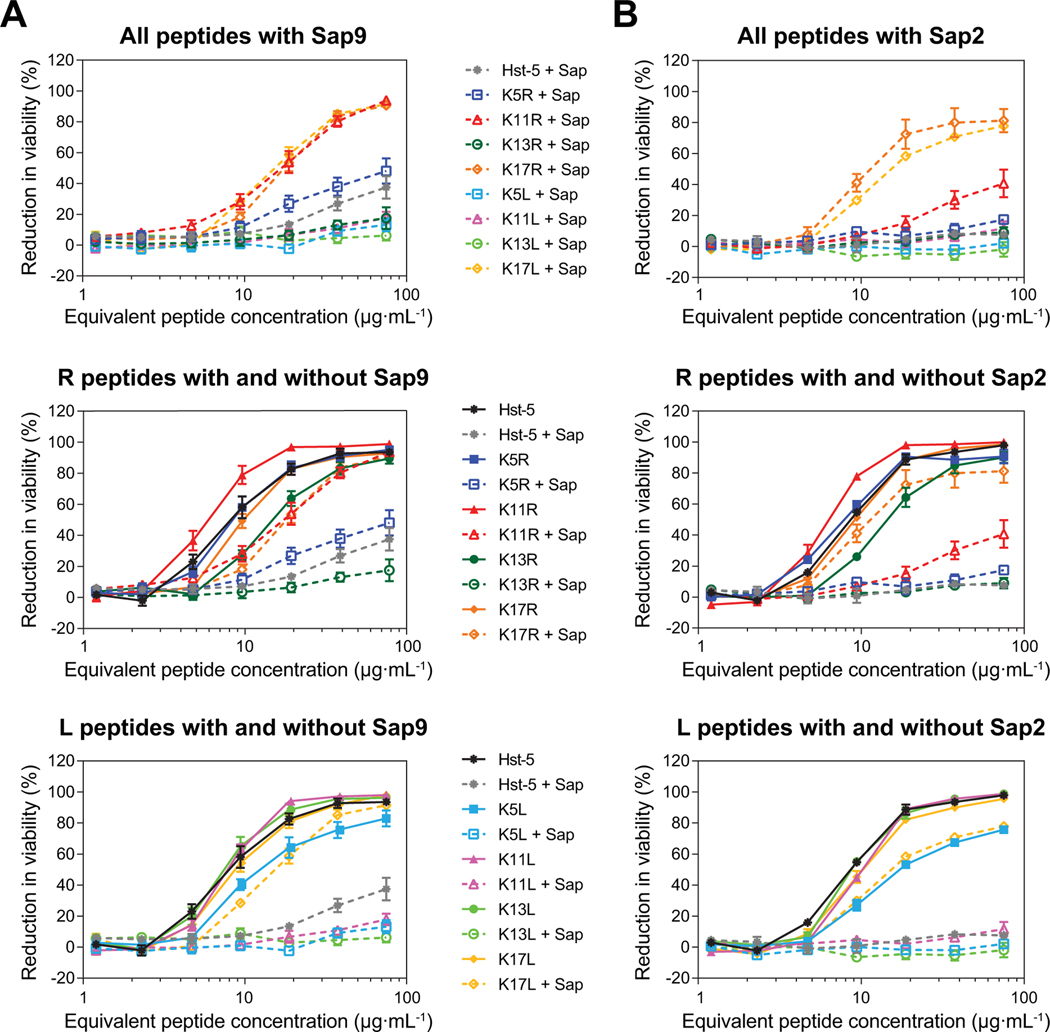
After confirming that substitutions to the lysine residues did not eliminate antifungal activity, we investigated whether the peptides retained their antifungal activity after exposure to purified Saps. To amplify differences in antifungal activity due to degradation by the Saps and to simulate an environment in which Saps are overexpressed, we increased the concentrations of the Saps by at least two-fold compared to the concentrations used for the gel electrophoresis data in Fig. 1 (to 6.25 μg·mL–1 for Sap9 and 18 μg·mL–1 for Sap2). Incubation with Sap9 led Hst-5 to lose over 60% of its antifungal activity, while incubation with Sap2 almost completely eliminated its activity (Fig. 6). In contrast, both K17 variants retained much of their antifungal activity after exposure to each Sap, which is consistent with the high level of intact peptide that remained following incubation with both Sap9 (over 60% remaining for each peptide) and Sap2 (over 90% remaining for K17R and over 70% remaining for K17L) (Fig. 7A). The K11R peptide also had a strong performance in the antifungal activity assay following exposure to Sap9, with a level of activity similar to that of the K17 variants. The improved activity following incubation indicates that resistance to proteolysis could prolong the antimicrobial potency in the presence of the Saps produced by C. albicans.
Fig. 6.
Antifungal activity of parent Hst-5 and the Hst-5 variants following incubation with Sap 9 and Sap2. The peptides (150 μg·mL–1) with (A) Sap9 (6.25 μg·mL–1) and (B) Sap2 (18 μg·mL–1) were incubated for 2 h at 37 °C. Samples were serially diluted and incubated with 2.5×105 cells·mL–1 C. albicans for 30 min at 30 °C. Error bars represent standard error of the mean (n = 6).
Fig. 7.
Degradation of parent Hst-5 and Hst-5 variants by purified (A) Sap9 and (B) Sap2. The peptides (150 μg·mL–1) and Saps (6.25 μg·mL–1 Sap9 and 18 μg·mL–1 Sap2) were incubated for 2 h at 37 °C in 1 mM NaPB. Samples were run on a gel to separate the intact peptide and peptide fragments. The amount of intact peptide was quantified by densitometry to compare the amount of intact peptide (upper band) to the peptide fragments. Error bars represent standard error of the mean (n = 3). The number of asterisks indicates the level of statistical significance against parent Hst-5 incubated with the Saps: * for p< 0.05 and **** for p< 0.0001. The lower band in the Hst-5 control lane in (B) is due to Coomassie dye.
Overall, following degradation by each of the Saps, the arginine-substituted peptides retained more antifungal activity compared to the analogous leucine-substituted peptides. For example, the Sap9- and Sap2-degraded K5R and K11R variants still displayed some antifungal activity, while the degraded K5L and K11L variants showed almost none. The K5R and K11R peptide also retained some intact peptide after Sap9 incubation (4.5% and 20% of peptide, respectively), while no statistically significant amount of these peptides remained after Sap2 incubation (Fig. 7B). The presence of intact K5R and K11R but not intact K5L and K11L after incubation with Sap9 further supports a preference for leucine at these sites by Sap9 and also shows a preference for lysine over arginine, since the parent Hst-5 is completely degraded under these conditions.
Although the variants with the largest amount of intact peptide remaining had the strongest antifungal activity, intact peptide was not required for antifungal activity. The parent Hst-5 peptide had no intact peptide remaining after incubation with Sap9, but it did show a measurable level of antifungal activity. Moreover, while less intact K11R peptide remained after incubation with Sap9 compared to K17R or K17L (Fig. 7A), the antifungal activity was at the same level as the K17 modified peptides. This suggests that the antifungal activity comes not only from the intact K11R peptide, but also from its proteolytic fragments. This is consistent with previous studies with Hst-5 that found varying levels of antifungal activity from Hst-5 truncation peptides [23, 30, 32].
Our results indicate that, while some peptide modifications make the intact peptide more robust (e.g., K17R, K17L), others can lead to degradation fragments with improved antifungal activity compared to the degradation fragments of parent Hst-5 (e.g., K11R). Consequently, even if the intact versions of Hst-5 variants do not have improved antifungal activity themselves, a longer half-life of intact peptide or a higher activity of degradation fragments leads to improved therapeutic potential of K11R, K17R, and K17L compared to Hst-5. We recently showed that Hst-5 delivered in a bioadhesive hydrogel could effectively prevent the development of an oral C. albicans infection in mice [33]. The modified peptides could be delivered directly to mucosal surfaces in the same manner, or they could be incorporated into coatings on implantable devices or prosthetics using thin films, which have been used previously to successfully create antimicrobial surfaces using other antimicrobial peptides [34–36].
In conclusion, we have demonstrated that changing a single lysine residue in the Hst-5 sequence can significantly alter its proteolysis by purified Sap9 and Sap2, as well as by C. albicans cells. We show that the effects of the modifications are site-, residue-, or Sap-dependent, and the substitutions affect not only cleavage at the substitution sites but also the degradation of the peptide as a whole. These findings can be used to design and test additional modifications to study the interaction between Hst-5 and both Saps and C. albicans cells. Additionally, our work provides peptide engineering approaches that can be used to design more robust peptides in the presence of aspartic proteases, which could have applications as potential therapeutics to treat or prevent C. albicans infections.
MATERIALS AND METHODS
Peptides and enzymes
The parent Hst-5 peptide and the variants in Table 1 were synthesized by GenScript with a purity ≥95% and trifluoroacetic acid salt removal to hydrochloride. Purified Sap2 and Sap9 were gifted by B. Hube from Friedrich Schiller University, Germany. The Saps were produced in Pichia pastoris, as previously described [37], and Sap9 was produced without its GPI anchor [16]. The proteolytic activities of the Saps were confirmed using the EnzChek Protease Assay Kit (ThermoFisher Scientific, Waltham, MA, USA).
Proteolytic degradation of the peptides
To determine the extent of degradation of the peptides by the Saps, Hst-5 and the Hst-5 variants were each mixed with Sap9 or Sap2 at final concentrations of 150 μg·mL–1 peptide and 3.13 μg·mL–1 or 6.25 μg·mL–1 protease for Sap9 or Sap2, respectively. Experiments were done in 1 mM NaPB. The mixtures were incubated at 37 °C for 2 h, and NaPB with no Sap was used as a control. An incubation time of 2 h was selected because it results in approximately 50% degradation of the parent Hst-5, allowing decreases and increases in degradation to be easily observed. After the incubation, the samples were mixed with tricine sample buffer (without Coomassie Blue G-250, except in control samples) containing β-mercaptoethanol and boiled for 5 min at 100 °C to inactivate the proteases. The degraded and non-degraded peptides were separated by gel electrophoresis in 10–20% Tris-tricine gels (Bio-Rad, Hercules, CA, USA), and the gels were then fixed in a 10% acetic acid/40% methanol/50% water mixture for 30 min. The fixed gels were stained with Bio-Safe Coomassie stain (Bio-Rad, Hercules, CA, USA) for 1 h and washed with fresh water three times, once overnight and then twice for at least 2 h each time. The gels were imaged on a ChemiDoc imager (Bio-Rad, Hercules, CA, USA), and densitometric analysis was done with Image Lab software (Bio-Rad, Hercules, CA, USA). In the analysis, the upper band was taken as intact peptide and the lower band was taken as degraded products. Three replicates of the assay were done.
For degradation with C. albicans and culture supernatant, a single colony of the ATCC 90028 strain (American Type Culture Collection, Manassas, VA, USA) was used to inoculate YPD. The culture was grown overnight, subcultured, and grown to an optical density of 1–1.2 at OD600. All cultures of C. albicans cells were grown at 30 °C. For degradation experiments with cells, the cells were washed three times in 100 mM NaPB and diluted to 2×109 cells·mL–1. Equal volumes of cells and peptides were mixed and incubated for 2 h at 37 °C. The cells were removed by centrifugation to yield the final degraded peptide samples. For degradation experiments with culture supernatant, the overnight subculture was diluted to OD600=0.1 and grown for 17.5 h. The cells were removed by centrifugation, and the supernatant was transferred to a 10 kDa molecular weight cutoff column (GE Healthcare Life Sciences, Pittsburgh, PA, USA), where the buffer was exchanged to 2 mM NaPB. Equal volumes of supernatant and peptides were mixed and incubated for 2 h at 37 °C to generate the degraded peptide samples. The degraded peptide samples generated by cells and the culture supernatant were mixed with tricine sample buffer and boiled for 10 min. The samples were then run on gels (10–20% Tris-tricine gels for samples incubated with cells and 16.5% Tris-tricine gels for samples incubated with supernatants) and analyzed as described above for degradation by the purified Saps. Three biological replicates were performed for each peptide with cells and with supernatant.
For statistical analysis, we performed one-way ANOVA tests with p< 0.05 and Dunett’s multiple comparison tests with the Hst-5 sample as the control. The number of asterisks indicates the level of statistical significance: * for p< 0.05, ** for p< 0.01, *** for p< 0.001, and **** for p< 0.0001.
Antifungal activity assay
The anti-Candida activities of the intact peptides were assessed by an antifungal activity assay. As described above for the degradation assay, an overnight culture of C. albicans was subcultured and grown in YPD media. Cells were washed three times in 2 mM NaPB and diluted to 5×105 cells·mL–1 or 5×107 cells·mL–1. Serial dilutions (0–100 μM for the lower cell density and 0–400 μM for the higher cell density) of parent Hst-5 and the Hst-5 variants were prepared in water, and 20 μL of the peptides and 20 μL of the cells were mixed and incubated in round-bottom 96-well culture plates for 30 min at 30 °C. After incubation, 320 μL of 1 mM NaPB was loaded into each well to stop additional killing of the cells by the peptides [31]. Mixtures were further diluted and approximately 250 cells were inoculated into round-bottom culture plates with equal volumes of YPD media and 1 mM NaPB at a total volume of 200 μL. Wells containing only YPD and NaPB were served as a sterility control and provided measurements for the background signal. The OD600 was measured after overnight incubation on a microplate shaker at 30 °C. The reduction in viability was calculated as
Three biological replicates were performed on separate days, with two replicates on each day.
To measure the antifungal activity of the peptides following degradation by the Saps, the antifungal activity assay was performed as described above, except the peptides were first exposed to Saps. The peptide fragments were prepared by incubating each peptide (150 μg·mL–1) with 6.25 μg·mL–1 Sap9 or 18 μg·mL–1 Sap2 for 2 h at 37 °C. The enzymes were then inactivated by heating at 100 °C for 5 min. As controls, each peptide was also incubated with only NaPB buffer. Samples were then stored at –20 °C until use in the antifungal activity assay.
MS/MS analysis
The cleavage sites of the Saps and the abundance of the fragments produced by cleavage were determined using mass spectrometry. After incubation with 3.13 μg·mL–1 Sap9 or 6.25 μg·mL–1 Sap2 and heat inactivation of the Saps, 25 μL of each sample was desalted using a C-18 TopTip micro-spin column (Glygen Corporation, Columbia, MD, USA) following the manufacturer’s protocol. The binding solution was 0.1% formic acid, and the releasing solution was 0.1% formic acid/60% acetonitrile (ACN). To ensure equal flow-through volume of each sample, 18 μL of desalted sample was aliquoted and 1 μL of the four-amino acid peptide MRFA at 0.1 mg·mL–1 was added. Samples were manually loaded through loop injection at 25 μL·min–1 with 40% ACN/0.1% formic acid. Mass spectra were acquired with an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, Waltham, MA, USA) with data-dependent analysis at a 5 s cycle time. Manufacturer recommended source parameters for a flow rate of 25 μL·min–1 were applied. Full scan mass spectra of m·z–1 350–1550 were acquired in the orbitrap at R=120000 (m·z–1 200) with fluoranthene ion as the internal calibrant. Collision-induced dissociation (CID) and electron-transfer dissociation (ETD) fragments of the most intense ions (z> 1) were recorded with the orbitrap at R=60000 (m·z–1 200). Dynamic exclusion was set at 30 s.
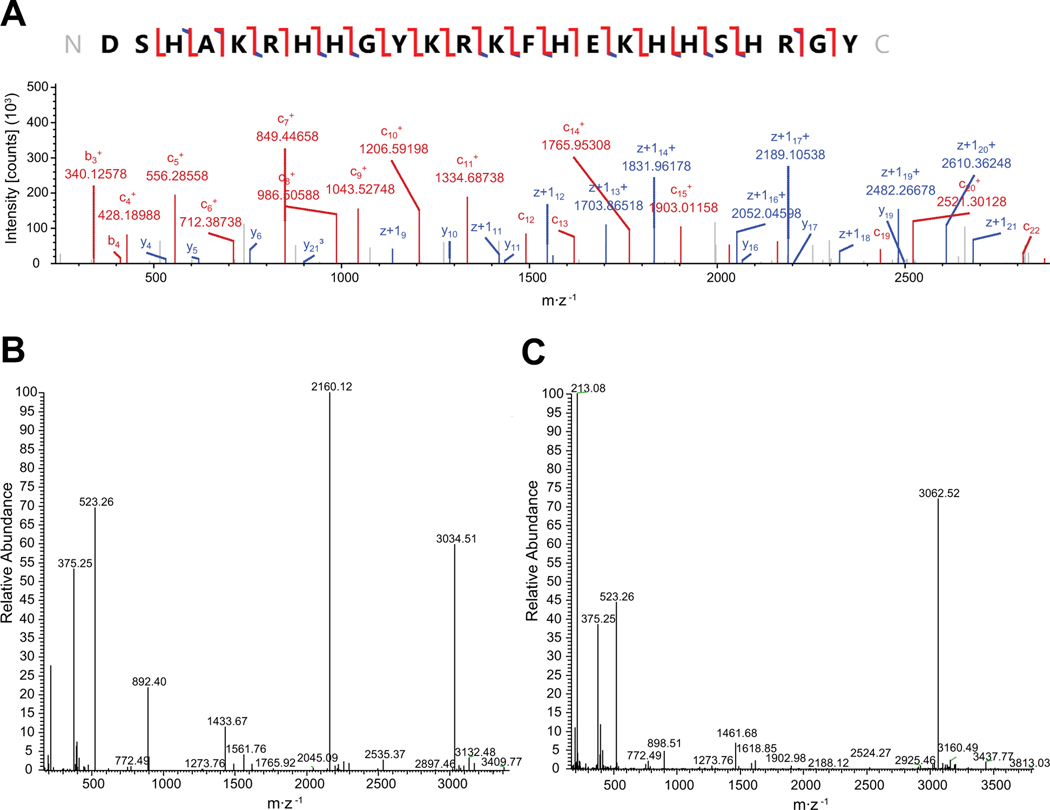
The molecular weights of the peptides and their fragments were calculated from full scan MS spectra using the XTract program in the XCalibur software (Thermo Scientific, Waltham, MA, USA). MS/MS spectra from ETD and CID fragmentation were processed using Proteome Discoverer (V2.1; Thermo Scientific, Waltham, MA, USA) with Prosight PD node to identify peptides and their degradation products (Fig. 8A). The database was the collection of Hst-5 and its analogs used in this study. Intensities of identified peptides and their degradation products relative to the internal standard MRFA were obtained from the deconvoluted full scan mass spectra (Fig. 8B and Fig. 8C).
Fig. 8.
Sample mass spectroscopy spectra. (A) Annotated electron-transfer and higher-energy collision dissociation (EThCD) spectrum of intact Hst-5. (B) & (C) Representative deconvoluted full scan mass spectra of Hst 5 (B) and K17R (C) after incubation with 3.13 μg·mL–1 Sap9.
ACKNOWLEDGMENTS
We thank Bernhard Hube for providing the purified Saps. This work was supported by a National Institutes of Health training grant in Host-Pathogen Interactions (T32AI089621B) and a University of Maryland Cross-Campus Seed Grant.
ABBREVIATIONS:
- Sap
secreted aspartic protease
- Hst-5
histatin-5
- NaPB
sodium phosphate buffer
Footnotes
CONFLICTS OF INTEREST: The authors have no conflicts of interest to declare.
REFERENCES
- 1.Xu J. & Mitchell TG (2003) Geographical differences in human oral yeast flora, Clin Infect Dis. 36, 221–224. [DOI] [PubMed] [Google Scholar]
- 2.Amerongen AV & Veerman EC (2002) Saliva−the defender of the oral cavity, Oral Dis. 8, 12–22. [DOI] [PubMed] [Google Scholar]
- 3.Oppenheim FG, Xu T, McMillian FM, Levitz SM, Diamond RD, Offner GD & Troxler RF (1988) Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans, J Biol Chem. 263, 7472–7. [PubMed] [Google Scholar]
- 4.Xu T, Levitz SM, Diamond RD & Oppenheim FG (1991) Anticandidal activity of major human salivary histatins, Infect Immun. 59, 2549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meiller TF, Hube B, Schild L, Shirtliff ME, Scheper MA, Winkler R, Ton A. & Jabra-Rizk MA (2009) A novel immune evasion strategy of candida albicans: proteolytic cleavage of a salivary antimicrobial peptide, PLoS One. 4, e5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan SA, Fidel PL, Thunayyan AA, Varlotta S, Meiller TF & Jabra-Rizk MA (2013) Impaired Histatin-5 Levels and Salivary Antimicrobial Activity against C. albicans in HIV Infected Individuals, Journal of AIDS & clinical research. 4, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helmerhorst EJ, van’t Hof W, Breeuwer P, Veerman ECI, Abee T, Troxler RF, Amerongen AVN & Oppenheim FG (2001) Characterization of histatin 5 with respect to amphipathicity, hydrophobicity, and effects on cell and mitochondrial membrane integrity excludes a candidacidal mechanism of pore formation, J Biol Chem. 276, 5643–5649. [DOI] [PubMed] [Google Scholar]
- 8.Puri S. & Edgerton M. (2014) How does it kill?: understanding the candidacidal mechanism of salivary histatin 5, Eukaryot Cell. 13, 958–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, Maccallum D, Odds FC, Schafer W, Klis F, Monod M. & Hube B. (2006) Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions, J Biol Chem. 281, 688–94. [DOI] [PubMed] [Google Scholar]
- 10.Naglik JR, Challacombe SJ & Hube B. (2003) Candida albicans secreted aspartyl proteinases in virulence and pathogenesis, Microbiol Mol Biol Rev. 67, 400–28, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monod M, Capoccia S, Léchenne B, Zaugg C, Holdom M. & Jousson O. (2002) Secreted proteases from pathogenic fungi, Int J Med Microbiol. 292, 405–419. [DOI] [PubMed] [Google Scholar]
- 12.Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, Schaller M. & Hube B. (2008) Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis, Microbiology. 154, 3266–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rapala-Kozik M, Bochenska O, Zawrotniak M, Wolak N, Trebacz G, Gogol M, Ostrowska D, Aoki W, Ueda M. & Kozik A. (2015) Inactivation of the antifungal and immunomodulatory properties of human cathelicidin LL-37 by aspartic proteases produced by the pathogenic yeast Candida albicans, Infect Immun. 83, 2518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bochenska O, Rapala-Kozik M, Wolak N, Kamysz W, Grzywacz D, Aoki W, Ueda M. & Kozik A. (2015) Inactivation of human kininogen-derived antimicrobial peptides by secreted aspartic proteases produced by the pathogenic yeast Candida albicans, Biol Chem. 396, 1369–75. [DOI] [PubMed] [Google Scholar]
- 15.Bochenska O, Rapala-Kozik M, Wolak N, Aoki W, Ueda M. & Kozik A. (2016) The action of ten secreted aspartic proteases of pathogenic yeast Candida albicans on major human salivary antimicrobial peptide, histatin 5, Acta Biochim Pol. 63, 403–410. [DOI] [PubMed] [Google Scholar]
- 16.Schild L, Heyken A, de Groot PW, Hiller E, Mock M, de Koster C, Horn U, Rupp S. & Hube B. (2011) Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10, Eukaryot Cell. 10, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki W, Kitahara N, Miura N, Morisaka H, Yamamoto Y, Kuroda K. & Ueda M. (2011) Comprehensive characterization of secreted aspartic proteases encoded by a virulence gene family in Candida albicans, J Biochem. 150, 431–8. [DOI] [PubMed] [Google Scholar]
- 18.Koelsch G, Tang J, Loy JA, Monod M, Jackson K, Foundling SI & Lin X. (2000) Enzymic characteristics of secreted aspartic proteases of Candida albicans, Biochimica et biophysica acta. 1480, 117–31. [DOI] [PubMed] [Google Scholar]
- 19.Laskay ÜA, Srzentić K, Monod M. & Tsybin YO (2014) Extended bottom-up proteomics with secreted aspartic protease Sap9, Journal of Proteomics. 110, 20–31. [DOI] [PubMed] [Google Scholar]
- 20.Driscoll J, Duan C, Zuo Y, Xu T, Troxler R. & Oppenheim FG (1996) Candidacidal activity of human salivary histatin recombinant variants produced by site-directed mutagenesis, Gene. 177, 29–34. [DOI] [PubMed] [Google Scholar]
- 21.Tsai H, Raj PA & Bobek LA (1996) Candidacidal activity of recombinant human salivary histatin-5 and variants, Infect Immun. 64, 5000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Situ H, Balasubramanian SV & Bobek LA (2000) Role of α-helical conformation of histatin-5 in candidacidal activity examined by proline variants, Biochimica et Biophysica Acta (BBA) - General Subjects. 1475, 377–382. [DOI] [PubMed] [Google Scholar]
- 23.Rothstein DM, Spacciapoli P, Tran LT, Xu T, Roberts FD, Dalla Serra M, Buxton DK, Oppenheim FG & Friden P. (2001) Anticandida activity is retained in P-113, a 12-amino-acid fragment of histatin 5, Antimicrob Agents Chemother. 45, 1367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruissen AL, Groenink J, Helmerhorst EJ, Walgreen-Weterings E, Van’t Hof W, Veerman EC & Nieuw Amerongen AV (2001) Effects of histatin 5 and derived peptides on Candida albicans, Biochem J. 356, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmerhorst EJ, Reijnders IM, van ‘t Hof W, Veerman EC & Nieuw Amerongen AV (1999) A critical comparison of the hemolytic and fungicidal activities of cationic antimicrobial peptides, FEBS Lett. 449, 105–10. [DOI] [PubMed] [Google Scholar]
- 26.Karlsson AJ, Pomerantz WC, Neilsen KJ, Gellman SH & Palecek SP (2009) Effect of sequence and structural properties on 14-helical β-peptide activity against Candida albicans planktonic cells and biofilms, ACS Chem Biol. 4, 567–79. [DOI] [PubMed] [Google Scholar]
- 27.Matejuk A, Leng Q, Begum MD, Woodle MC, Scaria P, Chou ST & Mixson AJ (2010) Peptide-based antifungal therapies against emerging infections, Drugs Future. 35, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang WS, Li XS, Sun JN & Edgerton M. (2008) The P-113 fragment of histatin 5 requires a specific peptide sequence for intracellular translocation in Candida albicans, which is independent of cell wall binding, Antimicrob Agents Chemother. 52, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruissen AL, Groenink J, Krijtenberg P, Walgreen-Weterings E, van ‘t Hof W, Veerman EC & Nieuw Amerongen AV (2003) Internalisation and degradation of histatin 5 by Candida albicans, Biol Chem. 384, 183–90. [DOI] [PubMed] [Google Scholar]
- 30.Raj PA, Edgerton M. & Levine MJ (1990) Salivary histatin 5: dependence of sequence, chain length, and helical conformation for candidacidal activity, J Biol Chem. 265, 3898–905. [PubMed] [Google Scholar]
- 31.Li XS, Reddy MS, Baev D. & Edgerton M. (2003) Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5, J Biol Chem. 278, 28553–28561. [DOI] [PubMed] [Google Scholar]
- 32.Helmerhorst EJ, Van’t Hof W, Veerman EC, Simoons-Smit I. & Nieuw Amerongen AV (1997) Synthetic histatin analogues with broad-spectrum antimicrobial activity, Biochem J. 326 ( Pt 1), 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong EF, Tsui C, Boyce H, Ibrahim A, Hoag SW, Karlsson AJ, Meiller TF & Jabra-Rizk MA (2016) Development and in vivo evaluation of a novel histatin-5 bioadhesive hydrogel formulation against oral candidiasis, Antimicrob Agents Chemother. 60, 881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla A, Fleming KE, Chuang HF, Chau TM, Loose CR, Stephanopoulos GN & Hammond PT (2010) Controlling the release of peptide antimicrobial agents from surfaces, Biomaterials. 31, 2348–57. [DOI] [PubMed] [Google Scholar]
- 35.Shi J, Liu Y, Wang Y, Zhang J, Zhao S. & Yang G. (2015) Biological and immunotoxicity evaluation of antimicrobial peptide-loaded coatings using a layer-by-layer process on titanium, Scientific reports. 5, 16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson AJ, Flessner RM, Gellman SH, Lynn DM & Palecek SP (2010) Polyelectrolyte multilayers fabricated from antifungal β-peptides: design of surfaces that exhibit antifungal activity against Candida albicans, Biomacromolecules. 11, 2321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borg-von Zepelin M, Beggah S, Boggian K, Sanglard D. & Monod M. (1998) The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages, Mol Microbiol. 28, 543–54. [DOI] [PubMed] [Google Scholar]