Abstract
In view of the restricted knowledge on the diversity of coronaviruses in poultry other than chicken, this study aimed to investigate the genetic diversity of coronaviruses in quail, pheasant, and partridge from two regions of Northern Italy. To this end, pools of tracheal and cloacal swabs from European quail (Coturnix Coturnix) and intestinal tract from pheasants (Phasianus Colchicus) and partridge (Perdix Perdix) flocks, with or without enteric signs, were collected during 2015. Avian coronavirus (Gammacoronavirus) was detected in quail not vaccinated against Infectious Bronchitis Virus (IBV) and in pheasants vaccinated with an IBV Massachusetts serotype. Based on DNA sequences for the gene encoding the S protein, the avian coronaviruses detected in the quail and pheasant are related to the IBV 793B and Massachusetts types, respectively. However, RNA-dependent RNA polymerase (RdRp) analyses showed the susceptibility of quail also to Deltacoronaviruses, suggesting that quail and pheasant avian coronaviruses share spike genes identical to chicken IBV spike genes and quail might host Deltacoronavirus.
Key words: Gammacoronavirus, Deltacoronavirus, quail, pheasant, infectious bronchitis virus
INTRODUCTION
Coronaviruses (CoV), positive-sense single stranded RNA viruses (Nidovirales: Coronaviridae: Coronavirinae), are currently classified in four genera. Alphacoronavirus (α-CoV) and Betacoronavirus (β-CoV) infect many mammalian species ranging from bats to humans (Chan, et al., 2013) whereas Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV) are largely established in birds and to a slightly degree in mammals (Woo et al., 2010; Woo et al., 2012; De Groot et al., 2012; Fehr and Perlman, 2015).
Avian coronavirus (Avian infectious bronchitis virus, Infectious Bronchitis Virus [IBV] in chickens), the causative agent of avian infectious bronchitis (IB), is a Gammacoronavirus that can replicate on epithelial tissues, affecting respiratory, reproductive, urinary, and enteric tracts (Cavanagh, 2003; Cavanagh, 2005; Villarreal et al., 2007). IBV and IBV-like strains can be detected in both gallinaceous and non-gallinaceous birds, such as geese, ducks, pigeons, pheasants (Cavanagh et al., 2002), and quail (Circella et al., 2007; Torres et al., 2013), asymptomatically in some cases (Cavanagh, 2005). This might suggest that these species would act as vectors spreading IBV strains over the world (De Wit et al., 2011).
Other coronaviruses, classified into the Deltacoronavirus genus (De Groot et al., 2012), were described by Woo et al. (2009), Chu et al. (2011) and Durães-Carvalho et al. (2015) in wild birds.
In the view of the restricted knowledge on coronaviruses in poultry other than chickens, this study aimed to investigate the genetic diversity of coronaviruses in farmed quail, pheasants and gray partridges from Northern Italy.
MATERIAL AND METHODS
Farms
The survey was performed on 10 farms including 17 quail, 8 pheasant, and 2 gray partridge flocks in 2015. Characteristics of flocks, vaccination programs applied, and sampling details are summarized in Table 1 . Farms included in the study were all located in Northern Italy, in particular in the Veneto and Emilia Romagna regions that are considered densely populated poultry areas.
Table 1.
Identification and characteristics of the Italian farms studied during April to July of 2015, regarding type of bird (quail, pheasant, and gray partridge), time of sampling (beginning, middle, end, or complete production cycle), type of sample, and IB vaccination programs applied.
| Farm ID | Bird type | Sampling time | Type of sample | IB vaccination program |
|---|---|---|---|---|
| F1 | Quail/Broiler | CPC# | CS/TS | Broilers vaccinated at hatchery = Mass and 793B |
| F2 | Quail | CPC | CS/TS | Non vaccinated |
| F3 | Quail | Beginning/middle | Intestine | Non vaccinated |
| F4 | Quail | CPC | CS/TS | Non vaccinated |
| F5 | Quail | CPC | Intestine | Quail breeders = IBV Mass |
| F6 | Pheasant | CPC | Intestine | One day old = IBV Mass |
| F7 | Pheasant | Middle | Intestine | Non vaccinated |
| F8 | Pheasant | Middle | Intestine | Non vaccinated |
| F9 | Pheasant | Middle/end | Intestine | Non vaccinated |
| F10 | Partridge | End | Intestine | Non vaccinated |
IB = infectious bronchitis.
CPC = Complete production cycle (beginning, middle and end).
CS = Cloacal swab.
TS = Tracheal swab.
Mass = Massachusetts serotype.
= farm 1 was sampled in three following production cycles.
For each flock, data regarding the occurrence of respiratory or enteric signs ongoing or experienced were recorded.
Sample Collection
Cloacal and tracheal dry swabs (10 birds/flock) or the intestinal tract (5 samples/flock) from necropsied birds, were collected for molecular investigation. The swabs for the molecular testing were dried at room temperature for approximately 15 minutes.
All samples were stored at −80°C until processing as a pool according to the sampling (dry swabs or intestinal tract).
RNA Extraction
Pools of tracheal and cloacal swabs were eluted in 2 mL of sterile PBS and pools of intestinal tissues and contents were prepared as a 50% (v/v) suspension in PBS and clarified at 12,000 × g for 15 min at 10°C. Total RNA was extracted from 200 μL of the supernatants and from elutions using High Pure RNA Isolation kit (Roche Diagnostics Italy) according to the manufacturer's instructions.
One Step Nested RT- PCR for Avian Coronavirus Screening
Each sample was screened for Avian coronavirus as described by Cavanagh et al. (2002) targeting the 3′-untranslated region (UTR) using SuperScript III RT/ Platinum Taq Mix (Invitrogen, Carlsbad, CA) as per manufacturer's instructions.
RNA-dependent RNA Polymerase (RdRp) and Spike Gene Analysis
3′UTR- Avian coronavirus positive samples were then tested using the following protocols: 1) a pan-coronavirus RT-PCR targeting the RNA- dependent RNA polymerase (RdRp) gene as described by Chu et al. (2011) (440 base of pairs [bp] amplicon); 2) a S-gene targeted multiplex RT-PCR for 793B, D274 and Massachusetts types (Cavanagh et al., 1999) and 3) finally a generic S gene RT-PCR described by Worthington et al. (2008) (393 bp amplicon). All RT-PCR were performed using SuperScript III RT/ Platinum Taq Mix (Invitrogen, Carlsbad, CA) as per manufacturer's instructions.
All RT-PCR amplicons were analyzed by electrophoresis in 2% (w/v) agarose stained with SYBR Safe DNA Gel Stain (Invitrogen, Carlsbad, CA) and visualized by ultraviolet transillumination.
DNA sequencing was performed at Macrogen (Macrogen Europe, Amsterdam, Netherlands). All chromatograms were manually checked with Finch TV program. 1.4.0 (2004–2006 Geospiza Inc) and submitted to quality evaluation by Phil's Read Editor (PHRED) online application http://asparagin.cenargen.embrapa.br/phph/ with a base-quality cut-off of 20. The final sequence of each sample was obtained with the Consensus application included in BioEdit 7.0.9.0 software (Tom Hall © 1997–2007) and aligned with homologous sequences retrieved from GenBank (see accession numbers in Figures Figure 1, Figure 2 ) with Clustal/W multiple alignment using the same software.
Figure 1.
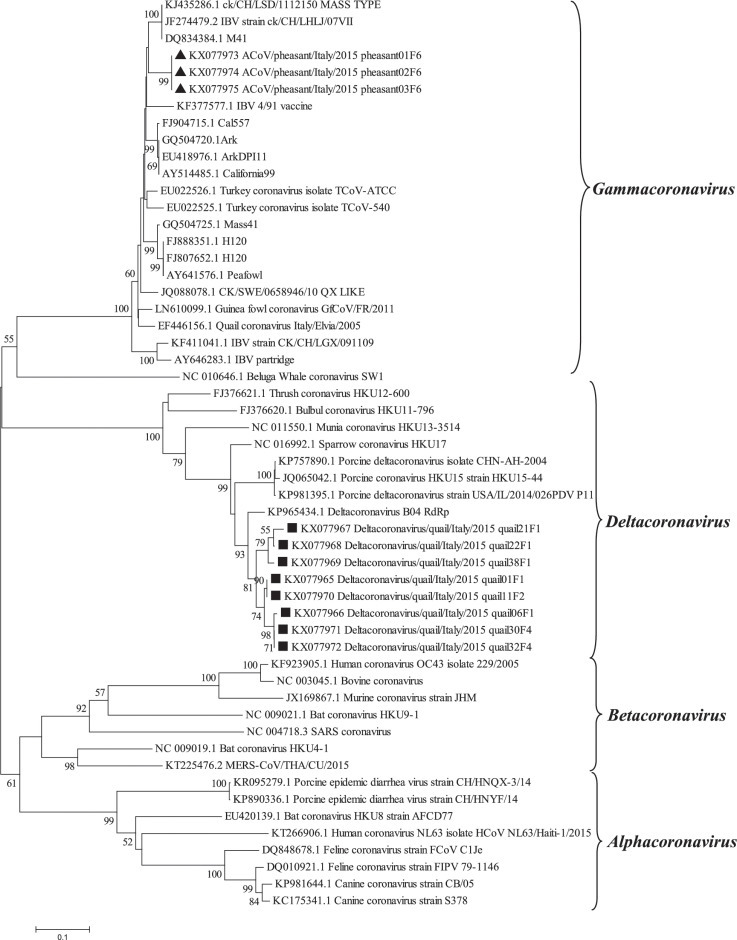
Phylogenetic tree based on the alignments of nucleotide sequences of RdRp gene (nt position 16,179 to 16,543) of samples sequenced in this study (square for quail and triangle for pheasant) and the prototype serotypes/genotypes (with Genbank accessing numbers) of the genera Alpha, Beta, Gamma, and Deltacoronavirus. The numbers above each node represent the bootstrap values for 1,000 replicates (only values greater than 50% are shown). The bar represents the number of nucleotide substitutions per site.
Figure 2.
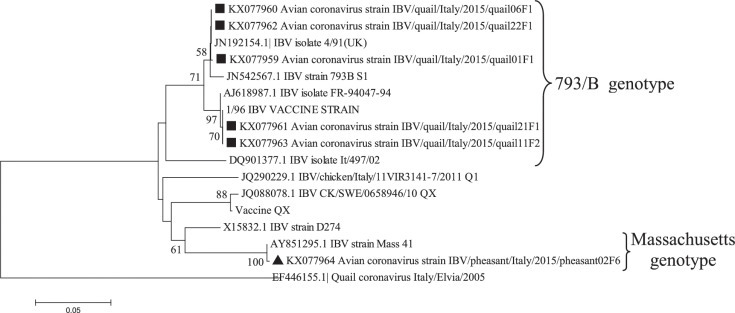
Phylogenetic tree based on the alignments of nucleotide sequences of partial S gene (nt position 726 to 1,073) of samples sequenced in this study (square for quail and triangle for pheasant) and the prototype serotypes/genotypes (with Genbank accessing numbers). The numbers above each node represent the bootstrap values for 1,000 replicates (only values greater than 50% are shown). The bar represents the number of nucleotide substitutions per site.
Nucleotide (Neighbor-joining, Maximum Composite Likelihood model, 1,000 bootstrap replications) trees for S and RdRp genes were built using MEGA 5.2.1 (Tamura et al., 2011).
RESULTS
Avian Coronavirus Screening
Fifteen flocks (9 of quail and 6 of pheasant) from 7 farms (F1, F2, F4, F5, F6, F7, and F8) were positive for Avian coronavirus UTR targeted RT-PCR. Gray partridge samples were negative.
IBVs detected by the multiplex typing PCR belonged to the 793B and Massachusetts genotypes in quail and pheasants, respectively. Strains amplified in RdRp and S genes are reported in Table 2 .
Table 2.
Strains included in the phylogenetic analysis of the partial RNA-dependent RNA polymerase (RdRp) and S genes according to species (quail or pheasant), clinical signs at the sampling time and sample.
| Strain | Host | Signs | Sample | Gene | GenBank accession number |
|---|---|---|---|---|---|
| Quail01/CS/F1 | Quail | N/S | Cloacal swab | RDRP | KX077965 |
| Quail01/CS/F1 | Quail | N/S | Cloacal swab | S | KX077959 |
| Quail06/CS/F1 | Quail | N/S | Cloacal swab | RDRP | KX077966 |
| Quail06/CS/F1 | Quail | N/S | Cloacal swab | S | KX077960 |
| Quail21/CS/F1 | Quail | N/S | Cloacal swab | RDRP | KX077967 |
| Quail21/CS/F1 | Quail | N/S | Cloacal swab | S | KX077961 |
| Quail22/TS/F1 | Quail | N/S | Tracheal swab | RDRP | KX077968 |
| Quail22/TS/F1 | Quail | N/S | Tracheal swab | S | KX077962 |
| Quail38/CS/F1 | Quail | Ent | Cloacal swab | RDRP | KX077969 |
| Quail11/TS/F2 | Quail | Ent | Tracheal swab | RDRP | KX077969 |
| Quail11/TS/F2 | Quail | Ent | Tracheal swab | S | KX077963 |
| Quail30/CS/F4 | Quail | Ent | Cloacal swab | RDRP | KX077971 |
| Quail32/CS/F4 | Quail | Ent | Cloacal swab | RDRP | KX077972 |
| Pheasant01/INT/F6 | Pheasant | Ent | Intestine | RDRP | KX077973 |
| Pheasant02/INT/F6 | Pheasant | Ent | Intestine | RDRP | KX077974 |
| Pheasant03/INT/F6 | Pheasant | Ent | Intestine | RDRP | KX077975 |
| Pheasant02/INT/F6 | Pheasant | Ent | Intestine | S | KX077964 |
Ent = Enteric.
N/S = Not signs.
The phylogenetic tree (Figure 1) for the RdRp gene indicates that all quail strains clustered in the δ-CoV genus. Mean RdRp nucleotide identities of the quail strains with Deltacoronavirus B04 genotype (Genbank KP965434) (Durães-Carvalho et al., 2015) and Porcine Deltacoronavirus, was 92.5% and 86%, respectively. Moreover, three subclusters were observed, with a mean nucleotide identity of 95.89%, suggesting the existence of different viral populations.
Strains detected in pheasants clustered in the Gammacoronavirus genus (Figure 1), with 100% identity amongst each other.
The phylogenetic tree based on the alignments of nucleotide sequences of partial S1 (Figure 2) confirmed the results of the multiplex RT-PCRs. All quail strains analyzed belonged to the 793B IBV genotype (mean nucleotide identity of 97%). Two strains (accession numbers KX077963 and KX077961) clustered with a commercial IBV vaccine based on 793B genotype widely used in Italy (mean nucleotide identity of 99.4%). The strains detected in pheasants clustered all with Massachusetts IBV genotype with 99% nucleotide identity.
DISCUSSION
The presence and molecular diversity of coronaviruses in quail, pheasants, and partridge from Northern Italy was investigated, allowing the detection of Avian coronavirus related in the spike gene to 793B and Massachusetts IBV genotypes in quail and pheasants, respectively.
The phylogenetic analysis of the S gene suggested circulation in quails of 793B IBV genotype. This genotype, after its first appearance in the 1990s (Gough et al., 1992), became one of the most widespread genotypes in Europe (Worthington et al., 2008); for its control homologous live vaccination is widely used.
Vaccine origin of IBV detections in quail could not be excluded because molecular tools currently available are unable to unequivocally distinguish between field and vaccine viruses. Furthermore, recent epidemiological study on broilers strongly suggests that 793B detected in Italy derived from the 793B vaccines in use (Franzo et al., 2014).
High homology of a 793B genotype, detected in quail, with the identical serotype used in the vaccine applied to a broilers flock housed in the same farm strongly supports this hypothesis.
Pheasants IBV spike detection clustered with Massachusetts genotype. Considering that the pheasants had been vaccinated with a Massachusetts live vaccine, this also suggests a vaccine origin for this detection, as IBV vaccines might be detected in vaccinated birds for weeks after vaccination (Cavanagh et al., 1999). The sequence of RdRp of the same sample clustered within Gammacoronaviruses.
Surprisingly the RdRp analysis showed that quail detections clustered with δ-CoV and not with γ-CoV as expected. The previously reported Quail coronavirus (QCoV) (Circella et al., 2007) based on the RdRp gene showed that QCoV was grouped along with γ-CoV and the S1 subunit of the S gene, showed 16 to 18% amino acid identity with IBV and 79 to 81% identity with TCoV (turkey coronavirus) strain, suggesting that specifically QCoV was not an IBV variant.
Taking into account the discrepant taxonomy of the quail coronaviruses detected herein regarding the RdRp and the spike genes (Deltacoronavirus and Gammacoronavirus, respectively) both a Gamma-Deltacoronavirus strain co-circulation or occurrence of a recombination event might be hypothesized. Nonetheless, recombination between-genera in coronaviruses, though initially considered but latter disregarded as a source of the SARS coronavirus, seems to be unlikely in nature (Thor et al., 2011, Woo et al., 2010).
Further efforts on full-genome sequencing using next generation sequencing of the quail coronaviruses detected herein will better clarify the characteristics and origin of the detected strains.
Isolation and inoculation of quails in experimental conditions will confirm the susceptibility of this avian species to δ-CoV.
Acknowledgments
We are grateful to the field veterinarians and farmers who submitted the samples.
REFERENCES
- Cavanagh D. Severe acute respiratory syndrome vaccine avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Welchmand Dde B., Britton P., Gough R.E. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (Infectious Bronchitis Virus) and turkeys. Avian Pathol. 2002;31:81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Britton P., Naylor C.J. Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specic polymerase chain reactions. Avian Pathol. 1999;28:593–605. doi: 10.1080/03079459994399. [DOI] [PubMed] [Google Scholar]
- Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Leung C.Y., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., Guan Y., Peiris J.S., Poon L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circella E., Camarda A., Martella V., Bruni G., Lavazza A., Buonavoglia C. Coronavirus associated with an enteric syndrome on a quail farm. Avian Pathol. 2007;36:251–258. doi: 10.1080/03079450701344738. [DOI] [PubMed] [Google Scholar]
- De Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Poon L, Rottier P.J.M., Talbot P.J., Woo P.C., Ziebuhr J. Family coronaviridae. In: King A., Adams M.J., Carstens E.B., Lefkowitz E.F., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier academic press; Amsterdam, Boston: 2012. pp. 806–820. [Google Scholar]
- De Wit J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durães-Carvalho R., Caserta L.C., Barnabé A.C.S., Martini M.C., Ferreira H.L., Felippe P.A.N., Santos M.B., Arns C.W. Coronaviruses Detected in Brazilian Wild Birds Reveal Close Evolutionary Relationships with Beta- and Deltacoronaviruses Isolated From Mammals. J. Mol. Evol. 2015;81:21–23. doi: 10.1007/s00239-015-9693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods. Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo G., Naylor C.J., Lupini C., Drigo M., Catelli E., Listorti V., Pesente P., Giovanardi D., Morandini E., Cecchinato M. Continued use of IBV 793B vaccine needs reassessment after its withdrawal led to the genotype's disappearance. Vaccine. 2014;32:6765–6767. doi: 10.1016/j.vaccine.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough R.E., Randall C.J., Dagless M., Alexander D.J., Cox W.J., Pearson D. A ‘new' strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet. Rec. 1992;130:493–494. doi: 10.1136/vr.130.22.493. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar M.S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutioanry distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S.W., Hilt D.A., Kissinger J.C., Paterson A.H., Jackwood M.W. Recombination in Avian Gamma-Coronavirus Infectious Bronchitis Virus. Viruses. 2011;3:1777–1799. doi: 10.3390/v3091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C.A., Villarreal L.Y.B., Ayres G.G.R., Richtzenhain L., Brandão P.E. An avian coronavirus in quail with respiratory and reproductive signs. Avian. Dis. 2013;57:295–299. doi: 10.1637/10412-100412-Reg.1. [DOI] [PubMed] [Google Scholar]
- Villarreal L.Y.B., Brandão P.E., Chacón J.L., Saidenberg A.B.S., Assayag M.S., Jones R.C., Ferreira A.J.P. Molecular characterization of infectious bronchitis virus strains isolated from the enteric contents of brazilian laying hens and broilers. Avian. Dis. 2007;51:974–978. doi: 10.1637/7983-041307.1. [DOI] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1805–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington K.J., Currie R.J., Jones R.C. A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37:247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]


