Abstract
Much of the mammalian genome is transcribed, generating long non-coding RNAs (lncRNAs) that can undergo post-transcriptional surveillance whereby only a subset of the non-coding transcripts is allowed to attain sufficient stability to persist in the cellular milieu and control various cellular functions. Paralleling protein turnover by the proteasome complex, lncRNAs are also likely to exist in a dynamic equilibrium that is maintained through constant monitoring by the RNA surveillance machinery. In this Review, we describe the RNA surveillance factors and discuss the vital role of lncRNA surveillance in orchestrating various biological processes, including the protection of genome integrity, maintenance of pluripotency of embryonic stem cells, antibody–gene diversification, coordination of immune cell activation and regulation of heterochromatin formation. We also discuss examples of human diseases and developmental defects associated with the failure of RNA surveillance mechanisms, further highlighting the importance of lncRNA surveillance in maintaining cell and organism functions and health.
Non-coding RNAs (ncRNAs) are a type of RNA that are not translated into protein and include small ncRNAs (such as microRNAs) in the range of 15–30 nucleotides1–3 and long ncRNAs (lncRNAs), which are generally considered as longer than 200 nucleotides4. LncRNAs include a variety of transcripts, all of which can modulate gene expression in specific ways based on cell type, developmental stage and function5–7 (Box 1). As only a fraction of lncRNAs may be biologically relevant to a given process, cells use RNA surveillance pathways to identify and degrade most lncRNAs.
Box 1 |. Functions of different long non-coding RNA species.
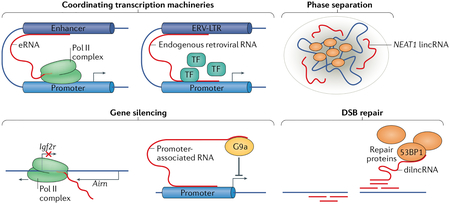
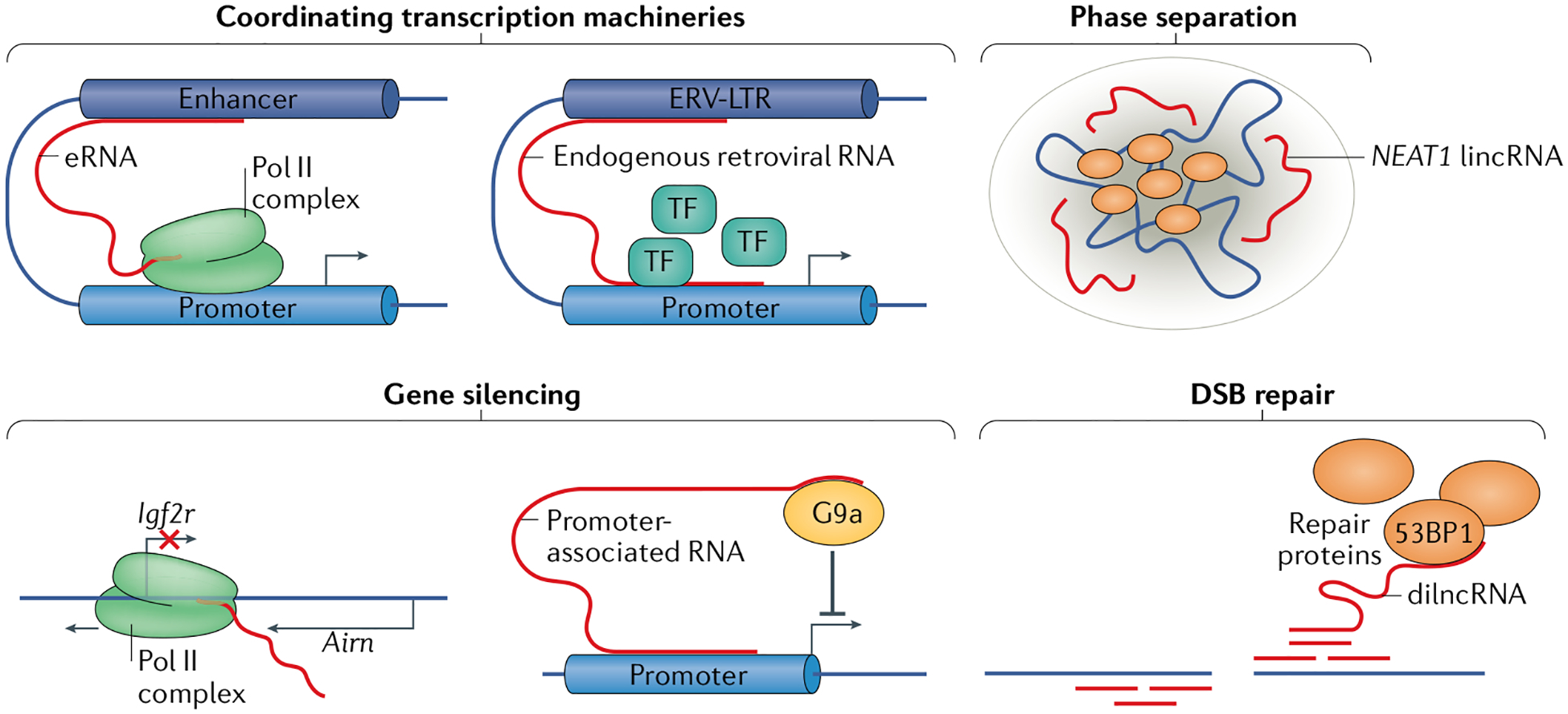
A major function of long non-coding rNas (lncrNas) is the coordination of gene-expression machinery. enhancer rNas (erNas) are lncrNas transcribed from enhancers; they are key to facilitating interactions between the enhancers and their target promoters through ‘chromatin looping’166 (see the figure). Transposable elements derived from endogenous retroviruses (ERVs) are remnants of ancient viral infections and are abundant in mammalian genomes167. although ERV transcripts are regulated by surveillance mechanisms, they can also have profound effects on gene expression, for example, through the assembly of transcription factors (TFs). ERV long terminal repeat (ERV-LTR) transcripts contain binding sites for several TFs, and studies in placental development have attributed a role for ERV-LTRs in enhancer-like recruitment and assembly of the RNA polymerase II (Pol II) complex at developmental genes168,169. Finally, some membraneless nuclear bodies, which can form through liquid–liquid phase separation, coordinate the transcription machinery around genes of interest170. For example, the long intergenic non-coding RNA (lincRNA) NEAT1 is required for the formation and structural integrity of paraspeckles by acting as a scaffold for paraspeckle proteins171,172.
Another vital function of lncRNAs is gene silencing. among the best characterized silencing non-coding RNAs (ncRNAs) is the lincRNA X inactive specific transcript (Xist), which is essential for X chromosome inactivation. Xist spreads in cis over the X chromosome, from which it is transcribed to initiate inactivation173. Promoter-associated transcripts are ncRNAs that have been identified for their role in gene silencing by recruitment of histone-modifying enzymes to gene promoters to activate or repress gene expression174. For example, in mice, the promoter-associated ncRNA Airn (antisense of IGF2R non-protein coding RNA) can recruit the histone methyltransferase G9a to the gene Igf2r and silence it175. Airn can also silence Igf2r by transcriptional interference of Airn with the Igf2r promoter64.
Aside from gene expression, lncRNAs also have roles in DNA damage repair. For example, transcription at sites of DNA double-stranded breaks (DSBs) results in production of DSB-induced lncRNA (dilncRNA), which can recruit DNA repair proteins such as p53-binding protein 1 (53BP1)176.
RNA surveillance is necessary for genome stability and gene expression8–10. Whereas mechanisms of mRNA surveillance have been extensively reviewed11,12, less is known about the surveillance of the diverse types of lncRNA. Although the various components of the RNA surveillance pathways are still being characterized, it is known that endoribonucleases, 5′-exoribonucleases and 3′-exoribonucleases can degrade nascent lncRNAs, often with the help of RNA helicases, which can unwind the non-coding transcripts. The many RNA surveillance factors are vital to the well being of the cell and the organism13–15.
This Review discusses the various mechanisms of nuclear lncRNA surveillance most tightly associated with transcription, initially giving an introduction to the major groups of lncRNA surveillance machinery. This is followed by a discussion of major molecular functions of lncRNA surveillance in the nucleus, such as regulation of antisense transcripts, genome integrity, chromatin state and nuclear architecture. Studies regarding the biological relevance of RNA surveillance machinery are then discussed in the context of immune–pathogen interplay and embryonic stem cell differentiation. Finally, the global importance of RNA surveillance is made clear in the discussion of human diseases and disorders associated with loss of or mutations in RNA surveillance components. The findings associated with RNA surveillance function at multiple levels of biology, from molecular mechanisms to human disease, highlight the importance of RNA surveillance machinery and of understanding the mechanisms of lncRNA turnover.
The RNA surveillance machinery
We first discuss the most prominent nuclear factors that have demonstrated ties to lncRNA processing and genome integrity. Their function is often related to handling harmful nucleic acid structures such as the R-loops (three-stranded structures consisting of an RNA–DNA hybrid and the remaining, displaced strand of DNA) that can form during transcription (Fig. 1), thereby underlying the importance of RNA surveillance to neutralize such threats to genome integrity.
Fig. 1 |. Transcription-associated RNA surveillance.
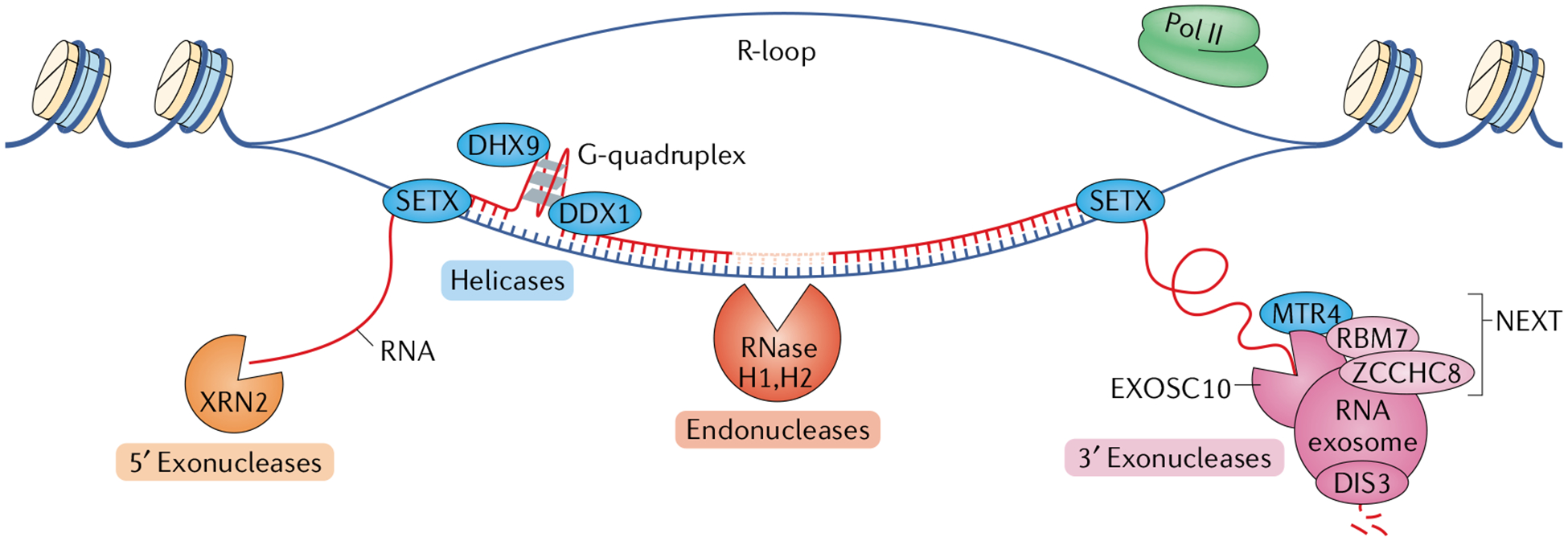
Several components of RNA surveillance are associated with the transcription of both coding and non-coding RNAs; these include factors for RNA degradation and for RNA–DNA and RNA–RNA unwinding that facilitate said degradation (not shown). The three major degradation machineries are 5′ exonucleases, endonucleases and 3′ exonucleases. Depicted here are the components that are most important for transcriptional surveillance. The proteins 5′–3′ exoribonuclease 1 (XRN1) and XRN2 are the primary 5′ exoribonucleases. XRN2 is located in the nucleus, making it most relevant for transcription-associated 5′ exonucleolytic RNA degradation. XRN2 and the helicase senataxin (SETX) function together in mediating transcription termination (not shown). The ribonuclease H (RNase H) enzymes are the primary endoribonucleases in mammalian cells. RNase H1 is located both in the mitochondria and in the nucleus, whereas RNase H2, which is the main source of transcription-associated endonuclease activity, is located only in the nucleus. The 3′ exoribonuclease most tightly associated with transcription of both coding and non-coding RNA is the RNA exosome complex. Its main catalytic subunit is DIS3, though exosome component 10 (EXOSC10) also has catalytic activity. The RNA exosome often associates with the nuclear exosome-targeting (NEXT) complex comprising RNA-binding protein 7 (RBM7), the scaffolding protein zinc finger CCHC domain-containing protein 8 (ZCCHC8) and the RNA helicase MTR4, which is required for efficient RNA exosome activity. In some cases, such as in the process of class switch recombination in B lymphocytes, SETX joins MTR4 in unwinding RNA–DNA and RNA–RNA substrates to promote RNA exosome activity. Several other RNA helicases function in transcription-associated RNA surveillance, including the DEAD-box (DDX) and DEAH-box (DHX) helicases, which help prevent DNA damage. For example, DDX1 and DHX9 can each unwind G-quadruplex structures to facilitate R-loop resolution. Pol II, RNA polymerase II.
Endonucleases.
Ribonuclease H1 (RNase H1) and RNase H2 are important regulators of genome stability (Table 1). Both enzymes process R-loops, as their depletion has been linked to R-loop accumulation in budding yeast, whereas their overexpression has been linked to R-loop depletion16,17. The RNase H enzymes are endonucleases that degrade the RNA strand of the RNA–DNA hybrid (Fig. 1) and, especially RNase H2, are required for the removal of harmful ribonucleotides from DNA strands18. The role of RNase H2 and RNase H1 in protecting genome integrity has been established by studies showing that they suppress hyper-recombination coupled with DNA breakage19–21. The use of catalytically inactive RNase H proteins as markers for R-loops22 has further validated their ubiquitous role in R-loop binding and resolution. Importantly, loss of this arm of RNA surveillance has been linked to disease, as discussed below.
Table 1 |.
Machineries of non-coding RNA surveillance
| Activity | Factor | Main function | Main subunits: related diseases |
|---|---|---|---|
| Endonuclease | RNase H1 | Mitochondrial RNA processing, DNA replication | – |
| RNase H2 | Nuclear R-loop processing, ribonucleotide excision repair | Aicardi-Goutieres syndrome150,151 | |
| Integrator complex | Transcription termination | – | |
| 3′−5′ Exonuclease | RNA exosome complex | Nuclear and cytoplasmic RNA degradation, R-loop processing | DIS3: multiple myeloma156; EXOSC2: retinitis pigmentosa140; EXOSC3: pontocerebellar hypoplasia type 1, spinal motor neuron disease141–144; EXOSC8: cerebellar and corpus callosum hypoplasia, spinal motor neuron disease139; EXOSC10 |
| PARN | Po1y(A)-specific ribonuclease | Dyskeratosis congenita177,178; familial pulmonary fibrosis179 | |
| 5′−3′ Exonuclease | XRN1 | Cytoplasmic RNA degradation | – |
| XRN2 | Nuclear RNA degradation, transcription termination | – | |
| Helicase | MTR4 | Processing double-stranded RNA and RNA–DNA hybrids | – |
| Senataxin (SETX) | R-loop processing, transcription termination | Ataxia with oculomotor apraxia type 2 (REF146); amyotrophic lateral sclerosis147 | |
| Aquarius | R-loop processing | – | |
| DDX1 | R-loop processing, G-quadruplex processing | – | |
| DHX9 | R-loop processing, G-quadruplex processing | – | |
| DDX5 | R-loop resolution with XRN2 | – | |
| DDX19 | RNA export, R-loop processing | – | |
| DDX21 | R-loop processing | – | |
| DDX23 | R-loop processing | Adenoid cystic carcinoma88 | |
| Cofactors | NEXT complex | Nuclear cofactor of the RNA exosome, primarily targeting nascent transcripts | MTR4, RBM7: pontocerebellar hypoplasia type 1-like and spinal muscular atrophy145; ZCCHC8: familial pulmonary fibrosis180 |
| PAXT complex | Nuclear cofactor of the RNA exosome, targeting poly(A)-tailed transcripts | MTR4, PABPN1, ZFC3H1 |
DDX, DEAD-box; DHX, DEAH-box; EXOSC, exosome component; NEXT, nuclear exosome targeting; PABPN1, poly(A) binding protein nuclear 1; PARN, poly(A)-specific ribonuclease; PAXT, poly(A)-tail exosome targeting; RBM7, RNA-binding protein 7; RNase, ribonuclease; XRN, 5′–3′ exoribonuclease; ZCCHC8, zinc finger CCHC domain-containing protein 8; ZFC3H1, zinc finger C3H1 domain-containing protein.
3′ Exoribonucleases.
The eukaryotic RNA exosome is a 3′→5′ exoribonuclease complex, which was initially discovered in budding yeast and shown to degrade ribosomal RNA and other small RNAs23,24. The 11-subunit exosome complex is highly conserved, with regard to structure and function, between budding yeast and mammals25,26. The RNA exosome is present in the cytoplasm and in the nucleus as a nine-subunit core complex, with two subunits — the mammalian proteins exosome component 10 (EXOSC10) and DIS3 (also known as RRP44) — bearing RNase activity27,28 (Fig. 1). The exosome is targeted to specific nascent and recently transcribed ncRNAs by nuclear targeting complexes comprising the DExH-box helicase MTR4 and associated accessory proteins. There are two mammalian RNA exosome-associated nuclear targeting complexes, one located in the nucleoplasm and the other in the nucleolus. In the nucleoplasm, MTR4, zinc finger CCHC domain-containing protein 8 (ZCCHC8) and RNA-binding protein 7 (RBM7) make up the nuclear exosome targeting (NEXT) complex, which is responsible for the RNA exosome-mediated degradation of various types of ncRNAs14,29,30 (Fig. 1). This role renders RNA surveillance a prominent factor in regulating gene expression and in the maintenance of genome integrity31–34.
5′ Exoribonucleases.
The major 5′→3′ exoribonucleases of RNA surveillance in mammals are the homologues 5′–3′ exoribonuclease 1 (XRN1) in the cytoplasm and XRN2 in the nucleus35 (Fig. 1). Both proteins were initially discovered and characterized in budding yeast36,37 and shown to function in ribosomal RNA processing38 and tRNA decay39 as well as in mammalian cells in mRNA turnover40–42. XRN2, in particular, has been associated with other nuclear RNA surveillance mechanisms in mediating transcription termination through the ‘torpedo model’43, wherein RNA polymerase II (Pol II) continues transcribing following cleavage of the mRNA beyond (3′ of) the poly(A) site43,44. XRN2 then degrades the superfluous transcript 5′→3′ until it catches up with Pol II and mediates its dissociation from the DNA template43–45. This has been demonstrated in the ACTB gene, where transcription through the poly(A) site and proximal genetic elements promote Pol II pausing and XRN2-mediated transcription termination46.
XRN2 also functions in transcription termination with other components of the RNA surveillance machinery. R-loops form at G-rich transcription pause sites, where the displaced DNA strand is more likely to form secondary structures such as the four-stranded G-quadruplexes, which are unwound by the RNA helicase senataxin (SETX) to facilitate XRN2-mediated transcription termination47 (Fig. 1). Other involved factors include the microprocessor complex (better known for its role in microRNA maturation) and EXOSC10, indicating the existence of a transcription termination complex that includes various components of the RNA surveillance machinery48. XRN2 and transcription termination are vital for the preservation of genome integrity; XRN2 and other factors, such as SETX, are recruited to R-loops for RNA degradation and prevention of DNA double-stranded breaks (DSBs)49. It remains unclear whether XRN2 also has a role in repairing DSBs. These observations highlight the functional importance of 5′ exonuclease activity in maintaining an effective DNA damage response and in the prevention of genome instability.
RNA helicases.
R-loops can present a threat to genome integrity by leaving a displaced single-stranded DNA (ssDNA) vulnerable to damage, and therefore degradation of the nascent transcripts is a crucial regulator of R-loop-mediated genome instability. The processing of R-loops by endoribonucleases and exoribonucleases requires the unwinding of the associated RNA and DNA strands by various helicases50 (Table 1). The human RNA helicase SETX has a role in XRN2-mediated transcription termination and transcription-associated replication fork integrity47,48,51. SETX and related DEAxQ-like domain-containing helicase Aquarius are vital for R-loop processing and genome stability52–54. The DExH helicase MTR4 is a nuclear cofactor of the RNA exosome in RNA surveillance; targeted RNA surveillance and optimal RNA exosome activity require association with MTR4 helicase and the NEXT complex55. Biochemical and genetic analyses of R-loop-associated somatic mutations, together with structural data generated by cryo-electron microscopy, show that MTR4 and the RNA exosome (along with SETX) form a complex that unwinds RNA–DNA hybrids and double-stranded RNA (dsRNA) to promote RNA exosome-mediated ncRNA degradation (discussed below). Several other DEAH-box (DHX) and DEAD-box (DDX) helicases have roles in the maintenance of genome stability56 (Table 1). For example, the RNA helicase DHX9 is involved in R-loop processing and prevention of DNA damage57,58 (Fig. 1), and DDX1 has been implicated in resolving RNA G-quadruplexes in immunoglobulin class switch recombination (CSR)59 (Fig. 2).
Fig. 2 |. RNA surveillance between deleterious and programmed DNA mutagenesis.
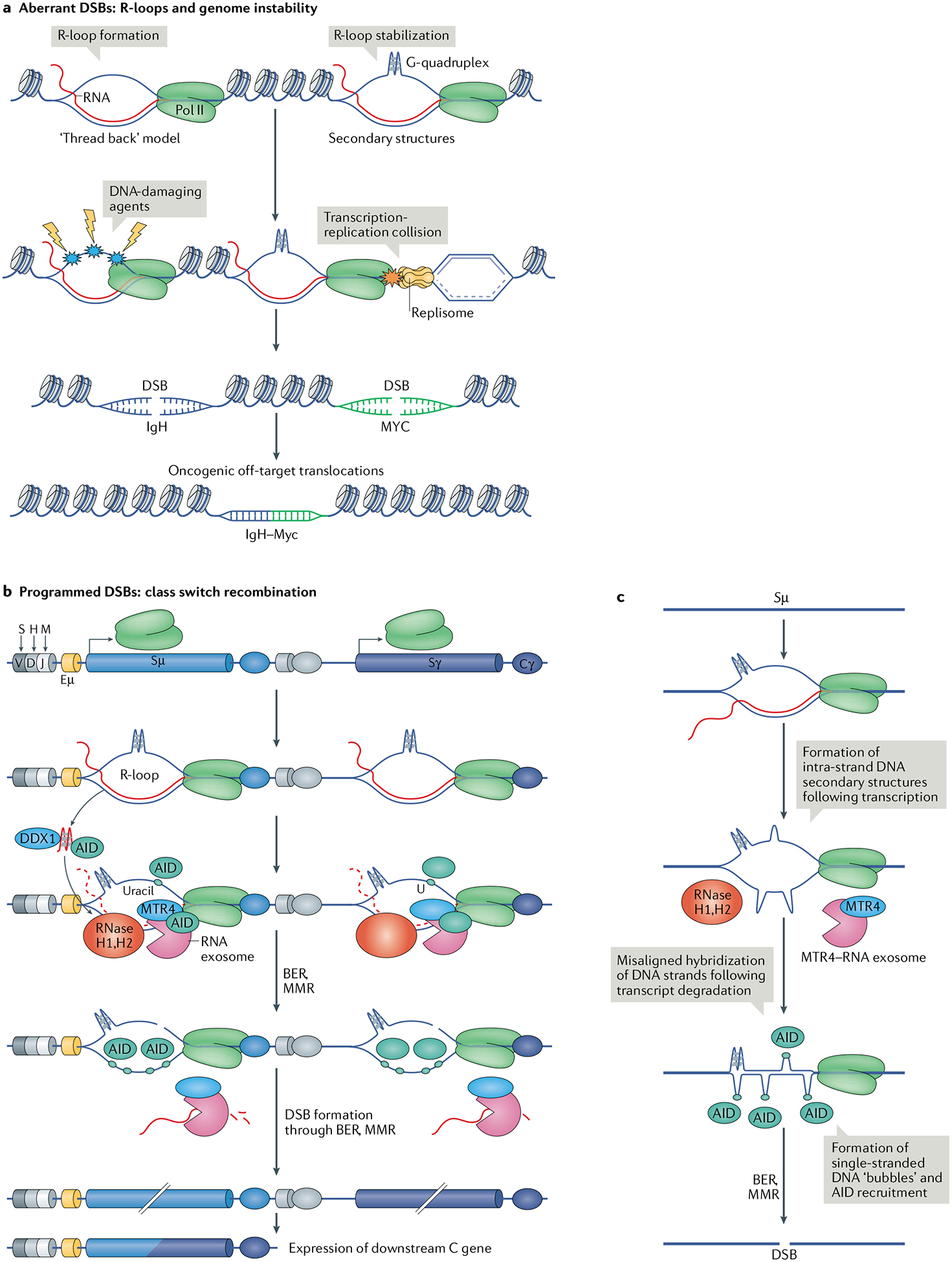
a | The current model of R-loop formation is ‘threading back’ of the nascent RNA into the DNA. R-loops are stabilized by the formation of secondary DNA structures, such as G-quadruplexes, on the exposed single-stranded DNA (ssDNA). R-loop formation and stabilization is a threat to DNA integrity because the ssDNA is vulnerable to DNA-damaging agents. R-loops and the associated RNA polymerase II (Pol II) can also pose a threat during DNA replication in the form of collisions between Pol II and the replisome. Consequently, R-loop persistence increases the threat of formation of aberrant DNA double-stranded breaks (DSBs), which can cause potentially oncogenic DNA translocations such as the common oncogenic translocation between the immunoglobulin heavy chain locus (IgH) and Myc. b | During class switch recombination of the immunoglobulin locus, transcription of non-coding RNA in switch sequences upstream of constant (C) genes leads to the formation of R-loops. The RNA exosome complex and other enzymes are recruited to these R-loops along with activation-induced cytidine deaminase (AID). R-loop formation allows AID to deaminate cytidines on the non-template strand. AID access to the template strand is enabled by the RNA exosome and ribonuclease (RNase) H1 and RNase H2, which degrade the nascent RNA strand of the RNA–DNA hybrid. Owing to the ability of the non-template DNA strand to form stable secondary DNA structures, the two DNA strands are slow to reanneal; this increases the availability of ssDNA substrates for AID and mutagenesis. G-quadruplex-forming, nascent non-coding RNAs at switch sequences are bound by AID, and this binding and RNA degradation by the RNA exosome are enhanced by the unwinding of the RNA by the helicase DEAD-box 1 (DDX1). Processing of AID-associated mutations in the switch sequence by the base excision repair (BER) or mismatch repair (MMR) pathway leads to DSB formation, ultimately resulting in recombination and expression of the downstream constant gene. c | Another possible mechanism of AID recruitment and DSB formation involves transcription in the Sμ segment, which is rich in dG–dC sequences, by Pol II and R-loop formation. The non-transcribed DNA strand, exposed as ssDNA, forms a G-quadruplex that ‘crunches’ the DNA strand and promotes misalignment of the two DNA strands. Upon degradation of the non-coding RNA transcript by the RNA exosome and RNase H enzymes, aberrant dG–dC pairing results in the formation of ‘bubbles’ of ssDNA, which act as AID substrates, thereby resulting in the formation of a DSB.
The activities of ncRNA surveillance
In this section, we discuss the molecular functions of lncRNA (and ncRNA) processing by RNA surveillance and the consequences of their absence.
Sense and antisense transcription.
A key regulator of protein-coding transcripts are antisense transcripts, many of which are associated with gene promoters or initiated from gene bodies (often from intragenic enhancers)32. Promoter-associated or gene body-associated antisense RNAs can modulate the expression of neighbouring or overlapping genes by multiple mechanisms such as histone modifications60, DNA methylation61, and co-transcriptional effects, such as RNA polymerase collisions and formation of secondary DNA structures32,62.
Antisense and sense transcripts can regulate each other in various manners to control gene expression. The mutual promotion of sense and antisense transcription is exemplified in antisense transcription start site RNAs (TSSa-RNAs), the transcription of which is divergent and the expression directly correlated with that of their sense mRNAs through the use of shared promoters63. Conversely, the expression of sense and antisense transcripts can be inversely correlated when the transcription of one transcript results in the suppression of the other. For example, transcription of the lncRNA Airn (antisense of IGF2R non-protein coding RNA) overlaps with the Igf2r gene, suppressing Igf2r transcription64. A recent study has suggested another possible relationship between sense and antisense transcripts, in which mRNA transcription can promote transcription of antisense RNA through an R-loop-dependent mechanism65.
The aforementioned correlations between sense and antisense transcripts indicate that RNA surveillance mechanisms could be important in regulating gene expression mediated by antisense transcription, particularly in relation to R-loops. Indeed, in the absence of RNA exosome activity, TSSa-RNAs accumulate at bidirectionally transcribed promoters (and enhancer RNAs (eRNAs; Box 1) accumulate at bidirectionally transcribed enhancers)32. The TSSa-RNA-accumulating bidirectional promoters show a marked increase in R-loop stability, suggesting that the RNA exosome is required for efficient degradation of antisense RNA in R-loops at divergent promoters.
In the absence of RNA surveillance, an antisense RNA-dependent R-loop structure was observed in the promoter and in the gene body of Myc32, which was also found to be extended in size in the absence of Integrator complex activity (measured by loss of the recruiter protein SPT6)66. The Integrator is an endoribonucleolytic complex that was shown to regulate transcription by cutting nascent mRNAs to mediate premature transcription termination and Pol II dissociation67. Loss of Integrator function results in the expression of extended transcription start site-associated lncRNAs owing to defects in transcription termination and an increase in R-loop size and accumulation at divergent promoters, similar to the previous findings in cells lacking RNA exosome function32,66. A similar correlation between the function of the RNA exosome and the Integrator complex has been identified also for eRNA33,68. The role of the RNA exosome and the Integrator complex in regulating the expression of antisense ncRNAs at gene promoters and other regulatory elements illuminates the importance of RNA surveillance for gene regulation.
Maintaining genome integrity.
Over the course of the cell cycle, genome integrity is constantly engendered by DNA damage, mutations, and chromatin breakage and rearrangements69. Such genomic insults can be stimulated by normal cellular processes, such as DNA replication and transcription, and are efficiently processed and cleared to prevent deleterious outcomes such as unregulated DNA break-associated recombination70. Genomic instability can lead to cancer development if repair and regulatory pathways are not intact71,72. DNA replication is a considerable source of genome instability, as the replisome encounters many ribonucleotides and protein barriers that can stall the replication fork and cause DNA vulnerability. Defects in pathways that monitor and aid replication result in replication-associated genome instability73–75. The mechanisms of replication regulation and maintenance have been described extensively in the literature76–79.
NcRNAs, especially nascent ncRNAs, are a considerable source of genome instability33,66. Likewise, R-loops are a common by-product of transcription and are widely implicated in transcription-associated genome instability80. The current model of R-loop formation during transcription is the ‘thread back’ model, whereby the nascent RNA strand exits the RNA polymerase and reanneals to the template DNA strand, leaving the displaced DNA strand vulnerable to damage80,81 (Fig. 2a). DNA features, such as high GC content, negative super-coiling and DNA nicks, stabilize R-loops and prolong the exposure of the ssDNA to damaging agents82,83. R-loops are also stabilized by G-rich sequences that can form G-quadruplexes, which inhibit reannealing of the DNA84,85. When not properly processed, transcription-associated R-loops can lead to genome instability in the form of DSBs and deleterious recombination86,87 (Fig. 2a).
Among the helicases relevant for maintaining genome stability are members of the DDX family59,88, the R-loop and G-quadruplex helicase DHX9 (reF.57), and SETX53,89. Loss of SETX in mouse spermatocytes resulted in the accumulation of R-loops and in apoptosis52. In neurons, SETX interacts with the RNA exosome to decrease R-loop burden; individuals with SETX mutations suffer from ocular degeneration, leading to ataxia with oculomotor apraxia 2 (AOA2; also known as spinocerebellar ataxia with axonal neuropathy 2) (Table 1). The molecular mechanism of AOA2 is not completely understood, but it is clear that the underlying SETX mutations uncouple the interaction between SETX and the RNA exosome90.
Programmed DNA mutagenesis.
B cells diversify their antibody gene repertoire through three remarkable DNA alteration processes. Immature B cells in the bone marrow undergo V(D)J recombination, and peripheral B cells undergo CSR and somatic hypermutation (SHM) following antigen exposure. Collectively, these phases of developmentally programmed DNA mutagenesis occur in the immunoglobulin (Ig) gene loci and involve processes that, in the absence of RNA surveillance, can cause genome instability at other regions of the mammalian genome. CSR and SHM require the activity of the ssDNA deaminase enzyme activation-induced cytidine deaminase (AID), which identifies ssDNA on the template and non-template strands of Ig switch regions to induce nicks that accumulate and form DSBs. Two such programmed DSBs are then synapsed to undergo DNA recombination, leading to isotype selection and CSR.
The R-loop-processing role of the RNA exosome in orchestrating CSR is arguably its most thoroughly characterized function in the immune system32,91. CSR requires transcription of ncRNA at switch sequences in the Ig locus of B cells, and AID recruitment and activity at this site facilitates DSB formation and recombination92 (Fig. 2b). Transcription-associated R-loops persist at transcribed switch sequences owing to the high guanine content of the non-transcribed DNA strand, which stabilizes DNA structures such as stem-loops and G-quadruplexes. The RNA exosome, which processes the switch region transcripts, recruits AID to the ncRNA template strand to deaminate and ultimately nick it91,92 (Fig. 2b). In addition to the activity of the RNA exosome complex, RNase H1 is capable of degrading the switch region transcript to facilitate R-loop resolution and CSR17. It has also been postulated that the AID protein is associated with a G-quadruplex in the nascent ncRNA93 that needs to be unwound by the G-quadruplex helicase DDX1 prior to AID recruitment to the switch sequences59 (Fig. 2b). The AID-associated G-quadruplex RNA could be an RNA exosome substrate upon its unwinding by DDX1, though the mechanism of AID–RNA interaction is currently being evaluated32,93,94. Following AID recruitment and catalysis of ssDNA nicks on both strands of switch sequences, the dU bases are then recognized and processed by either the base excision repair or mismatch repair pathways95. The ensuing DSB can then recombine with a downstream switch sequence that has undergone the same process (Fig. 2b).
Importantly, the switch region ncRNA must be degraded to efficiently process the R-loop and create a DSB13,32,96. It is also possible that unwinding and degradation of the switch region transcript leads to misalignment of the two switch region DNA strands owing to the formation of stable G-quadruplexes and other DNA structures. This may be followed by aberrant base pairing of long stretches of dG–dC sequences to create long stretches of ssDNA ‘bubbles’ that ultimately recruit AID (Fig. 2c). The functions of RNA surveillance factors at switch sequences are prime examples of the importance of RNA surveillance in maintaining the balance between programmed genetic instability and genome integrity. Although the mechanism of DNA mutagenesis by AID during SHM is still under investigation, it is already clear that the RNA surveillance machinery is important for programmed DNA mutagenesis and breaks during antibody gene diversification.
DNA double-stranded break repair.
RNA exosome-deficient embryonic stem cells (ESCs) accumulate DNA breaks, γH2AX foci (marker of DSB repair response) and aberrant chromosomal rearrangements33. Specifically, when the RNA exosome subunit EXOSC3 was depleted in ESCs, γH2AX foci appeared at sites of higher accumulation of exosome substrates such as enhancers. Analysis of metaphase chromosomes revealed an accumulation of translocation-dependent chromosomal alterations following RNA exosome depletion. These observations indicate that loss of the RNA exosome leads to deficiency in DNA damage repair.
A recent study97 has attributed a role for the RNA exosome subunit EXOSC10 in identifying R-loop-associated lncRNAs formed at DNA breaks as a result of pre-existing or DNA break-induced transcription, as well as in processing these lncRNAs to generate ssDNA available for the recruitment of replication protein A (RPA). RPA recruitment to ssDNA overhangs (that are free of exosome-sensitive lncRNAs) primes the strand-invasion step and DSB repair by homologous recombination (Fig. 3a). The targeting of the ssDNA-binding protein RPA to DSB sites is impaired, and DNA end resection is overstimulated in EXOSC10-depleted cells97. By contrast, defects in DNA end resection are restored by transcription inhibitors, and RNase H1 overexpression restores the RPA recruitment defect caused by EXOSC10 depletion. These observations compel us to suggest that clearance of newly synthesized lncRNAs at DSB sites is required for RPA recruitment (Fig. 3a). Moreover, it is likely that DNA end resection and assembly of the homologous recombination machinery depends on lncRNA transcription at DSB sites.
Fig. 3 |. Functions of RNA surveillance in DNA damage repair and chromatin biology.
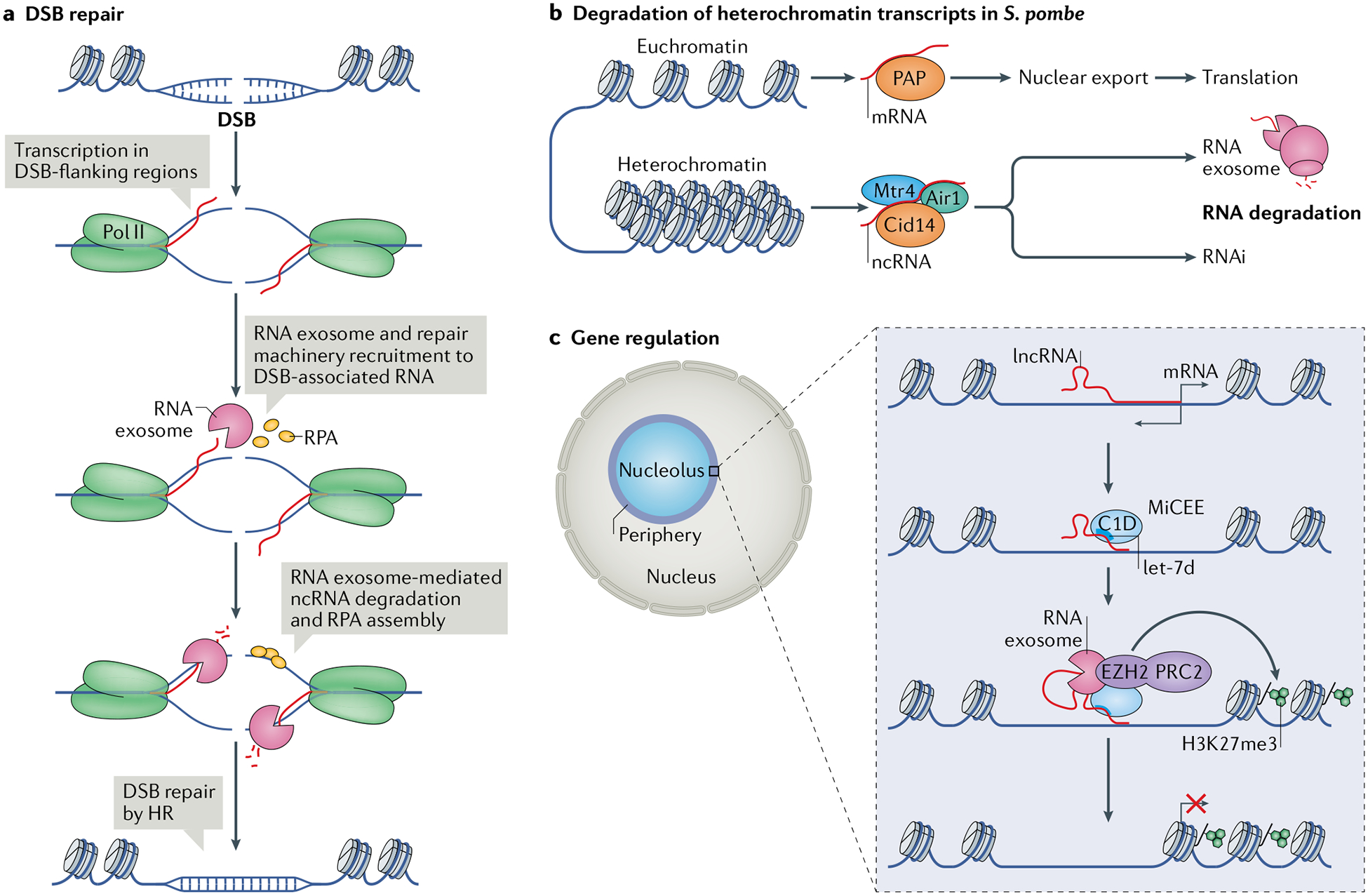
a | RNA surveillance factors such as the RNA exosome can function in DNA double-stranded break (DSB) repair97. Upon DSB formation, the DSB-flanking regions are transcribed and the RNA exosome is recruited to the DSB-associated nascent transcripts along with DNA repair factors such as replication protein A (RPA), which binds to single-stranded DNA (ssDNA). Degradation of the non-coding RNA (ncRNA) by the RNA exosome is coupled with ssDNA binding by RPA, which recruits the homologous recombination (HR) repair machinery and leads to repair of the DSB. b | In the yeast Schizosaccharomyces pombe, the RNA exosome has a role in degrading heterochromatin-derived transcripts in parallel to RNA interference (RNAi)-mediated degradation102. Euchromatin-derived mRNAs undergo processing by poly(A) polymerase (PAP), are exported from the nucleus and undergo translation in the cytoplasm. By contrast, heterochromatin transcripts are processed by the non-canonical poly(A) polymerase Cid14 in association with Mtr4 and Air1. The polyadenylated heterochromatin-derived transcripts are then degraded either by the RNA exosome, the RNAi pathway or both. c | Bidirectional transcription from gene promoters produces, in addition to mRNAs, long non-coding RNAs (lncRNAs) that can be bound by the microRNA let-7d–C1D–EXOSC10–EZH2 (MiCEE) complex. EZH2 is the catalytic component of Polycomb repressive complex (PRC2), which catalyses histone H3 Lys27 trimethylation (H3K27me3) and transcriptional silencing103. Pol II, RNA polymerase II.
Another prominent example of the role of RNA surveillance proteins in DNA damage repair is the role of SETX in resolving RNA–DNA hybrids at DSB sites and limiting aberrant DNA rearrangement. RNA–DNA hybrids were shown to accumulate on DSB-flanking chromatin and have a narrow overlapping SETX binding distribution53. Moreover, SETX promotes recruitment of the recombinase RAD51, thereby minimizing illegitimate re-joining of distant DNA ends and preventing chromosome translocations53. Taken together, these studies highlight different mechanisms by which RNA surveillance functions to regulate DSB repair and limit the number of deleterious chromosome translocations.
Chromatin regulation.
In the yeast Schizosaccharomyces pombe, lncRNAs generated from centromeric DNA by ‘pervasive transcription’ self-hybridize to form dsRNAs, which then associate with components of the heterochromatinization machinery to initiate transcription silencing98–101. RNA surveillance factors have a role in regulating centromeric silencing through RNA interference (RNAi)-dependent and RNAi-independent pathways102. Whereas transcripts synthesized in euchromatin regions are properly processed by the canonical poly(A) polymerases to be exported to the cytoplasm for translation, transcripts from heterochromatin regions are processed by a non-canonical poly(A) polymerase (Cid14) and are eventually identified and degraded by the RNA exosome complex or undergo degradation through RNAi102 (Fig. 3b). Deletion of the RNA exosome factor Rrp6 (the S. pombe homologue of EXOSC10) leads to accumulation of the antisense and sense centromeric transcripts by RNA-dependent RNA polymerase and formation of dsRNAs, which are processed to promote centromere silencing101.
An equivalent role of RNA surveillance proteins in regulating heterochromatin-associated transcripts is yet to be elucidated in mammalian cells. Nevertheless, RNA exosome function in heterochromatin has been evaluated in mammalian cells. Loss of EXOSC3 in ESCs has been shown to reduce the levels of the gene-silencing heterochromatin modification histone H3 Lys9 dimethylation (H3K9me2) at a selected set of silent enhancers, and loss of EXOSC10 can suppress the formation of facultative heterochromatin at certain genes owing to a decrease in H3K27 methylation levels33. The recently characterized MiCEE complex is composed of the microRNA let-7d, the ribosomal RNA processing factor C1D, EXOSC10 and EZH2 (the catalytic subunit of PRC2)103. In the process of degrading lncRNAs that are divergently transcribed from promoters of active genes, the RNA exosome and MiCEE recruit PRC2 to catalyse H3K27 trimethylation and gene silencing at the periphery of the nucleolus (Fig. 3c). Another recent study has also uncovered a link between the RNA exosome and PRC2-mediated chromatin modification through the RNA exosome cofactor poly(A)-tail RNA exosome targeting (PAXT) complex (Table 1). PAXT, which comprises MTR4, the zinc-finger protein ZFC3H1 and poly(A) binding protein nuclear 1 (PABPN1), binds polyadenylated nuclear transcripts and targets them for degradation by the RNA exosome30. ESCs lacking ZFC3H1 and PAXT function exhibit defective PRC2 function and aberrant gene expression104. The function of PRC2 was hindered by RNA accumulation, suggesting that the main role of PAXT in facilitating RNA exosome-mediated RNA degradation is also a crucial regulator of PRC2 function and chromatin status. These preliminary studies suggest that the RNA exosome and ncRNA surveillance have a central role in chromatin modification and gene expression.
Nuclear architecture.
The 3D organization of chromatin is linked with transcription regulation. The genome is organized into topologically associating domains (TADs), defined as genomic regions where regulatory elements and genes primarily interact with each other rather than with sequences in other TADs105,106. Structurally, chromatin is organized into loops that drive (and are driven by) promoter–enhancer interactions and gene expression. TAD borders are enriched in binding sites of the transcriptional repressor CTCF, the structural maintenance of chromosomes complex cohesin, the mediator of Pol II transcription (Mediator) complex and their associated proteins107,108. Transcription of RNA, and mainly of ncRNA, can coordinate chromatin topology. For example, a class of transcription-activating ncRNAs is thought to be necessary for chromosome looping, potentially facilitating interactions between CTCF–cohesin complexes and gene promoters109,110. In B cells, the RNA exosome is important for the interaction through chromatin looping of 3′ regulatory region super-enhancer (3′RR; which is the enhancer of the Ig locus) with other regions of the B cell genome, including a distal enhancer RNA-expressing element, lncRNA-CSRIgA (reF.33). In the absence of proper degradation of lncRNA-CSRIgA, its physiologically relevant topological association with 3′RR is perturbed, leading to decreased CSR (ref.33; G. Rothschild and U. Basu, unpublished results). Future studies should interrogate whether the RNA exosome and other RNA surveillance components have a role in the overall regulation of genome topology.
The organization of the nucleus is also shaped by liquid–liquid phase separation, which can drive the formation of membraneless nuclear bodies with various (often unknown) functions in gene regulation. LncRNAs, and RNA in general, are important for phase separation, suggesting the need to evaluate how RNA surveillance-sensitive lncRNAs can mediate phase separation to alter gene expression111. Though a definitive role for RNA surveillance in phase separation is yet to be determined, the finding that the PAXT complex (which is often associated with the RNA exosome) is required for the formation of specific nuclear foci112 suggests that the RNA exosome may be involved in the formation and function of membraneless nuclear bodies.
Biological roles of lncRNA surveillance
In this section, we discuss how ncRNA surveillance is important for orchestrating various physiological processes.
Controlling pathogens and the immune system.
LncRNAs regulate the expression of immune genes, the differentiation of immune cells and the ability to mount an inflammatory response to infection113,114. LncRNAs and other ncRNAs are also important in controlling bacterial and viral pathogenesis115. In Salmonella enterica serovar Typhimurium (Salmonella), the intergenic, antisense RNA AmgR is vital for growth in a low Mg2+ environment and for host control of Salmonella virulence116. LncRNAs also function in immune evasion and pathogen-mediated damage to the host. Kaposi’s sarcoma-associated herpesvirus (KSHV), a tumorigenic virus in epithelial cells, expresses a number of lncRNAs that facilitate its life cycle, the transformation of host cells and cancer development117–119. These and many other examples highlight the capacity of lncRNAs to dramatically affect the host–pathogen relationship.
Given the importance of lncRNA expression for the host immune system and pathogen infectivity, lncRNA surveillance is prominent in the host–pathogen interplay. RNA stability is linked with the functional dynamics of lncRNAs120,121. In the case of Salmonella infections, the accumulation of lncRNA in the nucleus was a direct consequence of Salmonella-associated loss of the host RNA helicase MTR4 and the RNA exosome subunit EXOSC10 (reFs122,123) (Fig. 4a). Upon Salmonella infection and cellular replication, normally unstable lncRNAs, likely involved in the immune response, were stabilized and expressed in the nucleus, ultimately resulting in Salmonella clearance122. The 3′→5′ RNA degradation machinery was primarily involved in this change in lncRNA stability, as loss of MTR4 mediated lncRNA accumulation owing to failure to recruit the RNA exosome122 (Fig. 4a). Although the full details of this mechanism are yet be elucidated, it is direct evidence for the involvement of RNA surveillance in host–pathogen interactions.
Fig. 4 |. The biological relevance of RNA surveillance.
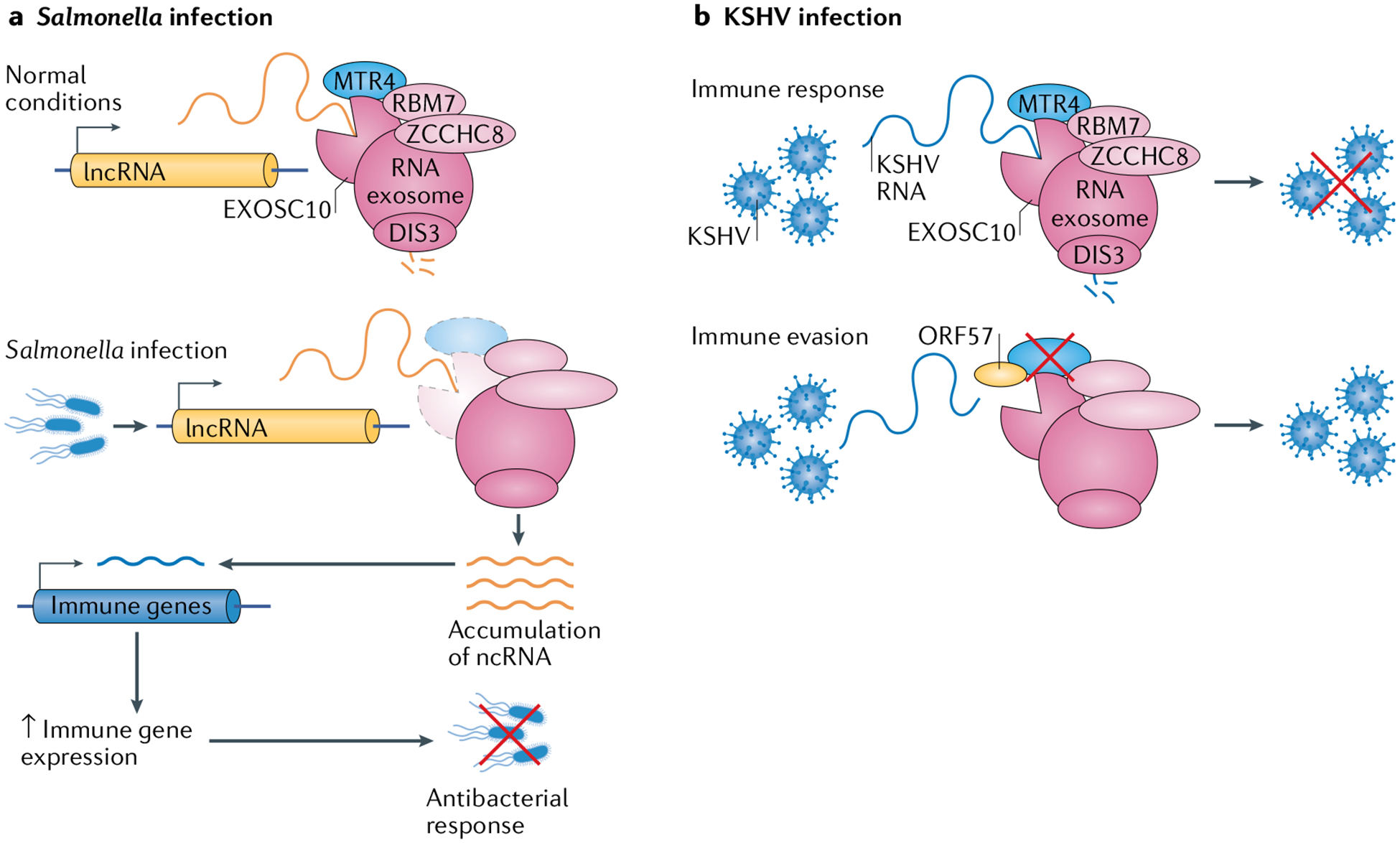
a | The role of RNA surveillance in bacterial infection is highlighted in the regulation of RNA exosome function in response to Salmonella enterica serovar Typhimurium (Salmonella) infection. The RNA exosome normally degrades many long non-coding RNA (lncRNA) transcripts. Following Salmonella infection, exosome component 10 (EXOSC10) and the RNA exosome-associated helicase MTR4 are degraded, resulting in lncRNA accumulation. These transcripts then activate transcription of immune genes and increase the immune response, thereby clearing the Salmonella infection. The specific mechanisms by which this process occurs are as yet unknown. b | A role for RNA surveillance in viral infection was uncovered in studies of host–pathogen interactions in Kaposi’s sarcoma-associated herpesvirus (KSHV) infection. KSHV infection is kept at bay by the degradation of viral transcripts by the RNA exosome. However, KSHV has a counter measure: the viral protein ORF57, which prevents access of RNA (including viral transcripts) to the RNA exosome, specifically by blocking access to MTR4. This allows the expression of viral proteins and viral persistence. ncRNA, non-coding RNA; RBM7, RNA-binding protein 7; ZCCHC8, zinc finger CCHC domain-containing protein 8.
Contrary to the mechanism seen in Salmonella infection, counteracting host lncRNA-associated inflammatory responses is a type of immune evasion employed by pathogens. In the case of KSHV infection, the viral protein ORF57 was found to prevent degradation of viral RNA in the nucleus, but the mechanism of RNA degradation by the host and the mechanism of ORF57-mediated protection against degradation remained unknown124. However, recent investigations have revealed that ORF57 protects viral transcripts from degradation by blocking their interactions with MTR4, potentially implicating the RNA exosome in antiviral responses125 (Fig. 4b). The RNA exosome and its cofactors have also been implicated in antiviral activity in Drosophila melanogaster126 and in HIV transcription in human cells127. Several other RNA surveillance factors are involved in counteracting viral infections128.
Maintenance of embryonic stem cells and differentiation.
The interplay between ESC pluripotency and differentiation is complex and relies heavily on transcription regulation and plasticity. ESCs are characterized by widespread, genome-wide transcription allowing them to quickly shift to any specific gene-expression programme upon differentiation to any cell type129. Several types of ncRNAs have a role in maintaining ESC pluripotency. Long non-coding eRNAs can regulate ESC differentiation and must be regulated themselves for pluripotency maintenance130. Endogenous retroviral elements, such as long interspersed nuclear element 1 (LINE1) and long terminal repeat (LTR) transcripts, can influence pluripotency, with evidence to support an association between LTRs and eRNAs131,132. Divergent transcription of lncRNAs and mRNAs can also mediate stem cell differentiation133,134, and lncRNAs can both be a product of pluripotency-maintenance transcription factors and profoundly affect their expression130,135.
The crucial role of lncRNAs in regulating ESC transcription and fate points to the importance of lncRNA surveillance in regulating ESC pluripotency and differentiation. The RNA exosome has been shown in multiple studies to control ESC gene expression and maintenance of the pluripotency state. Gene expression is globally deregulated in EXOSC3-deficient and in EXOSC10-deficient mouse ESCs, and the levels of silencing chromatin modifications decrease in regions of eRNA expression33. In RNA exosome-deficient mouse ESCs, genes with higher transcriptional activity in the differentiated state showed greater exosome sensitivity as measured by transcript accumulation in the deficient ESCs, suggesting a role for the RNA exosome in downregulating differentiation-associated genes in the ESC state136. Furthermore, upon depletion of the RNA exosome in human ESCs, the levels of mRNAs and of lncRNAs (such as LINE transcripts) that are normally expressed in later stages of embryonic development were increased, further supporting the role of the RNA exosome in regulating ESC differentiation137. In the future, the functional relationship between the expression of LINE transcripts and ESC pluripotency should be evaluated. Overall, these studies establish a role for RNA surveillance in regulating ESC pluripotency and differentiation.
RNA surveillance-related diseases
As RNA surveillance is crucial for essential cellular processes, it stands to reason that there would be a correlation between mutations in genes encoding RNA surveillance factors and human diseases. Indeed, defects in RNA surveillance have been identified in a number of human disorders and diseases, especially in neurodegenerative disorders, immunological disorders and cancer (Table 1).
Neurodegenerative disorders.
Several RNA surveillance mechanisms have been linked to neurodegenerative and neuromuscular disorders. Mutations in various subunits of the RNA exosome have been found to associate with neurodegenerative disorders in model organisms and in humans138. Missense mutations in the EXOSC8 gene, encoding a core component of the RNA exosome, cause cerebellar and corpus callosum hypoplasia, hypomyelination and spinal motor neuron disease139. Missense mutations in the exosome cap subunit EXOSC2 gene were associated with retinitis pigmentosa, progressive hearing loss, premature ageing and mild intellectual disability140. The first direct tie between the RNA exosome and human disease was the discovery that mutations in the integral cap subunit EXOSC3 gene correlate with pontocerebellar hypoplasia and associated spinal motor neuron degeneration141–144. Mutations in the RBM7 gene, a subunit of the exosome-associated NEXT complex, also cause pontocerebellar hypoplasia-like symptoms and spinal muscular atrophy145. The DNA/RNA helicase SETX has also been implicated in neurodegenerative disorders; mutations in the SETX gene are linked to AOA2 and with amyotrophic lateral sclerosis, both marked by muscle weakness and degeneration146,147. This phenotype is suggested to be caused by the crucial role of SETX in transcription termination and R-loop processing47,89,148.
Immunological disorders.
Aicardi–Goutières syndrome is both a neurological disorder and an autoimmune disease, which shares a certain resemblance with systemic lupus erythematosus (SLE)149; both diseases are marked by type I interferon (IFN)-mediated inflammation and loss of self-tolerance, resulting in an antibody response against nucleic acids associated with dying cells149. RNase H2 defects have been implicated in Aicardi–Goutières syndrome by studies using an RNase H2 knockout mouse model and patient samples150,151. RNase H2 mutants give rise to increased nucleic acid damage and accumulation, which activate the type I IFN response152–154. In the context of RNase H2 deficiencies, type I IFN-mediated inflammation is induced in response to damaged DNA that activates innate immunity through the nucleic-acid-sensing cGAS–STING pathway, which primarily functions as a viral DNA sensor154. Similarly, RNase H2 is important for removing aberrant ribonucleotides from DNA, and loss of RNase H2 results in the accumulation of damaged DNA, which stimulates inflammation and leads to systemic auto-immunity151. The RNA exosome-associated RNA helicase MTR4 is implicated in suppressing a similar inflammation signalling pathway, intracellular RNA-mediated RIG-I155. These studies highlight the role of RNA surveillance in modulating the activation of cell-intrinsic mechanisms of inflammatory responses.
Cancer.
RNA surveillance has an important role in balancing genomic integrity and dynamics, which, if lost, can quickly result in cancer development. The RNA exosome catalytic subunit DIS3 has specifically been implicated in cancer development. Loss-of-function mutations in the DIS3 gene are found in individuals with multiple myeloma, which is a mature B cell (plasma cell) malignancy156. Interestingly, DIS3 mutations coincide with IgH translocations in many individuals with multiple myeloma, suggesting that the role of the RNA exosome in regulating B cell translocations may also be implicated in cancer development32,157. RNA surveillance defects in R-loop and DSB processing are potential risks for cancer development as they have been shown to result in R-loop accumulation and to threaten genome stability158. The Ig locus is a prime example of the potential negative outcomes of deregulated R-loop and DSB processing. AID-mediated R-loop and DSB formation in appropriate switch regions is necessary for on-target CSR159, and development of cancers such as lymphomas is marked by off-target translocations between IgH and proto-oncogenes, such as MYC or BCL2, which support constitutive expression of the oncogene160. RNA surveillance mechanisms regulate AID targeting, R-loop processing and subsequent on-target recombination, thereby demonstrating their importance in cancer prevention. Ongoing research may identify additional mutations in RNA surveillance factors implicated in the development of a variety of malignancies.
Conclusions and future perspective
The role of lncRNA surveillance in preventing cellular and developmental defects and diseases highlights their biological importance. Currently, many studies of various physiological processes, such as immune system development and function, stress-associated apoptosis and developmental neurobiology, do not sufficiently consider the effects of lncRNA turnover and overlook the considerable cellular effects of ncRNAs. It is now becoming clear that many aberrations in mammalian gene regulation, cell function and development occur owing to overexpression of lncRNAs in the absence of RNA surveillance. Higher levels of lncRNAs interfere with gene regulation, and RNA surveillance has a role in titrating lncRNA levels down to non-existent or physiologically tolerable levels.
In addition to the RNA surveillance mechanisms discussed here, other RNA surveillance mechanisms exist, which are still less understood. For example, multiple types of RNA modifications could modulate RNA degradation, localization and other surveillance mechanisms. Most of these modifications have largely been studied in mRNA, the most widely studied being N6-methyladenosine, but much is yet to be characterized in terms of N6-adenosine methylation in lncRNAs161.
Similar to RNA modification is RNA editing, which is a post-transcriptional modification that introduces changes in the RNA sequence. One of the most prevalent types of RNA editing is mediated by adenosine deaminase acting on RNA (ADAR), which are enzymes that convert adenosine (A) into inosine (I) in dsRNA, mostly of non-coding origin162. The role of ADAR enzymes in RNA surveillance was first noted when a lack of ADAR1-mediated RNA editing in mammals was shown to cause symptoms that mimic viral infection163,164. Further investigations demonstrated that defective ADAR1 function leads to spontaneous activation of PKR and MDA5, which are pattern recognition receptors (PRRs) for viral dsRNA165. These studies suggest that ADAR1 editing of endogenous, mostly ncRNA prevents the activation of PRRs.
How A-to-I editing shields RNAs from PRR detection is not fully understood. In mammals, the vast majority of ADAR1 editing takes place in the abundant Alu repetitive elements residing in non-coding regions of mRNAs (untranslated regions, introns)162,165. One model is that A-to-I editing destabilizes the structure of the endogenous dsRNA formed by inverted Alu repeats, resulting in their evasion from detection by PRRs163. Alternatively, A-to-I editing might lead to the degradation of RNAs or may recruit other RNA binding proteins that shield RNAs from PRR detection. Taken together, the investigations into ADAR1 suggest that, alongside RNA degradation, mammals adapted the RNA editing machinery to ensure that cellular ncRNAs do not trigger an abnormal immune response.
There are many yet uncharacterized processes in which RNA surveillance mechanisms might be employed. The role of RNA and RNA surveillance in phase separation-driven mechanisms, for example, warrants immediate characterization. There is also potential for a shift or expansion to a broader conceptual understanding of the roles of ncRNAs and RNA surveillance in currently well-studied processes. Transcriptome diversity between single cells will be very informative about RNA surveillance in heterogeneous populations of cells undergoing development. Owing to the relatively short lifespan of ncRNAs and the established expression and turnover mechanisms associated with these, it is possible that, with further understanding of the underlying dynamics, ncRNAs may be recognized as central ‘on–off’ switches of gene expression. The challenge remains to uncover and characterize more ncRNA functions and further strengthen the burgeoning research of ncRNA surveillance.
Acknowledgements
The authors thank many colleagues in the field of RNA biology for exciting discussions that contributed to some of the ideas expressed in this article, and G. Rothschild for proofreading the manuscript. The authors apologize to the many colleagues whose work they were not able to cover in this article owing to limitations of space and scope. This work was supported by grants to U.B. (NIAID 1R01AI099195 and R01AI134988), Leukemia & Lymphoma Society, and the Pershing Square Sohn Cancer Research Alliance. L.N. is supported by an NIH grant (T32 AI106711).
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim VN, Han J & Siomi MC Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol 10, 126–139 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Ghildiyal M & Zamore PD Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matera AG, Terns RM & Terns MP Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol 8, 209–220 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Holoch D & Moazed D RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet 16, 71–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC & Chang HY Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatica A & Bozzoni I Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet 15, 7–21 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Batista PJ & Chang HY Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laffleur B & Basu U Biology of RNA surveillance in development and disease. Trends Cell Biol 29, 428–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolin SL, Sim S & Chen X Nuclear noncoding RNA surveillance: is the end in sight? Trends Genet 28, 306–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belair C, Sim S & Wolin SL Noncoding RNA surveillance: the ends justify the means. Chem. Rev 118, 4422–4447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenberg DR & Maquat LE Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet 13, 246–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh P, Saha U, Paira S & Das B Nuclear mRNA surveillance mechanisms: function and links to human disease. J. Mol. Biol 430, 1993–2013 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Lim J et al. Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell 169, 523–537.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lubas M et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Weick E-M et al. Helicase-dependent RNA decay illuminated by a cryo-EM structure of a human nuclear RNA exosome-MTR4 complex. Cell 173, 1663–1677.e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amon JD & Koshland D RNase H enables efficient repair of R-loop induced DNA damage. eLife 5, 115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maul RW et al. R-loop depletion by over-expressed RNase H1 in mouse B cells increases activation-induced deaminase access to the transcribed strand without altering frequency of isotype switching. J. Mol. Biol 429, 3255–3263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reijns MAM et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahba L, Amon JD, Koshland D & Vuica-Ross M RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huertas P & Aguilera A Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Aguilera A & Gómez-González B DNA-RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol 24, 439–443 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Chen L et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell 68, 745–757.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell P, Petfalski E & Tollervey D The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev 10, 502–513 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Allmang C et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18, 5399–5410 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell P, Petfalski E, Shevchenko A, Mann M & Tollervey D The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91, 457–466 (1997). [DOI] [PubMed] [Google Scholar]
- 26.van Hoof A & Parker R The exosome: a proteasome for RNA? Cell 99, 347–350 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Kilchert C, Wittmann S & Vasiljeva L The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol 17, 227–239 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Zinder JC & Lima CD Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev 31, 88–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubas M et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep 10, 178–192 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Meola N et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Andersson R et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pefanis E et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pefanis E et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuck AC & Tollervey D A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154, 996–1009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AW Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol 17, 6122–6130 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larimer FW, Hsu CL, Maupin MK & Stevens A Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120, 51–57 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Amberg DC, Goldstein AL & Cole CN Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev 6, 1173–1189 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Petfalski E, Dandekar T, Henry Y & Tollervey D Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol 18, 1181–1189 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ & Phizicky EM Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′−3′ exonucleases Rat1 and Xrn1. Genes Dev 22, 1369–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhlrad D, Decker CJ & Parker R Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′–>3′ digestion of the transcript. Genes Dev 8, 855–866 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Parker R & Song H The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol 11, 121–127 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Davidson L, Kerr A & West S Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J 31, 2566–2578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West S, Gromak N & Proudfoot NJ Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432, 522–525 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Proudfoot NJ Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fong N et al. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol. Cell 60, 256–267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gromak N, West S & Proudfoot NJ Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol 26, 3986–3996 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skourti-Stathaki K, Proudfoot NJ & Gromak N Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagschal A et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 150, 1147–1157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales JC et al. XRN2 links transcription termination to DNA damage and replication stress. PLoS Genet 12, e1006107–e1006122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamperl S & Cimprich KA The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair 19, 84–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alzu A et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151, 835–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becherel OJ et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet 9, e1003435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen S et al. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun 9, 533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sollier J et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puno MR & Lima CD Structural basis for MTR4-ZCCHC8 interactions that stimulate the MTR4 helicase in the nuclear exosome-targeting complex. Proc. Natl Acad. Sci. USA 115, E5506–E5515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jankowsky E RNA helicases at work: binding and rearranging. Trends Biochem. Sci 36, 19–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cristini A, Groh M, Kristiansen MS & Gromak N RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep 23, 1891–1905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakraborty P & Grosse F Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 10, 654–665 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Ribeiro de Almeida C et al. RNA helicase DDX1 converts RNA G-quadruplex structures into R-loops to promote IgH class switch recombination. Mol. Cell 70, 650–662.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Modarresi F et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol 30, 453–459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tufarelli C et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet 34, 157–165 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Hobson DJ, Wei W, Steinmetz LM & Svejstrup JQ RNA polymerase II collision interrupts convergent transcription. Mol. Cell 48, 365–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seila AC, Core LJ, Lis JT & Sharp PA Divergent transcription: a new feature of active promoters. Cell Cycle 8, 2557–2564 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Latos PA et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Tan-Wong SM, Dhir S & Proudfoot NJ R-loops promote antisense transcription across the mammalian genome. Mol. Cell 76, 600–616.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nojima T et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol. Cell 72, 970–984.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatomer DC et al. The integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev 33, 1525–1538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai F, Gardini A, Zhang A & Shiekhattar R Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aguilera A & Gómez-González B Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet 9, 204–217 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Mills KD, Ferguson DO & Alt FW The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev 194, 77–95 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Jeggo PA, Pearl LH & Carr AM DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer 16, 35–42 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Lee J-K, Choi Y-L, Kwon M & Park PJ Mechanisms and consequences of cancer genome instability: lessons from genome sequencing studies. Annu. Rev. Pathol 11, 283–312 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Heller RC & Marians KJ Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol 7, 932–943 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Cobb JA et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19, 3055–3069 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sogo JM, Lopes M & Foiani M Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Blumenfeld B, Ben-Zimra M & Simon I Perturbations in the replication program contribute to genomic instability in cancer. Int. J. Mol. Sci 18, 1138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaillard H, García-Muse T & Aguilera A Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Ganai RA & Johansson E DNA replication-a matter of fidelity. Mol. Cell 62, 745–755 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Zhu W, in Genome Instability in Cancer Development (ed. Niggs EA) 249–279 (Springer, 2005). [Google Scholar]
- 80.Aguilera A & García-Muse T R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012). [DOI] [PubMed] [Google Scholar]
- 81.Roy D, Yu K & Lieber MR Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol 28, 50–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roy D & Lieber MR G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol 29, 3124–3133 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roy D, Zhang Z, Lu Z, Hsieh C-L & Lieber MR Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol 30, 146–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lipps HJ & Rhodes D G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19, 414–422 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Millevoi S, Moine H & Vagner S G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA 3, 495–507 (2012). [DOI] [PubMed] [Google Scholar]
- 86.Aguilera A & Gaillard H Transcription and recombination: when RNA meets DNA. Cold Spring Harb. Perspect. Biol 6, a016543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim N & Jinks-Robertson S Transcription as a source of genome instability. Nat. Rev. Genet 13, 204–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sridhara SC et al. Transcription dynamics prevent RNA-mediated genomic instability through SRPK2-dependent DDX23 phosphorylation. Cell Rep 18, 334–343 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Yüce O & West SC Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol 33, 406–417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richard P, Feng S & Manley JL A SUMO-dependent interaction between senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev 27, 2227–2232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Basu U et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu Z, Zan H, Pone EJ, Mai T & Casali P Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol 12, 517–531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng S et al. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 161, 762–773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen J et al. The RNA-binding protein ROD1/PTBP3 cotranscriptionally defines AID-loading sites to mediate antibody class switch in mammalian genomes. Cell Res 28, 981–995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rada C, Di Noia JM & Neuberger MS Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16, 163–171 (2004). [DOI] [PubMed] [Google Scholar]
- 96.Sun J et al. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes Dev 27, 1821–1833 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Domingo-Prim J et al. EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat. Commun 10, 2135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chalamcharla VR, Folco HD, Dhakshnamoorthy J & Grewal SIS Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl Acad. Sci. USA 112, 15548–15555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reyes-Turcu FE, Zhang K, Zofall M, Chen E & Grewal SIS Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol 18, 1132–1138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamanaka S et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493, 557–560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang K et al. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331, 1624–1627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bühler M & Moazed D Transcription and RNAi in heterochromatic gene silencing. Nat. Struct. Mol. Biol 14, 1041–1048 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Singh I et al. MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization. Nat. Genet 50, 990–1001 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Garland W et al. A functional link between nuclear RNA Decay and transcriptional control mediated by the polycomb repressive complex 2. Cell Rep 29, 1800–1811.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nora EP et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Handoko L et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet 43, 630–638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kagey MH et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ørom UA & Shiekhattar R Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lai F et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Silla T, Karadoulama E, Mąkosa D, Lubas M & Jensen TH The RNA exosome adaptor ZFC3H1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep 23, 2199–2210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen YG, Satpathy AT & Chang HY Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol 18, 962–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y & Cao X Long noncoding RNAs in innate immunity. Cell. Mol. Immunol 13, 138–147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spurlock CF 3rd, Crooke PS 3rd & Aune TM Biogenesis and transcriptional regulation of long noncoding RNAs in the human immune system. J. Immunol 197, 4509–4517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee E-J & Groisman EA An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol 76, 1020–1033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li W et al. Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218–5p network. PLoS Pathog 15, e1007578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chandriani S, Xu Y & Ganem D The lytic transcriptome of Kaposi’s sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes. J. Virol 84, 7934–7942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Campbell M et al. A lytic viral long noncoding RNA modulates the function of a latent protein. J. Virol 88, 1843–1848 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Clark MB et al. Genome-wide analysis of long noncoding RNA stability. Genome Res 22, 885–898 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tani H et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res 22, 947–956 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Imamura K et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J 37, e97723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Munschauer M & Vogel J Nuclear lncRNA stabilization in the host response to bacterial infection. EMBO J 37, e99875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sahin BB, Patel D & Conrad NK Kaposi’s sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog 6, e1000799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ruiz JC, Hunter OV & Conrad NK Kaposi’s sarcoma-associated herpesvirus ORF57 protein protects viral transcripts from specific nuclear RNA decay pathways by preventing hMTR4 recruitment. PLoS Pathog 15, e1007596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Molleston JM et al. A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation. Genes Dev 30, 1658–1670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Contreras X et al. Nuclear RNA surveillance complexes silence HIV-1 transcription. PLoS Pathog 14, e1006950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Molleston J & Cherry S Attacked from all sides: RNA decay in antiviral defense. Viruses 9, 2–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Efroni S et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P & Lipovich L Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16, 324–337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fort A et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet 46, 558–566 (2014). [DOI] [PubMed] [Google Scholar]
- 132.Garcia-Perez JL et al. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet 16, 1569–1577 (2007). [DOI] [PubMed] [Google Scholar]
- 133.Sigova AA et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl Acad. Sci. USA 110, 2876–2881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luo S et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18, 637–652 (2016). [DOI] [PubMed] [Google Scholar]
- 135.Dinger ME et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18, 1433–1445 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lloret-Llinares M et al. The RNA exosome contributes to gene expression regulation during stem cell differentiation. Nucleic Acids Res 46, 11502–11513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Belair C et al. The RNA exosome nuclease complex regulates human embryonic stem cell differentiation. J. Cell Biol 218, 2564–2582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morton DJ et al. The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boczonadi V et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun 5, 4287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Di Donato N et al. Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J. Med. Genet 53, 419–425 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Wan J et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet 44, 704–708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rudnik-Schöneborn S et al. Pontocerebellar hypoplasia type 1: clinical spectrum and relevance of EXOSC3 mutations. Neurology 80, 438–446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eggens VR et al. EXOSC3 mutations in pontocerebellar hypoplasia type 1: novel mutations and genotype-phenotype correlations. Orphanet J. Rare Dis 9, 23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Biancheri R et al. EXOSC3 mutations in isolated cerebellar hypoplasia and spinal anterior horn involvement. J. Neurol 260, 1866–1870 (2013). [DOI] [PubMed] [Google Scholar]
- 145.Giunta M et al. Altered RNA metabolism due to a homozygous RBM7 mutation in a patient with spinal motor neuropathy. Hum. Mol. Genet 25, 2985–2996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moreira M-C et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet 36, 225–227 (2004). [DOI] [PubMed] [Google Scholar]
- 147.Chen Y-Z et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet 74, 1128–1135 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suraweera A et al. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet 18, 3384–3396 (2009). [DOI] [PubMed] [Google Scholar]
- 149.Lee-Kirsch MA, Wolf C & Günther C Aicardi-Goutières syndrome: a model disease for systemic autoimmunity. Clin. Exp. Immunol 175, 17–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rabe B Aicardi-Goutières syndrome: clues from the RNase H2 knock-out mouse. J. Mol. Med 91, 1235–1240 (2013). [DOI] [PubMed] [Google Scholar]
- 151.Günther C et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest 125, 413–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bartsch K et al. RNase H2 loss in murine astrocytes results in cellular defects reminiscent of nucleic acid-mediated autoinflammation. Front. Immunol 9, 587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bartsch K et al. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet 26, 3960–3972 (2017). [DOI] [PubMed] [Google Scholar]
- 154.Mackenzie KJ et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35, 831–844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Eckard SC et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat. Immunol 15, 839–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chapman MA et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lionetti M et al. A compendium of DIS3 mutations and associated transcriptional signatures in plasma cell dyscrasias. Oncotarget 6, 26129–26141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Groh M & Gromak N Out of balance: R-loops in human disease. PLoS Genet 10, e1004630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Manis JP, Tian M & Alt FW Mechanism and control of class-switch recombination. Trends Immunol 23, 31–39 (2002). [DOI] [PubMed] [Google Scholar]
- 160.Küppers R Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer 5, 251–262 (2005). [DOI] [PubMed] [Google Scholar]
- 161.Frye M, Harada BT, Behm M & He C RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tan MH et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liddicoat BJ et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rice GI et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet 44, 1243–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chung H et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Furlong EEM & Levine M Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Löwer R, Löwer J & Kurth R The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl Acad. Sci. USA 93, 5177–5184 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chuong EB, Rumi MAK, Soares MJ & Baker JC Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet 45, 325–329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sundaram V et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24, 1963–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yamazaki T et al. Functional domains of NEAT1 architectural lncRNA Induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 172.Clemson CM et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Penny GD, Kay GF, Sheardown SA, Rastan S & Brockdorff N Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996). [DOI] [PubMed] [Google Scholar]
- 174.Guenther MG, Levine SS, Boyer LA, Jaenisch R & Young RA A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nagano T et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 (2008). [DOI] [PubMed] [Google Scholar]
- 176.Michelini F et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol 19, 1400–1411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tummala H et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest 125, 2151–2160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Moon DH et al. Mutations in the poly(A)-specific ribonuclease (PARN) impair telomerase RNA 3’ end maturation in dyskeratosis congenita patients. Blood 126, 669–669 (2015). [Google Scholar]
- 179.Stuart BD et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet 47, 512–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gable DL et al. ZCCHC8, the nuclear exosome targeting component, is mutated in familial pulmonary fibrosis and is required for telomerase RNA maturation. Genes Dev 33, 1381–1396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


