Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) participates in adipocyte differentiation and maintenance, including the promotion of lipid storage in mammals. In the present study, 3 duck PPARγ small interfering RNA (siRNA) expression plasmids were constructed to investigate the effect of downregulating the expression of PPARγ on adipogenesis and fat accumulation in ducks. The results indicate that the 3 siRNA specific for conserved regions of PPARγ can effectively inhibit expression of PPARγ. It was demonstrated that the expression of lipoprotein lipase and adipocyte fatty acid-binding protein in duck adipose tissue is repressed when the expression of PPARγ is downregulated by siRNA. At the same time, the weight of abdominal fat at 21 and 35 d of age is decreased significantly (P < 0.05) compared with the control. However, the triglyceride levels in serum and muscle are not affected when the mRNA of PPARγ is repressed. The current study indicates that the suppression of PPARγ reduces abdominal fat deposition and regulates adipogenesis in ducks.
Key words: duck, peroxisome proliferator-activated receptor γ, adipogenesis, small interfering ribonucleic acid
INTRODUCTION
Adipose tissue, a major energy reservoir in animals, is associated with nutrient intake and endocrine signaling. It stores excess energy in the form of triglycerides (TG) and releases the stored TG as free fatty acids and glycerol when necessary (Mandrup and Lane, 1997; Anghel et al., 2007). This is true in food animals as well. Modern selective breeding has been successful in improving both egg and meat production in recent years, and the target of intensive selection in modern commercial meat lines is to increase growth rate and muscle mass (Huang et al., 2007; Zhang et al., 2007; Brun et al., 2008). However, excessive fat deposition has also accompanied these selective progresses. Excessive fat in ducks has been one of the major problems for modern duck meat production. It not only reduces carcass yield and the efficiency of feed but also leads to difficulties in processing. Unfortunately, it is likely that breeding techniques will not be able to reduce fat deposition while improving meat production because of the adverse correlation between these traits (Baeza et al., 2002). Furthermore, little is known regarding lipid metabolism in ducks. It is therefore imperative to study adipogenesis and the genes that regulate the deposition and release of lipids in ducks to discover an approach to decrease excessive fat deposition and maintain the fast growth of ducklings.
In mammals, peroxisome proliferator-activated receptor γ (PPARγ), an important regulator in fat metabolism, participates in adipocyte differentiation and maintenance, including the promotion of lipid storage (Rosen and Spiegelman, 2001). Previous research indicates that the expression of PPARγ increases during adipocyte differentiation and induces the accumulation of oil droplets in mouse embryonic fibroblast cells, whereas the knockout of PPARγ impairs adipocyte differentiation (Rosen et al., 1999). Peroxisome proliferator-activated receptor γ acts as a nuclear receptor and transactivates multiple target genes involved in lipid metabolic pathways, such as lipoprotein lipase (LPL) and adipocyte fatty acid-binding protein 4 (FABP4) (Nakachi et al., 2008). Meanwhile, the high affinity of PPARγ to thiazolidinediones, insulin-sensitive drugs, suggests that PPARγ is also involved in the insulin signaling pathway in mammals (Miles et al., 2003). However, there is little evidence to prove that PPARγ plays a role in modulating fat deposition in ducks, but this will be clarified in the present study.
Ribonucleic acid interference is a sequence-specific mechanism to knock down the expression of target genes. This technique has been successfully used both in vivo and in vitro (Singh and Hajeri, 2009). For instance, specific small interfering RNA (siRNA) targeting the severe acute respiratory syndrome and influenza viruses have been shown to efficiently inhibit viral replication in nonhuman primates and mice (Li et al., 2005; Zhou et al., 2007). Further, siRNA targeting of vascular endothelial growth factor receptor-1 to inhibit ocular neovascularization in mice has also been successful (Shen et al., 2006). In the present study, 3 PPARγ siRNA expression plasmids were constructed and used to determine if siRNA that target PPARγ can modulate PPARγ expression and fat accumulation in ducks.
MATERIALS AND METHODS
Target Sequence Selection and Vector Construction
The complete coding sequence of duck PPARγ (EF546801) was derived from the GenBank database. Three target sites at position 168, 271, and 273 in the coding region (Table 1 ) were selected according to the siRNA program of the Sfold Web-based Criteria (Ding et al., 2004), and the 3 selected sequences were submitted to a BLAST search against the chicken and duck genomes to ensure their specificity. To obtain short hairpin RNA, a typical oligonucleotide that has 5 bases containing a restriction site at its 5′ end, 19 bases of sense strand, 7 to 9 bases of hairpin loop, 19 bases of antisense strand, 6 bases of terminator, and 6 bases corresponding to a unique restriction site (resulting in a total length of 65 bases) and 2 complementary oligonucleotides were synthesized. These were annealed and inserted into the Bam HI and Eco RI sites of the RNAi-Ready pSIREN-RetroQ-ZsGreen Vector (BD Biosciences, Clontech, Mountain View, CA). The recombinant plasmids were designated as pS-P167, pS-P271, and pS-P273, respectively. A plasmid (pS-negative) encoding a hairpin siRNA comprising a nonsense sequence that has not been found in the mouse, human, or duck genomes was used as the negative control.
Table 1.
Target sequences of duck peroxisome proliferator-activated receptor γ
| siRNA name1 | Position on CDS2 | Target sequence (5′→3′) |
|---|---|---|
| siPPARγ167 | 167 | AACTGATCAGACAAGCAT |
| siPPARγ271 | 271 | GTTCAGTTGTACAATAAAC |
| siPPARγ273 | 273 | TCAGTTGTACAATAAACCT |
| Negative control | TGGACATAGGCGACGTGAT |
siRNA = small interfering RNA; siPPARγ = small interfering peroxisome proliferator-activated receptor γ.
CDS = coding sequence.
Antibodies
Rabbit anti-PPARγ polyclonal antibodies, β-actin monoclonal antibodies, goat anti-rabbit IgG (L+H)-horseradish peroxidase antibodies, and rabbit anti-mouse IgG (L+H)-horseradish peroxidase antibodies were purchased from Boster Biological Technology Ltd. (Wuhan, China).
Ducks
Healthy 7-d-old Cherry Valley ducklings were raised in floor pens. A commercial diet and water were provided ad libitum throughout the experiment.
Bird Treatment and Sample Collection
A total of 80 Cherry Valley ducklings (aged 15 d) were divided randomly into 5 groups (16 in each group), of which three were testing groups, one was the negative control group, and one was the blank group. The siRNA expression plasmids pS-P167, pS-P271, and pS-P273 were injected (100 μg) intravenously within 5 s into each duck in the test groups, the pS-negative plasmid (same dosage) was injected into each duck of the negative control group, and an equivalent volume of PBS was injected into the blank group. Eight ducks from each group were killed at age 21 and 35 d, respectively. Blood, abdominal fat, muscle, and liver samples were collected, and the abdominal fat and liver were weighed. Serum samples were separated from blood and stored at −20°C.
TG Measurements of Serum and Muscle Samples
Triglycerides in the serum were measured with a human testing kit (Nanjing Jiancheng, Nanjing, China; Yang et al., 2008) using a serum biochemical autoanalyzer. The TG levels in the muscles of all ducks were measured using a Buchi B-811 device (Buchi Labortechnik AG, Flawil, Switzerland) according to the protocols of the manufacturer.
RNA Analysis
Total RNA was extracted from the abdominal fat, purified using an RNeasy mini kit (Qiagen, Valencia, CA; Zou et al., 2009), and reverse-transcribed into cDNA using AMV reverse transcriptase (Takara, Beijing, China). Real-time PCR was carried out using Light-Cycler FastStart DNA MasterPLUS SybrGreen I (Roche Applied Science, Indianapolis, IN) to monitor the reaction amplification on a 7500 real-time thermal cycler (Applied Biosystems, Foster City, CA). Peroxisome proliferator-activated receptor γ, LPL, and FABP4 were studied, and the primers used are listed in Table 2 . Gene expression levels were normalized using β-actin amplified from the same sample, and the expression values are reported as the fold change.
Table 2.
Primer sequences and product sizes of real-time PCR
| Primer name1 | GenBank accession number | Sequence2 | Product size (bp) |
|---|---|---|---|
| PPARγ | EF546801 | F: 5′-TAACGCTCCTGAAATACGGT-3′ | 198 |
| R: 5′-GAACTTCACAGCGAACTCAA-3′ | |||
| LPL | EU598456 | F: 5′ -TGACGCTGAGTTTATGGA-3′ | 248 |
| R: 5′-TTTCTTCGTAGAGGAGGG-3′ | |||
| FABP4 | DQ358123 | F: 5′-CCAAGCACAATGTCGCCA-3′ | 198 |
| R: 5′-TCCCTTTGCCATCCCACT-3′ | |||
| β-Actin | AY251275 | F: 5′-CGCAAATGCTTCTAAACC-3′ | 166 |
| R: 5′-AGACTGCTGCTGATACCTT-3′ |
PPARγ = peroxisome proliferator-activated receptor γ; LPL = lipoprotein lipase; FABP4 = fatty acid-binding protein 4.
F = forward; R = reverse.
Western Blot Analysis
Tissue explants were homogenized in 10 mL of lysis buffer (1% SDS in PBS containing protease inhibitors) and placed on ice for 2 h with vortexing every 10 min. Samples were centrifuged at 20,627 × g for 30 min. The top fat layer was discarded, and the middle clear layer of solubilized proteins was collected. After protein quantification via the Bradford method, Western blots were performed. Briefly, 60 μg of total protein from the tissue lysates was subjected to SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 1% BSA in buffer containing 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.05% Tween-20 for 1 h and then incubated with a rabbit anti-PPARγ antibody and a monoclonal antibody against β-actin as a control. The membranes were visualized with an enhanced chemiluminescence reagent (New England Nuclear, Boston, MA).
Statistical Analysis
The SAS Application Package (SAS Institute Inc., Cary, NC) was used for statistical analysis. Data for multiple comparisons were analyzed using ANOVA, followed by Duncan's multiple range test. The level of significance in the current study was P < 0.05.
RESULTS
Verification of siRNA Expression Plasmids
There is a Hin dIII site at position 2456 in the pSIREN-RetroQ-ZsGreen plasmid, and we inserted a Hin dIII site in the hairpin fragment of the siRNA. Thus, the production of 2 fragments (2,400 and 4,000 bp, respectively) from recombinant plasmids digested with Hin dIII revealed that the siRNA were correctly inserted. This cloning was further confirmed by sequencing the recombinant plasmids; no mutations were found in the 3 designed hairpin fragments (Figure 1 ).
Figure 1.
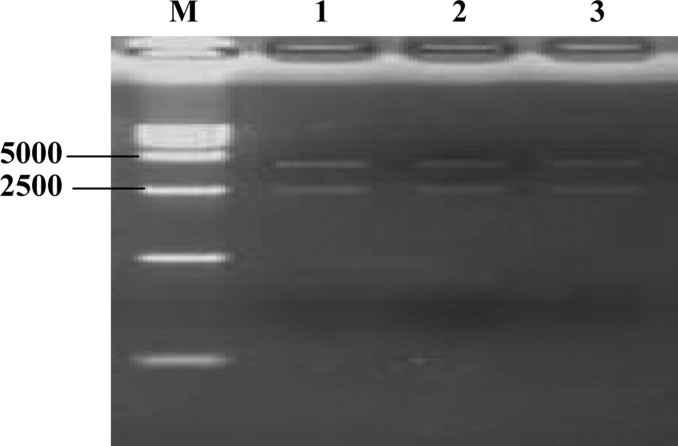
Identification of pS-P167, pS-P271, and pS-P273 plasmids by Hin dIII digestion. Lane 1 = pS-P167 plasmid digest; lane 2 = pS-P271 plasmid digest; lane 3 = pS-P273 plasmid digest; M = DL15000 DNA marker.
Effect of the siRNA Expression Plasmids on the Expression of PPARγ
Downregulation of PPARγ due to the different siRNA is shown in Figure 2 . It was demonstrated that the expression of PPARγ mRNA could be significantly inhibited by the 3 specific siRNA expression plasmids (P < 0.05) but not by pS-negative (P > 0.05). The silencing efficiencies of pS-P167, pS-P271, pS-P273, and pS-negative, compared with the blank control, were 52.26, 30.4, 67.58, and 3%, respectively. To further confirm the alterations of protein expression after treatment with siRNA plasmids, Western blot analysis was carried out to investigate PPARγ protein levels (Figure 3 ). The results show that the PPARγ protein was significantly inhibited by pS-P167 and pS-P273 but only moderately inhibited by pS-P271. No significant difference in the expression of β-actin was observed among the different groups, and the expression of PPARγ was not changed in the negative control group. These data agree with the mRNA expression changes shown by reverse transcription-PCR analysis and suggested that the inhibitory effects occurred at the translational level.
Figure 2.
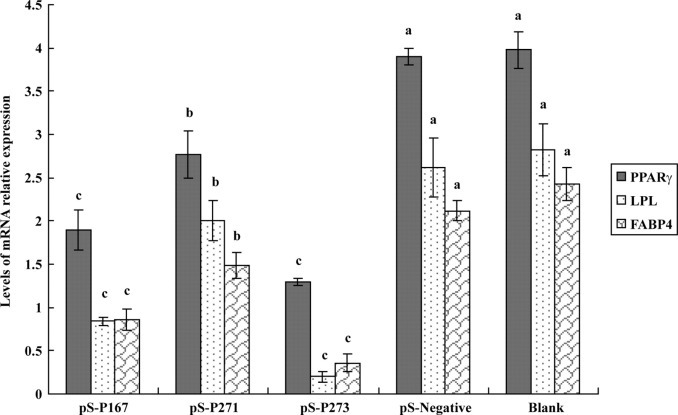
Peroxisome proliferator-activated receptor γ (PPARγ), lipoprotein lipase (LPL), and fatty acid-binding protein 4 (FABP4) mRNA expression in the adipose tissue of ducks from the pS-P167, pS-P271, pS-P273, pS-negative, and blank groups at 21 d. Data are presented as the means ± SD, n = 8 in each group. a–c Different letters indicate significant difference (P < 0.05).
Figure 3.
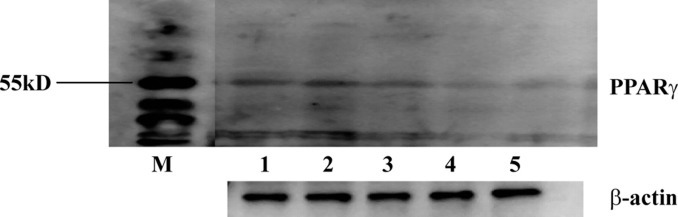
Peroxisome proliferator-activated receptor γ (PPARγ) protein expression in the adipose tissue of ducks from the pS-P167, pS-P271, pS-P273, pS-negative, and blank groups at 21 d. Lane 1 = pS-negative group; lane 2 = the blank group; lane 3 = the pS-P271 group; lane 4 = the pS-P167 group; lane 5 = the pS-P273 group; M = protein marker.
Effect of PPARγ Downregulation on the Expression of LPL and FABP4 in Abdominal Fat
There was a significant decrease of LPL and FABP4 mRNA in abdominal fat when the expression of PPARγ was downregulated by pS-P167, pS-P271, and pS-P273, respectively (but not by pS-negative), compared with normal individuals (blank, Figure 2). It was discovered that pS-P167 and pS-P273 were more effective at decreasing the expression of FABP4 and LPL than pS-P271, and this mirrors the results of the PPARγ protein levels under these treatments.
Effect of PPARγ Downregulation on the Weights of Abdominal Fat and Liver
The weights of the abdominal fat and liver from the experimental ducks are summarized in Figure 4A and B . There were no obvious differences in the liver weights between the 3 experimental groups and the control groups at either 21 or 35 d (Figure 4B). However, significant (P < 0.05) differences in the weight of abdominal fat between the siRNA groups (pS-P167 and pS-P273) and the control groups (pS-negative and blank) at 21 and 35 d were recorded (Figure 4A). The weight of the abdominal fat was greatly decreased in the 2 siRNA groups. The abdominal fat weight of the pS-P271 siRNA group was also decreased relative to the control groups, but this difference was not statistically significant.
Figure 4.
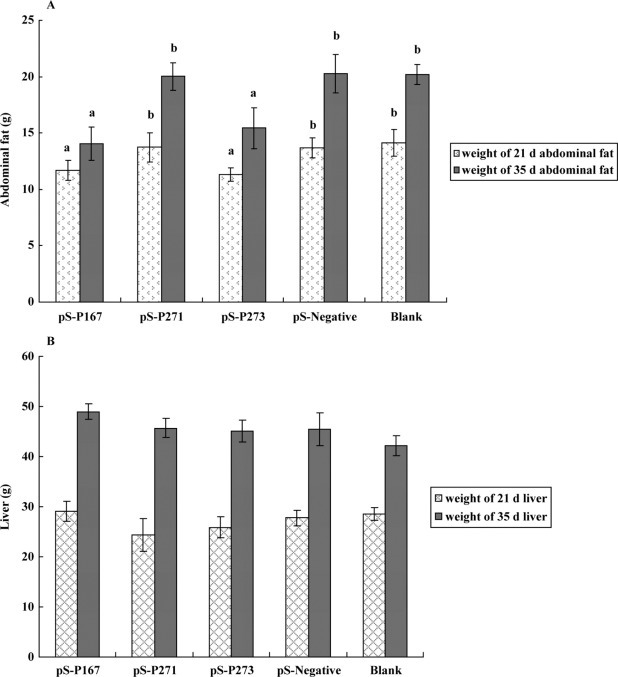
The abdominal fat (A) and liver weights (B) of ducks from the pS-P167, pS-P271, pS-P273, pS-negative, and blank groups at 21 and 35 d. Data are presented as the means ± SD, n = 8 in each group. a,b Different letters indicate significant difference (P < 0.05).
Effect of PPARγ Downregulation on TG Content in Muscle and Serum
The TG levels in serum and muscle from the siRNA groups were lower than in the control groups at both 21 and 35 d. However, none of these differences were statistically significant compared with the controls (Figure 5A and B ).
Figure 5.
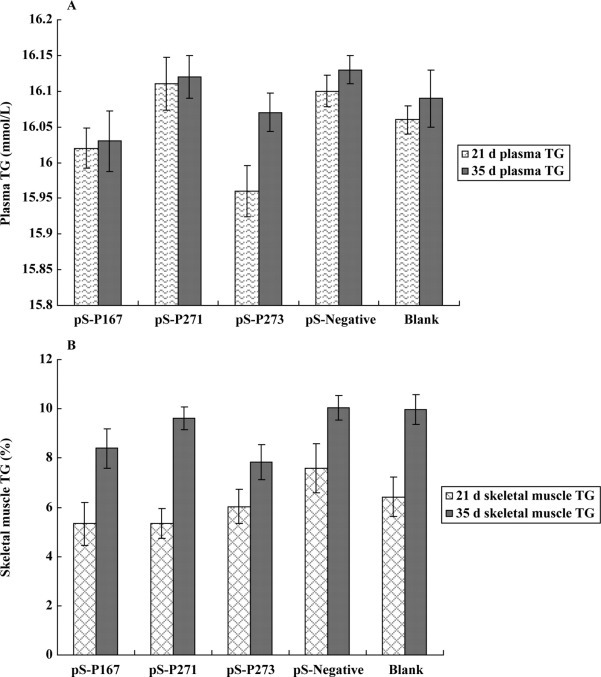
The triglyceride (TG) content of serum (A) and muscle (B) of ducks from the pS-P167, pS-P271, pS-P273, pS-negative, and blank groups at 21 and 35 d. Data are presented as the means ± SD, n = 8 in each group.
DISCUSSION
The function(s) of PPARγ in white adipose tissue has been studied by several groups, and their results have demonstrated that PPARγ is an essential transcription factor during the process of adipogenesis and adipocyte differentiation in mammals (Barak et al., 1999; Desvergne and Wahli, 1999; Kubota et al., 1999; Michalik et al., 2002). It is also known that a moderate reduction of PPARγ activity in PPAR+/− mice decreases the TG content of white adipose tissue (Matsui et al., 2004). Further, Wang et al. indicated that the suppression of PPARγ led to the inhibition of chicken preadipocyte proliferation (Wang et al., 2008). At present, there are 2 mRNA sequences of duck PPARγ in the National Center for Biotechnology Information GenBank database and both sequences encode the PPARγ 1. The Western blot analysis of duck PPARγ only displays 1 band (Figure 3). Therefore, we are not sure if there is PPARγ 2 in ducks. The 3 siRNA of PPARγ interfered against both duck PPARγ 1 and 2 if PPARγ 2 exists. Most of the sequences of PPARγ 1 and 2 are identical, except the PPARγ 2 encodes 11 more amino acids in the N-terminal sequence.
In China, the Cherry Valley duck markets at 35 d after hatching and it is divided into 2 periods [i.e., brooding period (0 to 21d) and finishing period (22 d to marketing)]. It is thought that the ducks deposit fat during the finishing period (start at 21st day), so the diet is changed into a kind of feed with higher metabolic energy at the 21st day to meet the requirement for fat deposition. Because it is reported that the PPARγ was expressed at a relatively low level except in fat tissues, we only measured the expression of PPARγ in fat tissue. In the current study, we found that after PPARγ gene expression is decreased by siRNA in vivo, the weight of abdominal fat is significantly decreased in siRNA-treated ducks compared with the negative control group at ages 21 and 35 d (P < 0.05). This indicates that suppression of PPARγ reduces abdominal fat deposition in ducks.
Peroxisome proliferator-activated receptor γ, a nuclear receptor, binds to the promoter regions of LPL and FABP4 via PPAR response elements and eventually promotes TG synthesis in mammals (Nakachi et al., 2008). Lipoprotein lipase, which is secreted by adipocytes, triggers the release of fatty acids from lipoprotein-bound TG in the extracellular space (Wu et al., 2008). Fatty acid-binding protein 4 is involved in shuttling fatty acids to cellular compartments, modulating intracellular lipid metabolism, and regulating gene expression (Mohlig et al., 2007). In the current study, the expression of the LPL and FABP4 genes was significantly decreased (P < 0.05) when PPARγ protein levels were also decreased. This indicates that LPL and FABP4 may also be the downstream target genes that are regulated by PPARγ in duck adipose tissue. We speculate that the downregulation of PPARγ decreases the abdominal fat deposition in ducks by directly decreasing the expression of LPL and FABP4 mRNA.
In birds, lipogenesis largely occurs in the liver, but storage occurs in extrahepatic tissues, mainly in s.c. tissues and muscles. Hepatic lipids are synthesized into very low density lipoproteins (VLDL), which are secreted into the blood, leading to lipid uptake and storage in adipose and other tissues (O'Hea and Leveille, 1969). Plasma TG are the major components of VLDL in Tsaiya ducks, and increases in the plasma VLDL level can reflect increased liver assembly and secretion of VLDL (Lien et al., 2005; Andre et al., 2007). A low content of LPL protein causes a decreased TG degradation rate in laying chickens than in the growing chickens (Griffin et al., 1990). In the present study on ducks, the TG levels did not obviously decrease in serum at 21 or 35 d of age after the expression of PPARγ mRNA was downregulated. This indicates that downregulating PPARγ decreased the liver lipogenesis rate and also decreased the rate of VLDL-TG degradation by LPL protein by decreasing the expression of LPL mRNA. Thus, the combination of decreased TG secreted into blood and decreased storage in tissues could maintain the content of VLDL-TG in serum.
Triglycerides are transported into tissues via fatty acid transporters, such as CD36, which is regulated by PPARγ. The PPAR+/− mice show markedly reduced CD36 expression in adipose tissue and skeletal muscle, and therefore, these tissues show reduced TG uptake and reduced TG content in PPAR+/− mice (Matsui et al., 2004). In the present study, as the expression of PPARγ mRNA was downregulated, the TG levels did not obviously decrease in duck muscle tissue at 21 or 35 d of age. According to previous studies on broilers, the genetic correlation between abdominal fat weight and i.m. fat percentage is not very close, and changes in the size of adipose tissue are not accompanied by i.m. fat changes (Zerehdaran et al., 2004). In poultry, the i.m. fat content is low (Chartrin et al., 2006), and this may explain why the variation in muscle TG did not reach significantly different levels in ducks in our study.
The efficiency of gene silencing, which is associated with the local mRNA structure at the targeted site, varies significantly according to the different gene positions targeted by siRNA (Luo and Chang, 2004). As we know, off-target is major factor that can confound the expected results. There are far more than 3 siRNA designed when we search siRNA using sFold software (Ding et al., 2004). Off-target effects will be minimized when as few siRNA as possible are needed to knock down their targets, so we chose three of the best of them. These 3 sequences barely have sequence homology with other genes when compared with chicken and duck genomes. Our results show that the PPARγ pS-P167 and pS-P273 siRNA can effectively suppress the expression of PPARγ. The reason for the lower efficiency of the pS-P271 siRNA may be due to positional effects.
To our knowledge, this study is the first to report that the duck PPARγ gene can regulate adipogenesis. Our results indicate that suppression of the duck PPARγ gene reduces abdominal fat deposition and also regulates the expression of LPL and FABP4 mRNA. Thus, the duck PPARγ gene is a key transcription factor affecting fat deposition in ducks.
Acknowledgments
The current research was financially supported by the Project for Critical Technology during the Eleventh 5-Year Plan in the Hubei Province (2006AA202A04), the Key Project for Industrialization during the Eleventh 5-Year Plan in Wuhan (20062001017), and the National Project for the Support of Science and Technology (2008BADB2B08).
REFERENCES
- Andre J.M., Guy G., Gontier-Latonnelle K., Bernadet M.D., Davail B., Hoo-Paris R., Davail S. Influence of lipoprotein-lipase activity on plasma triacylglycerol concentration and lipid storage in three genotypes of ducks. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007;148:899–902. doi: 10.1016/j.cbpa.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Anghel S.I., Bedu E., Vivier C.D., Descombes P., Desvergne B., Wahli W. Adipose tissue integrity as a prerequisite for systemic energy balance: A critical role for peroxisome proliferator-activated receptor γ. J. Biol. Chem. 2007;282:29946–29957. doi: 10.1074/jbc.M702490200. [DOI] [PubMed] [Google Scholar]
- Baeza E., Dessay C., Wacrenier N., Marche G., Listrat A. Effect of selection for improved body weight and composition on muscle and meat characteristics in Muscovy duck. Br. Poult. Sci. 2002;43:560–568. doi: 10.1080/0007166022000004471. [DOI] [PubMed] [Google Scholar]
- Barak Y., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M. PPAR γ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Brun J.M., Mialon-Richard M.M., Sellier N., Batellier F., Brillard J.P. Duration of fertility and hatchability of the common duck (Anas platyrhynchos) in pure- or crossbreeding with Muscovy drakes (Cairina moschata) Theriogenology. 2008;69:983–989. doi: 10.1016/j.theriogenology.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Chartrin P., Meteau K., Juin H., Bernadet M.D., Guy G., Larzul C., Remignon H., Mourot J., Duclos M.J., Baeza E. Effects of intramuscular fat levels on sensory characteristics of duck breast meat. Poult. Sci. 2006;85:914–922. doi: 10.1093/ps/85.5.914. [DOI] [PubMed] [Google Scholar]
- Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- Ding Y., Chan C.Y., Lawrence C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin H.D., Windsor D., Zammit V.A. Regulation of carnitine palmitoyltransferase I in chick liver. Biochem. Soc. Trans. 1990;18:981–982. doi: 10.1042/bst0180981. [DOI] [PubMed] [Google Scholar]
- Huang Y., Haley C.S., Wu F., Hu S., Hao J., Wu C., Li N. Genetic mapping of quantitative trait loci affecting carcass and meat quality traits in Beijing ducks (Anas platyrhynchos) Anim. Genet. 2007;38:114–119. doi: 10.1111/j.1365-2052.2007.01571.x. [DOI] [PubMed] [Google Scholar]
- Kubota N., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Eto K., Tsubamoto Y., Okuno A., Murakami K., Sekihara H., Hasegawa G., Naito M., Toyoshima Y., Tanaka S., Shiota K., Kitamura T., Fujita T., Ezaki O., Aizawa S., Kadowaki T. PPAR γ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q., Xu J., Liu Y., Zheng B.J., Woodle M.C., Zhong N., Lu P.Y. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien T.F., Jan D.F., Chen K.L. Lipoprotein profiles and components in Tsaiya ducks under ad libitum feeding and fasting. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005;142:325–330. doi: 10.1016/j.cbpa.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Luo K.Q., Chang D.C. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 2004;318:303–310. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- Mandrup S., Lane M.D. Regulating adipogenesis. J. Biol. Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- Matsui J., Terauchi Y., Kubota N., Takamoto I., Eto K., Yamashita T., Komeda K., Yamauchi T., Kamon J., Kita S., Noda M., Kadowaki T. Pioglitazone reduces islet triglyceride content and restores impaired glucose-stimulated insulin secretion in heterozygous peroxisome proliferator-activated receptor-γ-deficient mice on a high-fat diet. Diabetes. 2004;53:2844–2854. doi: 10.2337/diabetes.53.11.2844. [DOI] [PubMed] [Google Scholar]
- Michalik L., Desvergne B., Dreyer C., Gavillet M., Laurini R.N., Wahli W. PPAR expression and function during vertebrate development. Int. J. Dev. Biol. 2002;46:105–114. [PubMed] [Google Scholar]
- Miles P.D., Barak Y., Evans R.M., Olefsky J.M. Effect of heterozygous PPARγ deficiency and TZD treatment on insulin resistance associated with age and high-fat feeding. Am. J. Physiol. Endocrinol. Metab. 2003;284:E618–E626. doi: 10.1152/ajpendo.00312.2002. [DOI] [PubMed] [Google Scholar]
- Mohlig M., Weickert M.O., Ghadamgadai E., Machlitt A., Pfuller B., Arafat A.M., Pfeiffer A.F., Schofl C. Adipocyte fatty acid-binding protein is associated with markers of obesity, but is an unlikely link between obesity, insulin resistance, and hyperandrogenism in polycystic ovary syndrome women. Eur. J. Endocrinol. 2007;157:195–200. doi: 10.1530/EJE-07-0102. [DOI] [PubMed] [Google Scholar]
- Nakachi Y., Yagi K., Nikaido I., Bono H., Tonouchi M., Schonbach C., Okazaki Y. Identification of novel PPARγ target genes by integrated analysis of ChIP-on-chip and microarray expression data during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2008;372:362–366. doi: 10.1016/j.bbrc.2008.05.037. [DOI] [PubMed] [Google Scholar]
- O'Hea E.K., Leveille G.A. Lipid biosynthesis and transport in the domestic chick (Gallus domesticus) Comp. Biochem. Physiol. 1969;30:149–159. doi: 10.1016/0010-406x(69)91309-7. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. PPARγ: A nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- Shen J., Samul R., Silva R.L., Akiyama H., Liu H., Saishin Y., Hackett S.F., Zinnen S., Kossen K., Fosnaugh K., Vargeese C., Gomez A., Bouhana K., Aitchison R., Pavco P., Campochiaro P.A. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Hajeri P.B. siRNAs: Their potential as therapeutic agents—Part II. Methods of delivery. Drug Discov. Today. 2009;14:859–865. doi: 10.1016/j.drudis.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Wang Y., Mu Y., Li H., Ding N., Wang Q., Wang Y., Wang S., Wang N. Peroxisome proliferator-activated receptor-γ gene: A key regulator of adipocyte differentiation in chickens. Poult. Sci. 2008;87:226–232. doi: 10.3382/ps.2007-00329. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang H.L., Wang J., Liu X.L. Discovery of a SNP in exon 7 of the lipoprotein lipase gene and its association with fatness traits in native and Cherry Valley Peking ducks. Anim. Genet. 2008;39:564–566. doi: 10.1111/j.1365-2052.2008.01761.x. [DOI] [PubMed] [Google Scholar]
- Yang R.L., Shi Y.H., Hao G., Li W., Le G.W. Increasing oxidative stress with progressive hyperlipidemia in human: Relation between malondialdehyde and atherogenic index. J. Clin. Biochem. Nutr. 2008;43:154–158. doi: 10.3164/jcbn.2008044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerehdaran S., Vereijken A.L., van Arendonk J.A., van der Waaijt E.H. Estimation of genetic parameters for fat deposition and carcass traits in broilers. Poult. Sci. 2004;83:521–525. doi: 10.1093/ps/83.4.521. [DOI] [PubMed] [Google Scholar]
- Zhang T.J., Li H.F., Chen K.W., Chang H., Tang Q.P., Zhang J.X. Genetic diversity and systematic evolution of Chinese domestic ducks along the Yangtze-Huai River. Biochem. Genet. 2007;45:823–837. doi: 10.1007/s10528-007-9121-y. [DOI] [PubMed] [Google Scholar]
- Zhou H., Jin M., Yu Z., Xu X., Peng Y., Wu H., Liu J., Liu H., Cao S., Chen H. Effective small interfering RNAs targeting matrix and nucleocapsid protein gene inhibit influenza A virus replication in cells and mice. Antiviral Res. 2007;76:186–193. doi: 10.1016/j.antiviral.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Zou W., Yu Z., Zhou H., Tu J., Jin M. Genetic characterization of an H5N1 avian influenza virus with neurovirulence in ducks. Virus Genes. 2009;38:263–268. doi: 10.1007/s11262-008-0319-9. [DOI] [PubMed] [Google Scholar]


