Abstract
Avian coronavirus infectious bronchitis virus (IBV) poses a major threat to the global poultry industry. New IBV geno- and serotypes are continually reported. However, information on IBV prevalence is not frequently addressed in these reports. This study reports on a viral surveillance program in Taiwan from 2005 to 2006 with sampling conducted in poultry slaughterhouses. The genetic features of the obtained field isolates were investigated using sequence analysis and SimPlot analysis. A 1-directional neutralization test was performed to examine the antigenic variations among the collected viruses. The selection pressures that may contribute to the evolution of Taiwan IBV during recent decades were assessed. The surveillance program revealed that 8 out of 47 flocks (17%) were IBV-infected, from which 13 IBV isolates were recovered. Based on the phylogenetic analysis of the S1 gene, 11 of 13 isolates (84.6%) clustered with Taiwan group I. One IBV isolate showed evidence of frequent recombination events with China-like IBV in the spike glycoprotein (S) gene. Another isolate demonstrated the incorporation of China-like and H120-like genome fragments within the S2 gene and the membrane protein (M) gene region, respectively. Some antigenic changes were found in the 1-directional neutralization test. However, no positive selection pressures were related to those variations in the S1 genes among Taiwan IBV. Based on our work, we suggest that sampling chickens in poultry slaughterhouses is an effective and valuable means of compiling viral prevalence data, particularly in situations where there is subclinical infection. Infectious bronchitis viruses from slaughtered chickens revealed intertypic genetic recombination and antigenic diversity.
Key words: infectious bronchitis virus, poultry slaughterhouse, sequence analysis, serotype, viral surveillance
INTRODUCTION
Infectious bronchitis virus (IBV) is the best-known avian coronavirus (Cook, 2002). This disease caused by this virus was first reported in 1931 and was soon distributed worldwide (Cavanagh and Naqi, 2003). Infectious bronchitis virus is a highly infectious and contagious agent in chickens. The respiratory tract is the primary target of virus infection, whereas the urogenital system, including the kidneys and the oviducts, is also affected. Young chickens infected with IBV can acquire permanent damage to their oviducts. Older infected chickens may exhibit poor feed efficiency (Cavanagh and Naqi, 2003; Cavanagh, 2007). Infectious bronchitis virus has great economic importance owing to its effects on egg-laying performance and meat production (Cook, 2002).
The genome of avian coronaviruses is made up of a long single-stranded RNA (approximately 27.6 kb), which encodes 4 structural proteins. The spike glycoprotein (S), which forms the surface projections of the virion, is thought to be a determinant of the host range and pathogenicity (Cavanagh, 2007). The posttranslationally cleaved subunits of S, S1, and S2 carry the epitopes for inducing virus-neutralizing antibodies (Koch et al., 1990). The S1 subunit is involved in virus entry and hemagglutination activity (Cavanagh and Davis, 1986; Koch and Kant, 1990). The envelope (E) and membrane (M) proteins are required for virion assembly. The nucleocapsid (N) protein is closely associated with the RNA genome packaging (Lai and Holmes, 2001).
Like most RNA viruses, IBV undergoes mutations at a high frequency owing to the error-prone RNA polymerase (Wege et al., 1982). Moreover, IBV possesses a unique discontinuous transcription system and a polymerase jumping phenomenon, which contribute to the high frequency of RNA recombination (Lai and Holmes, 2001). Over the last decade, the continuing emergence of variant strains has complicated disease control. A circulating serotype may experience antigenic shift via slight variations in the S1 gene (Cavanagh et al., 1992). Poor cross-protection is conferred among the dozens of serotypes (Cavanagh, 2007).
Vaccination has been implemented to control IBV for several decades. In developing live vaccines, virulent viruses are attenuated by passage in chicken embryos (Cavanagh, 2007). Massachusetts (Mass)-type strains such as M41 and H120 have been widely used for immunization (Cavanagh and Naqi, 2003). However, exposure of animals to live vaccines can facilitate the spread of the viruses in large poultry populations (Farsang et al., 2002). By acting as a heterologous RNA donor template, vaccine viruses may contribute to the genetic evolution of IBV (Wang et al., 1993); that is, genomic recombination with vaccine strains can occur (Jia et al., 1995; Bochkov et al., 2007; Mondal and Cardona, 2007).
Infectious bronchitis virus field outbreaks still occur despite routine vaccination (Cavanagh and Naqi, 2003). New IBV geno- and serotypes are continually reported (Liu and Kong, 2004; Dolz et al., 2006). However, information on IBV prevalence is not frequently addressed in these reports. In Taiwan, in addition to 2 local progeny strain types, Taiwan group I (TW-I) and Taiwan group II (TW-II), the heterologous Mass serotype has been used as a vaccine for over a decade. A recent molecular investigation using the 6.8-kb structural gene region of IBV revealed that Taiwan IBV undergo recombination (Chen et al., 2009). In such a vulnerable epizootiological situation, viruses may evolve rapidly under selection pressure. Therefore, monitoring disease status and analyzing the circulating viruses is critical.
In the present study, an intensive IBV viral investigation program was performed in chickens. Samples were collected from poultry slaughterhouses. Disease prevalence was calculated using the virus isolation rate. Obtained IBV field isolates were molecularly characterized, and the putative recombinants were further analyzed using the 6.8-kb fragment sequence. The selection pressures that may contribute to the evolution of Taiwan IBV were assessed. In addition, a serological method was employed to study the antigenic varieties among the field isolates and reference strains. Through this study, IBV from slaughtered chickens were characterized to reveal intertypic genetic recombination and antigenic diversity.
MATERIALS AND METHODS
Sample Collection and Virus Isolation
Four hundred seventy tracheal samples from chickens originating from 47 flocks in Taiwan were collected in 2 poultry slaughterhouses during 2005 and 2006. Five to 10 samples from each flock were pooled and homogenized in tryptose phosphate broth (Difco Labs, Detroit, MI). The homogenates were clarified using centrifugation at 3,000 × g for 20 min and were sterilized by passing through 0.45-μm syringe filters (Pall Corp., Ann Arbor, MI). Virus isolation was performed using the allantoic route in 9- to 11-d-old specific-pathogen-free (SPF) chicken embryos (Animal Health Research Institute, Tamsui, Taiwan). Each egg received a 0.1- to 0.2-mL inoculation and was incubated at 37°C for 48 h. After 2 blind passages, allantoic fluid was harvested. Reverse transcription-PCR (RT-PCR) detection of the S1 and N genes of IBV (described below) was used to confirm virus isolation.
Viral RNA Extraction, RT-PCR, and DNA Sequencing
Viral RNA was extracted from infected allantoic fluid using the QIAamp Viral RNA kit (Qiagen, Valencia, CA) following the instructions of the manufacturer. Genes of interest were amplified with previously published primers (Huang and Wang, 2007) using a 1-step RT-PCR. The obtained DNA products were submitted to a commercial service (Tri-I Biotech, Taipei, Taiwan) for sequencing in both directions. Each nucleotide was confirmed from 4 identical results.
Sequence Analyses and Recombination Analyses
Sequences were compiled and aligned using the Lasergene software package (DNASTAR, Madison, WI). Phylogenetic trees were constructed with the neighbor-joining method using MEGA version 4.0 (Tamura et al., 2007). SimPlot version 3.5.1 (Lole et al., 1999) was employed to detect recombination events and to calculate informative sites among the reference IBV and field isolates. The recombination breakpoints among the alignments were determined by the maximization of χ2 analysis (Robertson et al., 1995; Lole et al., 1999). The P-values for the divisions of the resulting informative sites were calculated by the χ2 test.
Selection Pressure Analyses of the S1 Gene
The selective pressures that contributed to the variation in the whole S1 gene of previously identified (from 1992 to 2007) local IBV, TW-I and TW-II, were assessed. The average ratio of number of synonymous substitutions per synonymous site to number of nonsynonymous substitutions per nonsynonymous site (dS/dN) was calculated with the web-based SNAP program (http://www.hiv.lanl.gov). A ratio lower than 1 indicates that a specific gene region may be under positive pressures (Korber, 2000). In addition, the presence of positive selection (dN > dS) and purifying selection (dN < dS) was tested using a 1-tailed z-test with MEGA version 4.0 (Tamura et al., 2007). All possible pairs of sequences were analyzed, and the variance of the difference between dS and dN was computed using the bootstrap method with 1,000 replicates.
Viruses and Antisera Titration
Viruses were 10-fold serially diluted with PBS and were used to inoculate 9- to 11-d-old SPF chicken embryos. After 1 wk, embryos were evaluated for IBV infection by the observation of death, dwarfing, and urate deposition. The 50% viral embryo infectious dose was calculated by the method of Reed and Muench (1938). Antisera against strains 2575/98 (TW-I) and 2296/95 (TW-II) were prepared in 3-wk-old SPF chickens (Wang and Huang, 2000). The Mass-type strain antiserum was purchased from Charles River Laboratories (North Franklin, CT). Each antiserum was 4-fold serially diluted with PBS and titrated against the homologous virus in 9- to 11-d-old SPF chicken embryos. The last dilution that protected 50% of the embryos was considered to be 1 antiserum unit (Cowen and Hitchner, 1975).
1-Directional Neutralization Test
A 1-directional neutralization test was performed by a constant virus-constant antiserum procedure as described in previous studies (Cowen and Hitchner, 1975; Wang and Huang, 2000). Briefly, 100 μL of virus (containing 100 50% viral embryo infectious dose) was incubated with 100 μL of antiserum (containing 20 units) at room temperature for 1 h. The infectivity of the virus-antiserum mixture was assayed in ten 9- to 11-d-old SPF chicken embryos. After 1 wk, embryos were evaluated for IBV infection by the observation of death, dwarfing, and urate deposition as described for virus titration. In the control groups, antisera were replaced by PBS. A virus was considered to match the serotype of the antiserum if the antiserum protected 5 or more of the embryos.
Accession Numbers
The nucleotide sequence data reported in this paper have been submitted to GenBank and have been assigned accession numbers: GQ229232 and GQ229237 to GQ229258 (Table 1 ). The accession numbers and genes of interest in the IBV reference strains included in this study are as follows: 1171/92, DQ646406 (S1-N); 1211/92, AF250006 (S1); 2296/95, DQ646404 (S1); 2575/98, DQ646405 (S1); 2993/02, AY606316 (S1); 3025/02, AY606317 (S1); 3051/02, AY606318 (S1); 3071/03, AY606319 (S1); 3263/04, EU822338 (S1); 3468/07, EU822336 (S1); T03/01, AY606315 (S1); T07/02, AY606322 (S1); CK/CH/LDL/97I, EF030996 (S1) and EF602445 (S2-N); Gray, L18989 (S1) and M85245 (N); H120, EU822341 (S1-N); and JMK, L14070 (S1).
Table 1.
Infectious bronchitis viruses isolated in this study
| Strain | Year of isolation | Chicken type | Location | Genotype1 | Accession no. of S1 gene | Accession no. of N gene |
|---|---|---|---|---|---|---|
| 3339/05 | 2005 | Taiwan country chicken | Yunlin | TW-I | GQ229237 | GQ229248 |
| 3368/05 | 2005 | Broiler | Yilan | TW-I | GQ229238 | GQ229249 |
| 3369/05 | 2005 | Broiler | Yilan | TW-I | GQ229239 | GQ229250 |
| 3370/05 | 2005 | Broiler | Yilan | TW-I | GQ229240 | GQ229251 |
| 3371/05 | 2005 | Broiler | Yilan | TW-I | GQ229241 | GQ229252 |
| 3372/05 | 2005 | Broiler | Yunlin | TW-I | GQ229242 | GQ229253 |
| 3373/05 | 2005 | Broiler | Yunlin | TW-I | GQ229243 | GQ229254 |
| 3374/05 | 2005 | Broiler | Changhua | China-like | EU8223372 | EU8223372 |
| 3376/06 | 2006 | Broiler | Taoyuan | TW-I | GQ229244 | GQ229255 |
| 3381/06 | 2006 | Broiler | Taoyuan | JMK-like | GQ229245 | GQ229256 |
| 3382/06 | 2006 | Broiler | Taoyuan | TW-I | GQ2292322 | GQ2292322 |
| 3384/06 | 2006 | Broiler | Taoyuan | TW-I | GQ229246 | GQ229257 |
| 3385/06 | 2006 | Broiler | Taoyuan | TW-I | GQ229247 | GQ229258 |
Genotype was determined based on the S1 gene. TW-I = Taiwan group I.
The sequences from the S1 gene to the N gene of isolates 3374/05 and 3382/06 were submitted.
RESULTS
IBV Prevalence
Among the 47 flocks sampled, 8 were positive for IBV infection after virus isolation. Thirteen IBV were isolated (Table 1). The flock prevalence was 17%. The chicken prevalence ranged from 1.8 to 3.7%, as calculated from the pooled samples using the binomial distribution.
Sequence Analyses of the S1 Gene
The S1 genes of the 13 isolates obtained in this study were directly sequenced. Based on the phylogenetic analysis of the full S1 gene (Figure 1 ), 11 of the 13 isolates (84.6%) clustered with the TW-I group. These TW-I isolates shared 92.1 to 100% homology in the S1 gene. Of the other 2 isolates, the S1 gene of one isolate, 3374/05, was genetically related to the China CK/CH/LDL/97I-type strains. The other isolate, 3381/06, shared 97.9 and 97.2% homology with reference strains JMK and Gray, respectively.
Figure 1.
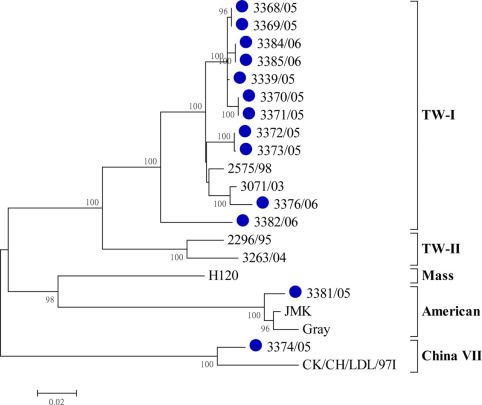
Phylogenetic analysis of the nucleotide sequences of the whole spike glycoprotein 1 (S1) gene from the infectious bronchitis virus isolates recovered in this study (●). The phylogenetic trees were constructed using MEGA version 4.0 by the neighbor-joining method (bootstrapping for 1,000 replicates, bootstrap value >70%). TW-I = Taiwan group I; TW-II = Taiwan group II; Mass = Massachusetts. Color version available in the online PDF.
Sequence Analyses of the Partial N Gene
Partial N gene fragments (nt 178–755) of the 13 isolates were sequenced, which showed that 86.9 to 100% homology was shared among the 13 field isolates. Interestingly, the N gene from isolate 3382/06 was more similar (97.4% homology) to that of the Mass type H120 than to that of the TW-I isolates (86.6 to 89.1% homology). As illustrated in Figure 2 , 3382/06 clustered with the Mass group based on the N gene. The N gene sequence of isolate 3381/06 showed 97.5% identity with reference strain Gray.
Figure 2.
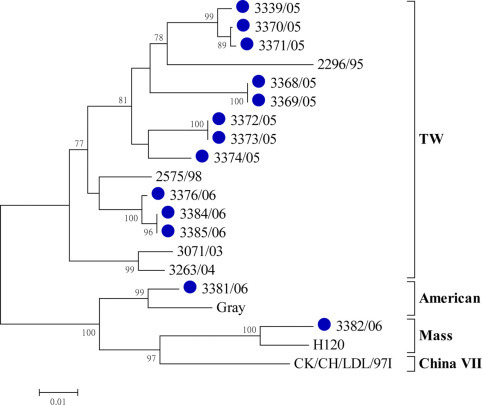
Phylogenetic analysis of the nucleotide sequences of the partial nucleocapsid (N) gene from the infectious bronchitis virus isolates recovered in this study (●). The phylogenetic trees were constructed using MEGA version 4.0 by the neighbor-joining method (bootstrapping for 1,000 replicates, bootstrap value >70%). TW = Taiwan group; Mass = Massachusetts. Color version available in the online PDF.
Identification of Intertypic Recombination
A 3′ 6.8-kb gene fragment from isolate 3382/06 was further sequenced. SimPlot analysis was used to display the consecutive nucleotide identity and illustrate the crossover events among the queried strain and the parental strains. In this case, reference strains 1171/92 (TW-I, an older local strain isolated in 1992), CK/CH/LDL/97I (China genotype VII), and H120 (Mass) served as the parental strains. As shown in Figure 3 , two crossover events were suggested by the similarity plot. The first recombination breakpoint (nt 2267) was located in the S2 gene, and the second one was located in the M gene (nt 4498). There were significant differences (P < 0.01) between the resultant divisions of informative sites. Isolate 3382/06 was therefore identified as an intertypic recombinant among the 3 putative parental strains.
Figure 3.
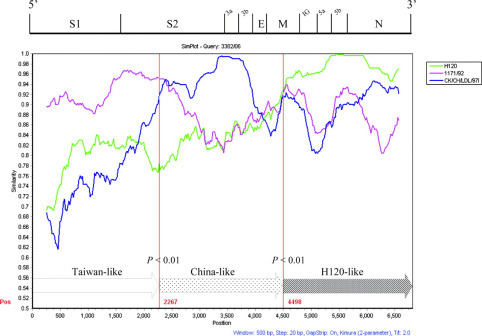
SimPlot analysis of isolate 3382/06. The 1171/92 strain (pink), the H120 strain (green), and the China strain CK/CH/LDL/97I (deep blue) were used as putative parental strains. The similarity plot displays the consecutive nucleotide identity (%) from the spike glycoprotein (S) gene to the nucleocapsid (N) gene among the queried strain and parental strains. The genomic scale was given at the top of the plot. Isolate 3382/06 was identified as an intertypic recombinant among the 3 putative parental strains. The 2 recombination breakpoints were indicated in red. P-values were calculated by the χ2 test. The 6.8-kb genome of isolate 3382/06 was schematically assembled using Taiwan-like, China-like, and H120-like sequence regions. IG = intergenic region; E = envelope protein gene; M = membrane protein gene. Color version available in the online PDF.
Selection Pressure Analyses of the S1 Gene
The average dS/dN ratios obtained from the TW-I group (19 strains, including the 11 recovered in this study) and the TW-II group (4 strains) were 4.27 and 3.34, respectively. Both values were higher than 1. No statistical evidence for positive selection of TW-I and TW-II strains was observed (P > 0.05). However, 84% (144/171) of the pairwise comparisons of TW-I strains and 100% (6/6) of the pairwise comparisons of TW-II strains showed purifying selection (P < 0.05). Thus, the results showed no positive selection pressures but did show purifying selection related to the variations in the S1 gene from the 2 Taiwan IBV genogroups.
Neutralization Test
Based on the molecular characterization results, the 2 viral recombinants identified in this study, 3374/05 and 3382/06, were selected for use in performing the neutralization assays. Antisera from the 3 main serotypes circulating in Taiwan (TW-I, TW-II, and Mass type) were employed. The results (Table 2 ) showed that the China-like recombinant 3374/05 possessed a serotype distinct from all of the tested antisera. The H120-like recombinant 3382/06 was of the same serotype as TW-I.
Table 2.
One-directional neutralization test of infectious bronchitis virus isolates against antisera of TW-I, TW-II, and Mass types, respectively
| Isolate | No. of protected embryos/no. of total embryos |
|||
|---|---|---|---|---|
| TW-I1 antiserum | TW-II2 antiserum | Mass3 antiserum | PBS control | |
| 3374/05 | 1/10 | 2/10 | 2/10 | 0/3 |
| 3382/06 | 9/10 | 1/10 | 0/10 | 0/5 |
TW-I = Taiwan group I.
TW-II = Taiwan group II.
Mass = Massachusetts.
DISCUSSION
This work indicated that the IBV prevalence was 17% in Taiwanese chicken flocks during 2005 and 2006, based on virus isolation. This surveillance program was the first collection of IBV prevalence data in Taiwan. Compared with previous surveillance reports, the prevalence found in Taiwan was not relatively high. According to Roussan et al. (2008), IBV was detected in up to 60% of the examined broiler flocks in Jordan, and 59% of submitted clinical samples were positive for IBV in Western Europe (Worthington et al., 2008). The RT-PCR assays employed in most surveillance studies, which amplify sequences directly from tissues or samples, are sensitive. However, this assay may not be able to discriminate between disease and vaccine strains. A higher IBV detection rate may be found in investigations with samples or flocks that were analyzed because of suspected IBV infection. In this study, the tracheas of slaughtered chickens were collected in poultry slaughterhouses, where sampled chickens were inspected and found to be apparently normal. Biased sampling was therefore avoided. The data obtained from our study may cover subclinical infection circumstances and may represent a more factual disease status. The virus isolation-based diagnosis performed in this work also made it possible to recover the live virulent viruses.
Infectious bronchitis virus infection in Taiwan was recognized in 1968. A decade ago, local IBV were molecularly characterized and found to belong to 2 populations (TW-I and TW-II) based on the RFLP of the S1 gene (Wang and Tsai, 1996). However, local IBV are continually evolving. Infectious bronchitis virus 3374/05 was previously characterized as a natural recombinant between the Taiwan- and China CK/CH/LDL/97I-type strains, and frequent recombination events have been observed in several progeny strains (Chen et al., 2009). The 13 reported IBV in this work were molecularly characterized. Among them, most isolates (11/13, 84.6%) were closely related to TW-I strains, suggesting that TW-I is a dominant genogroup in Taiwan. However, phylogenetic analyses revealed that isolate 3382/06 clustered with the TW-I group based on the S1 gene but not based on the N gene. To identify the recombination site, a fragment covering the 3′ end of the structural protein gene region, about 6.8 kb in length, was further investigated. We determined the viral sequences using at least 4 identical results from separate RT-PCR products. The results obtained from the interpretation programs were supported by statistical evidence. Surprisingly, in addition to the first crossover with the CK/CH/LDL/97I-type strains in the S gene, there was an abrupt shift in nucleotide identities among the M, 5a, 5b, and N genes, strongly indicating another recombination event. This second recombination event was between isolate 3382/06 and strain H120. Two putative H120-like recombinants have also been reported in Italy (Bochkov et al., 2007), from which different phylogeny profiles between the S1 and N gene regions with respect to H120 were described. In this study, we schematically present the crossover events in the plot, and the genome positions of the putative recombination sites are readily observed.
This was the first observation of vaccine virus genome incorporation into wild-type IBV in Taiwan. For a decade, due to the lack of proper homologous protection against local viruses, disease prevention in Taiwan relied on a cross-protective effect conferred by heterologous Mass-type strains such as M41 and H120. In particular, live attenuated viruses are widely used to vaccinate many chicken types. Viruses were likely shed and now circulate in the field. Furthermore, isolate 3381/06 was closely related to the American group strains, another heterologous vaccine type approved in Taiwan. More challenging problems may arise if multiple genome incorporation events occur among viruses. Thus, the crucial observation from this study alerts the need for reconsideration of the policy of IBV control. Indeed, 2 local IBV vaccines have been developed for this purpose (Huang and Wang, 2006).
During virus evolution, higher mutation rates may be observed when viruses are subjected to host immune defenses (Duffy et al., 2008). Mutations in the S1 gene may alter the antigenicity and pathogenicity of the virus (Lai and Holmes, 2001). To address the relationship between the selection pressures and the long-term genetic variations in Taiwan IBV, nearly all available S1 gene sequences from local viruses were collected and analyzed. Compared with the average dS/dN ratio of 1.6 previously obtained in the molecular characterization of Italy 02 genotypes in Spain (Dolz et al., 2006), our data, which showed average dS/dN ratios of 4.27 (TW-I) and 3.34 (TW-II), were not suggestive of a high rate of nonsynonymous changes in Taiwan IBV. No positive selection pressures might drive the evolution of the 2 Taiwan IBV genogroups. Alternatively, there was evidence of purifying selection. These observations may be explained by the fact that current heterologous vaccine strains offer only partial protection and may not drive many nonsynonymous substitutions in amino acid sequences in local viruses.
In addition to the molecular characterization of the field isolates, examination of the antigenicity of these isolates is also important, particularly when a new variant is reported. The 2 recombinant variants 3374/05 and 3382/06 were selected for further analysis of their antigenic serotypes. The results showed that no cross-neutralization effect was achieved between isolate 3374/05 and 2 groups of local viruses as well as the Mass type, suggesting that isolate 3374/05 is a new serotype in Taiwan. Isolate 3382/06 possessed the same serotype as its S1 gene-homologous TW-I strains. Therefore, chickens infected with those variants were probably not protected by the current IBV prevention program. However, the disease history of the infected flocks from which variants 3374/05 and 3382/06 originated was unavailable. The pathogenicity of those variants awaits further study.
Based on our work, we suggest that sampling chickens in poultry slaughterhouses is an effective and valuable means of compiling viral prevalence data. We further provide information on the viral evolution in a population of IBV isolates, which exhibited genetic and antigenic diversity.
ACKNOWLEDGMENTS
The financial support from the National Science Council and Council of Agriculture, Executive Yuan, Taiwan, is greatly appreciated.
REFERENCES
- Bochkov Y.A., Tosi G., Massi P., Drygin V.V. Phylogenetic analysis of partial S1 and N gene sequences of infectious bronchitis virus isolates from Italy revealed genetic diversity and recombination. Virus Genes. 2007;35:65–71. doi: 10.1007/s11262-006-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: Removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67:1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K., Li D., Kant A., Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Naqi S.A. Infectious bronchitis. In: Saif A.M., Fadly Y.M., McDougald L.R., Swayne D.E., editors. Diseases of Poultry. Iowa State University Press; Ames: 2003. pp. 101–119. [Google Scholar]
- Chen H.W., Huang Y.P., Wang C.H. Identification of Taiwan and China-like recombinant avian infectious bronchitis viruses in Taiwan. Virus Res. 2009;140:121–129. doi: 10.1016/j.virusres.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. Coronaviridae. In: Jordan F., Pattison M., Alexander D., Faragher T., editors. Poultry Diseases. W. B. Saunders; New York, NY: 2002. pp. 298–306. [Google Scholar]
- Cowen B.S., Hitchner S.B. Serotyping of avian infectious bronchitis viruses by the virus-neutralization test. Avian Dis. 1975;19:583–595. [PubMed] [Google Scholar]
- Dolz R., Pujols J., Ordonez G., Porta R., Majo N. Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype. Avian Pathol. 2006;35:77–85. doi: 10.1080/03079450600597295. [DOI] [PubMed] [Google Scholar]
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renstrom L.H., Baule C., Soos T., Belak S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31:229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.P., Wang C.H. Development of attenuated vaccines from Taiwanese infectious bronchitis virus strains. Vaccine. 2006;24:785–791. doi: 10.1016/j.vaccine.2005.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.P., Wang C.H. Sequence changes of infectious bronchitis virus isolates in the 3′ 7.3 kb of the genome after attenuating passage in embryonated eggs. Avian Pathol. 2007;36:59–67. doi: 10.1080/03079450601110015. [DOI] [PubMed] [Google Scholar]
- Jia W., Karaca K., Parrish C.R., Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G., Hartog L., Kant A., van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: Correlation with biological functions. J. Gen. Virol. 1990;71:1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Koch G., Kant A. Binding of antibodies that strongly neutralise infectious bronchitis virus is dependent on the glycosylation of the viral peplomer protein. Adv. Exp. Med. Biol. 1990;276:143–150. doi: 10.1007/978-1-4684-5823-7_21. [DOI] [PubMed] [Google Scholar]
- Korber B. HIV signature and sequence variation analysis. In: Rodrigo A.G., Learn G.H., editors. Computational Analysis of HIV Molecular Sequences. Kluwer Academic Publishers; Dordrecht, the Netherlands: 2000. pp. 55–72. [Google Scholar]
- Lai M.M.C., Holmes K.V. Coronaviridae: The virus and their replication. In: Knipe D.M., Howley P.M., editors. Field Virology. Lippincott Williams Wilkins; Philadelphia, PA: 2001. pp. 1163–1185. [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S.P., Cardona C.J. Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes. 2007;34:327–341. doi: 10.1007/s11262-006-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent end points. Am. J. Hyg. 1938;27:493–496. [Google Scholar]
- Robertson D.L., Hahn B.H., Sharp P.M. Recombination in AIDS viruses. J. Mol. Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- Roussan D.A., Totanji W.S., Khawaldeh G.Y. Molecular subtype of infectious bronchitis virus in broiler flocks in Jordan. Poult. Sci. 2008;87:661–664. doi: 10.3382/ps.2007-00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Huang Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.H., Tsai C.T. Genetic grouping for the isolates of avian infectious bronchitis virus in Taiwan. Arch. Virol. 1996;141:1677–1688. doi: 10.1007/BF01718291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Junker D., Collisson E.W. Evidence of natural recombination within the S1 gene of infectious bronchitis virus. Virology. 1993;192:710–716. doi: 10.1006/viro.1993.1093. [DOI] [PubMed] [Google Scholar]
- Wege H., Siddell S., ter Meulen V. The biology and pathogenesis of coronaviruses. Curr. Top. Microbiol. Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- Worthington K.J., Currie R.J., Jones R.C. A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37:247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]


