Abstract
Chicken astrovirus (CAstV) is one of many viruses related to enteric diseases in poultry that are associated with Runting-Stunting Syndrome (RSS), which affects young chickens. CAstV was also recently associated with an unusual condition in chicks called “white chicks.” Some hatcheries in certain states of Brazil have reported several incubation problems, mortality, and the presence of chicks with white plumages over the past several months. These chicks were termed locally as “white chicks.” The present work investigated 30 chicks with this unusual condition using a multidisciplinary approach. Postmortem examination of each chick showed enlarged livers and intestines that were full of liquid and gas (30/30). The pancreas, kidneys, and spleen were pale (30/30). The other organs did not show any macroscopic alterations. CAstV, chicken parvovirus (ChPV), avian nephritis virus (ANV), avian rotavirus (ARtV), avian reovirus (AReoV), infectious bronchitis virus (IBV), and fowl adenovirus group I (FAdV-1) were tested in the intestines, pancreas, proventriculus, gizzard, liver, spleen, bursa, kidneys, thymus, lung, heart, brain, and yolk sac in each chick. All organs and yolk sacs were positive for CAstV in different titres and negative for the other tested viruses. The partial molecular characterization of the ORF 1b gene of CAstV using 28 sequences revealed a high similarity of the nucleotides and amino acids with sequences of CAstV from North America, Europe, and Asia, and our CAstV sequences clustered into a unique group that was separate from the other sequences. These results demonstrated that CAstV was associated with the white chick condition in Brazil. The virus was distributed in most organs, including the brain and yolk sac. These results suggest that the virus could be transmitted vertically. The molecular characterization also revealed that the CAstV associated with white chick condition was molecularly related to other CAstV sequences found worldwide.
Key words: chicken, astrovirus, white chicks, detection, characterization
INTRODUCTION
Enteric diseases are of paramount importance to the national and global poultry industry (Jindal et al., 2012). Runting-Stunting Syndrome (RSS) is an enteric syndrome with mild clinical signs, such as diarrhea, that causes flocks to mature in a non-uniform pattern, which causes the body weights at slaughter to differ broadly (Otto et al., 2006). Olsen first reported RSS in the broiler industry in 1977 (Olsen, 1977). This syndrome is a transmissible disease of uncertain etiology that affects chickens early in life (Goodwin et al., 1993; Kang et al., 2012b). Enteric viruses, such as CAstV, ChPV, ANV, ARtV, AReoV, and FAdV-1 are related and have been detected in chickens with signs of RSS (Day et al., 2007; Pantin-Jackwood et al., 2008; Pantin-Jackwood et al., 2011; Mettifogo et al., 2014). However, CAstV is recognised as the causal agent of enteritis in chickens, primarily young chicks, and it has been detected in one-day-old chicks (Pantin-Jackwood et al., 2008; Mettifogo et al., 2014). There are actually 2 astroviruses in chickens that are genetically characterised as ANV and CAstV (Imada et al., 2000; Baxendale and Mebatsion, 2004; Todd et al., 2009a). Astroviruses are small, spherical viruses that are 25 to 35 nm in diameter and possess single-stranded, positive-sense RNA genomes approximately 7 kb in length. The genome encodes for 3 proteins: the non-structural (NS) polyprotein, the RNA-dependent RNA polymerase (RdRp), and the capsid protein. The NS polyprotein and capsid protein are encoded by individual open-reading frames (ORF), ORF1a and ORF2, and RdRP (ORF 1b) acts as a fusion protein to the NS protein (Kang et al., 2012a). The genome begins with a 5′ untranslated region (UTR), followed by the 3 ORFs, a 3′ UTR, and a poly A tail (Koci and Schultz-Cherry, 2002; Nuñez and Piantino Ferreira, 2013a). ORF1a encodes the viral protease and is followed by ORF1b, which encodes the RNA polymerase (RdRp). ORF2 encodes the precursor of the capsid protein, and it is located downstream of ORF1b and prior to the untranslated region of the genome (Kang et al., 2012a). An experimental infection with CAstV in one-day-old chickens showed mild diarrhea and distension of the small intestine (De Wit et al., 2011). Clinical signs, including diarrhea, decreased food consumption, and nervousness, developed between one and 3 wk of age (Moser and Schultz-Cherry, 2005). CAstV has been detected in the United Kingdom, United States, Korea, India, Netherlands, Croatia (Day et al., 2007; Pantin-Jackwood et al., 2007; Todd et al., 2009a,b; Smyth et al., 2010; Kang et al., 2012b; Koo et al., 2013). And, more recently, in Brazil (Mettifogo et al., 2014), which demonstrates a worldwide distribution (De Benedictis et al., 2011). Most of the CAstV reported worldwide was detected in chickens with enteric disease. However, more recent reports have indicated an association of CAstV with a low hatching rate, weakness, and white plumage, which is characterised as an abnormal condition termed white chicks (Smyth et al., 2013).
The molecular characterization of CAstV is very important in understanding the relationship and genetic similarity among the different isolates worldwide. This study examined the viral agent that is associated with the condition of white chicks in Brazil and described the partial molecular characterization of the agent related with chicks with this unusual condition and its presence in Brazilian poultry.
MATERIAL AND METHODS
After 2014, some states of Brazil began reporting problems in their hatcheries of an increase in pipped eggs and chick mortality immediately after hatching. The chicks presented white plumage with discolored and pale beaks and legs. This uncommon condition was termed “white chicks” (Smyth et al., 2013). Thirty chicks with this unusual condition were sent to the Laboratory of Avian Diseases at the School of Veterinary Medicine, University of São Paulo, to determine the agent that is related to this condition (Table 1 ). The chicks were subjected to postmortem examination and molecular screening for CAstV, ANV, ARtV, AReoV, IBV, FAdV-1, and ChPV.
Table 1.
Description of samples’ characteristics obtained from poultry companies with hatching of white chicks, in Brazil.
| External characteristics of chicks with unusual condition of white chicks |
||||||
|---|---|---|---|---|---|---|
| Sample designation | Origin of sample | White plumage | Pale beak and legs/feet | Age of chicks | Brazilian state localization | GenBank accession number |
| 541–1 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013267 |
| 541–2 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013275 |
| 541–3 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013276 |
| 541–4 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013268 |
| 541–5M | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013271 |
| 541–6M | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013274 |
| 541–7F | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013269 |
| 541–8 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013272 |
| 541–9 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013270 |
| 541–10 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013273 |
| 541–11 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013254 |
| 541–12 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013255 |
| 541–13 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013251 |
| 541–14 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013253 |
| 541–15 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013249 |
| 541–16 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013252 |
| 541–17 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013250 |
| 541–18 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013256 |
| 541–19 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013258 |
| 541–20 | Broiler chicks | Yes | Yes | 1 day | São Paulo | Not performed |
| 541–21 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013262 |
| 541–22 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013257 |
| 541–23 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013264 |
| 541–24 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013259 |
| 541–25 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013263 |
| 541–26 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013260 |
| 541–27 | Broiler chicks | Yes | Yes | 1 day | São Paulo | Not performed |
| 541–28 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013265 |
| 541–29 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013261 |
| 541–30 | Broiler chicks | Yes | Yes | 1 day | São Paulo | KR013266 |
Postmortem Examination
The chicks were subjected to postmortem examination. Each organ was collected separately, and weighed. The organs analyzed in the present survey were brain, intestines (duodenum, jejunum, ileum, and ceca), pancreas, liver, kidney, heart, lung, thymus, bursa, spleen, gizzard, proventriculus, and yolk sac.
Molecular Detection
Each organ was subjected to the molecular detection of the above-mentioned enteric viruses using PCR and RT-PCR (Table 2 ). An aliquot of each organ or the entire organ, depending on organ weight, was placed into a 1.5 mL sterile microfuge tube containing 0.1 M phosphate-buffered saline (PBS, pH 7.2), in equivalent volume to the organ weight or the aliquot placed in the microfuge tube. Macerated thymus, pancreas, and spleen were placed into a 1.5 mL sterile microfuge tube with 300 μL of 0.1 M PBS (pH 7.2). An aliquot of yolk (750 μL) was also placed into a 1.5 mL sterile microfuge tube with the same quantity of 0.1 M PBS (pH 7.2). These suspensions were subjected to 3 cycles of freezing to −80°C for 10 min and thawing for one min to 56°C, homogenised, and centrifuged to 12.000 x g for 20 min to 4°C. The supernatants (250 μL) were treated with TriZOL reagent for the extraction of RNA and DNA (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instruction. Ultra-pure water was used as a negative control.
Table 2.
Primers sequences used in the PCR and RT-PCR used in the present work, gene target, amplicon generated, and references for each virus.
| Virus | Gene target | Primer designation | Sequences | Amplicon | Reference |
|---|---|---|---|---|---|
| CAstV | ORF-1b | CAS pol 1F | 5′GAYCARCGAATGCGRAGRTTG3′ | 362 bp | Day et al., 2007 |
| CAS pol 1R | 5′TCAGTGGAAGTGGGKARTCTAC3′ | ||||
| ANV | ORF-1b | ANV Pol 1F | 5′GYTGGGCGCYTCYTTTGAYAC3′ | 473 bp | Day et al., 2007 |
| ANV Pol 1R | 5′CRTTTGCCCKRTARTCTTTRT3′ | ||||
| Avian rotavirus | NSP4 | NSP4-F30 | 5′GTGCGGAAAGATGGAGAAC3′ | 630 bp | Day et al., 2007 |
| NSP4-R660 | 5′GTTGGGGTACCAGGGATTAA3′ | ||||
| Avian reovirus | S4 | S4-F13 | 5′GTGCGTGTTGGAGTTTCCCG3′ | 1120 bp | Pantin-Jackwood et al., 2008 |
| S4-R1133 | 5′TACGCCATCCTAGCTGGA3′ | ||||
| IBV | UTR | UTR 11 | 5′ATGTCTATCGCCAGGGAAATGTC3′ | 179 bp | Cavanagh et al., 2002 |
| UTR 44 | 5′GGGCGTCCAAGTGCTGTACCC3′ | ||||
| UTR 31 | 5′GCTCTAACTCTATACTAGCCTA3′ | ||||
| FAdV-1 | Hexon | Hexon A | 5′CAARTTCAGRCAGACGGT3′ | 897 bp | Meulemans et al., 2001 |
| Hexon B | 5′TAGTGATGMCGSGACATCAT3′ | ||||
| ChPV | NS | PVF1 | 5′TTCTAATAACGATATCACT3′ | 561 bp | Zsak et al., 2009 |
| PVR1 | 5′TTTGCGCTTGCGGTGAAGTCTGGCTCG3′ |
Reverse Transcriptase
The RNA obtained was submitted to a reverse transcription reaction to obtain complementary DNA (cDNA). A volume of 3.5 μL of extracted RNA was denatured at 95°C for 5 min in a 200-μL microtube, and 6.5 μL of a mixture containing 2 μL 5X buffer, one μL dithiothreitol (DTT), 10 mM deoxynuclotides triphosphates (dNTPs), one μL of each forward and reverse primers, and 0.5 μL M-MLV enzyme (Invitrogen Life Technologies, Carlsbad, CA) was added. A reverse transcription reaction was performed under the following conditions: 45°C for 60 min and 72°C for 10 min. The cDNA obtained was submitted to PCR.
Polymerase Chain Reaction
The PCR reaction used 24 μL of a mixture that contained 0.5 μM of each forward and reverse primers, 2.5 μL 10X buffer, 5 mM of each dNTP, 37.5 mM of MgCl2, one U of Platinum DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA), and one μL cDNA. PCR amplification was performed under the following conditions: a cycle of 94°C for 3 min to completely denature the cDNA; 30 cycles of a denaturation temperature at 94°C for one min; hybridisation of primers at 56°C for one min; extension at 72°C for one min; and a final incubation at 72°C for 10 min. The reaction was maintained at 4°C for an undetermined period until storage at 20°C. RT-PCR for CAstV amplification was performed using similar protocols as Day et al. (2007) (Table 2). Chicks with the unusual condition white chicks were tested for other enteric viruses. PCR for ChPV was performed according to Zsak et al. (2009). PCR for FAdV-1 was performed according to Meulemans et al. (2001). RT-PCR for ANV and avian rotavirus was also performed according to Day et al. (2007). RT-PCR/NESTED for IBV was performed according to Cavanagh et al. (2002), and RT-PCR for reovirus was performed according to Pantin-Jackwood et al. (2008). Table 2 describes the primer sequences used. The amplified products (362-bp CAstV, 473-bp ANV, 179-bp IBV, 630-bp ARtV, 1120-bp AReoV, 561-bp ChPV, and 897-bp FAdV-1) were submitted to electrophoresis in a 1.5% agarose gel. The samples were stained with Sybr Safe (Invitrogen by Life Technologies, Carlsbad, CA) and compared with a 100-bp molecular ladder (Invitrogen by Life Technologies, Carlsbad, CA). The results were analyzed on a transilluminator and photographed using an Alpha Imager Mini Analysis System (Alpha Innotech by Protein Symple, Santa Clara, CA).
DNA Sequencing and Phylogenetic Analyses
The amplified products of the ORF1b gene of CAstV from each animal were purified using a GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare, Piscataway, NJ) as described by the manufacturer. Each purified product was sequenced in the forward and reverse direction using a BigDye® Terminator v3.1 Cycle Sequencing kit (Applied Biosystems by Life Technologies, Carlsbad, CA). Sequencing reactions were run in an ABI 3730 DNA Analyzer (Applied Biosystems by Life Technologies, Carlsbad, CA). Nucleotide sequences were edited using the CLC Main WorkBench 7.5.1 package software (CLC Bio, Qiagen, Waltham, MA) and aligned with other sequences using the CLUSTAL W method available in the ClustalX 2.0.11software package (UCD, Dublin, Ireland). The nucleotide phylogenetic tree was inferred using the neighbour-joining, maximum composite-likelihood method with 1,000 bootstrap replicates that were integrated in the MEGA version 5 software (Tamura et al., 2011).
GenBank Accession Number
The accession number of ORF 1b sequences of CAstV were obtained here are as follows: KR013249 to KR013276. Figure 3 shows the GenBank accession numbers of the sequences used for molecular analyses in the present work.
Figure 3.
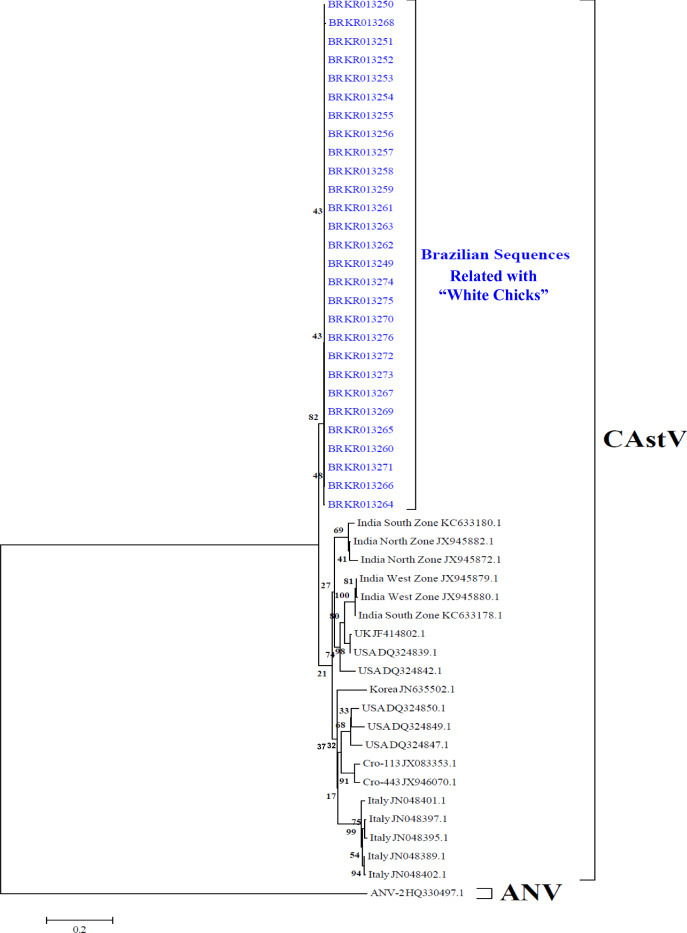
Neighbor-joining tree based on alignments of the partial ORF 1b sequences of CAstV. Phylogenetic relationships of the Brazilian (BR) CAstV sequences from white chicks compared with other sequences from Croatia (CRO), India, Italy, Korea, United Kingdom (UK), and United States (USA). Sequences were aligned using the Clustal W method in ClustalX software. The tree was constructed using MEGA 5 software package. Numbers along the branches refer to bootstrap values for 1,000 replicates. The scale bar represents the number of substitutions per site. ANV-2 sequence was used as the out-group.
RESULTS
Postmortem Examination
White chicks exhibited discolored beaks and legs/feet and white plumage (30/30; 100%), as shown in Figure 1 . The abdominal organs showed several abnormalities. Livers were enlarged and yellow and surpassed the last ribs by approximately 0.5 cm (30/30; 100%), and the gallbladders were full. Intestines were filled with liquid and gas bubbles (30/30; 100%) with several intestines containing meconium (25/30; 83.33%), yellowish with a yolk-like consistency, and all yolk sacs were filled with yolk (30/30; 100%), as presented in Figure 2 . The pancreas and spleen were pale, and the spleen had an oval form (30/30; 100%). Some chicks showed a dilatation of the proventriculus (12/30; 40%). The kidneys were pale with some yellow foci (30/30; 100%). The gizzard was apparently normal without erosions or hemorrhage. The brain, thymus, bursa, heart, and lungs did not exhibit any macroscopic alterations.
Figure 1.
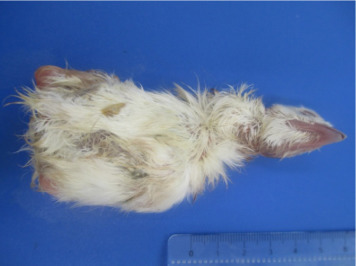
Chicks showing white plumage.
Figure 2.
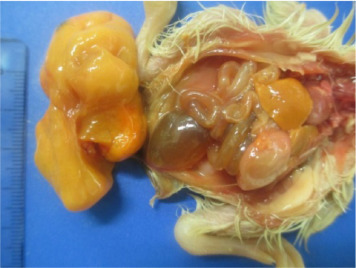
Postmortem examination of white chicks showed enlarged liver, and intestines filled with gas and liquid. Also, chick is presenting white feet.
Molecular Detection
RT-PCR amplified the ORF1b gene of CAstV and obtained an amplicon of 362 bp. All of the collected organs were subjected to the molecular detection of enteric viruses. At least one organ from each bird was positive for CAstV in this study. The intestines, liver, spleen, thymus, bursa, pancreas, kidneys, heart, and lungs were positive for CAstV, and no other enteric viruses were detected (Table 3 ). CAstV was primarily detected in samples of the gizzard (96.67%), intestines (93.33%), lungs (93.33%), kidneys (83.33%), pancreas (80%), spleen (80%), and yolk (60%), but the virus was less detected in the liver (13.13%), proventriculus (10%), heart (6.67%), brain (6.67%), and thymus (6.67%), as shown in Table 3. The present study demonstrated that CAstV was distributed in all organs tested and the yolk.
Table 3.
Molecular detection using RT-PCR of CAstV in the organs from chickens with unusual condition of white chicks.
| Sample designation | Proventriculus | Intestine | Pancreas | Spleen | Kidney | Liver | Yolk sac | Lung | Bursa | Thymus | Brain | Heart | Gizzard |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 541—1 | – | + | + | + | + | – | – | + | – | – | – | – | + |
| 541—2 | – | + | + | + | + | – | – | + | – | – | + | – | + |
| 541–3 | – | + | + | + | – | – | – | – | – | – | – | – | + |
| 541–4 | – | + | + | + | + | – | – | + | – | – | – | – | + |
| 541–5M | – | + | – | – | + | – | + | + | – | – | – | – | + |
| 541–6M | – | + | + | + | + | – | + | + | – | – | – | – | + |
| 541–7F | – | + | – | + | + | + | + | + | – | – | – | – | + |
| 541–8 | – | + | + | + | + | – | + | + | – | – | – | – | + |
| 541–9 | – | – | + | – | + | – | + | + | – | – | – | – | + |
| 541–10 | – | + | + | + | + | – | + | + | – | – | – | – | + |
| 541–11 | – | + | + | + | + | – | + | + | – | – | – | – | + |
| 541–12 | – | + | + | + | + | – | + | + | – | – | – | – | + |
| 541–13 | – | – | – | + | + | – | – | + | – | – | – | – | + |
| 541–14 | – | + | + | + | + | – | – | + | – | – | – | – | + |
| 541–15 | – | + | + | + | – | – | – | + | – | – | – | – | + |
| 541–16 | – | + | + | + | + | – | – | + | – | – | – | – | + |
| 541–17 | – | + | – | + | + | – | + | + | + | – | – | – | + |
| 541–18 | – | + | + | + | + | + | + | + | + | – | – | – | + |
| 541–19 | – | + | + | – | + | – | + | + | + | – | – | – | + |
| 541–20 | – | + | – | – | – | – | – | + | – | – | – | – | – |
| 541–21 | – | + | – | – | + | – | + | + | + | – | – | – | + |
| 541–22 | – | + | + | + | + | – | – | + | – | – | – | – | + |
| 541–23 | + | + | + | + | – | – | + | + | – | – | + | – | + |
| 541–24 | – | + | + | + | + | – | + | + | – | + | + | + | + |
| 541–25 | + | + | + | + | + | – | – | + | – | – | – | + | + |
| 541–26 | – | + | + | + | + | – | + | + | – | – | – | – | + |
| 541–27 | – | + | + | + | + | + | + | + | – | – | – | – | + |
| 541–28 | – | + | + | + | – | + | + | + | – | – | – | – | + |
| 541–29 | + | + | + | + | + | – | – | + | – | – | – | – | + |
| 541–30 | – | + | + | – | + | – | + | – | – | + | – | – | + |
| Total of positives | 3/30 | 28/30 | 24/30 | 24/30 | 25/30 | 4/30 | 18/30 | 28/30 | 4/30 | 2/30 | 3/30 | 2/30 | 29/30 |
| (10%) | 93.33%) | (80%) | (80%) | (83.33%) | (13.13%) | (60%) | (93.33%) | (13.33%) | (6.67%) | (10%) | (6.67%) | (96.67%) |
DNA Sequencing and Phylogenetic Tree
The sequences of CAstV from white chicks were edited and compared with other sequences from GenBank using the BLAST tool. The results showed high similarity with other CAstV. A fragment of approximately 360 bp from sequences of the CAstV from white chicks in the present work was compared with other sequences. The Brazilian sequences of CAstV obtained here revealed a high similarity to nucleotide (nt) (100 to 99.1%) and amino acid (aa) (100 to 98.3%) between the sequences. Comparisons with sequences from other countries showed a high similarity of nt (90.3 to 85.6%) and aa (97.5 to 95%) with sequences of the United States; a high similarity of nt (88.9 to 88.6%) and aa (96.6 to 95%) with sequences of the India West Zone; a high similarity of nt (89.7 to 88.1%) and aa (97.5 to 93.3%) with sequences of the India North Zone; a high similarity of nt (89.2 to 88.3%) and aa (97.5 to 95.8%) with sequences of the India South Zone; a high similarity of nt (86.7 to 85.3%) and aa (96.6 to 95.8%) with sequences of Italy; a high similarity of nt (87.8 to 87.2%) and aa (98.3 to 96.6%) with sequences of Croatia; a high similarity of nt (86.1 to 85.9%) and aa (94.1 to 93.3%) with sequences of Korea; and a high similarity of nt (90 to 89.7%) and aa (97.5 to 96.6%) with sequences of the United Kingdom. The phylogenetic tree clustered the Brazilian sequences in a separate group with a bootstrap value of 82% and 0.01451 substitutions per site. The sequences of other countries were clustered into another group, with a bootstrap value of 21% and 0.04038 substitutions per site. The sequences of CAstV from other countries clustered accordingly to their geographic origin, except for the sequence from the United Kingdom, which clustered with the sequences of the United States (Figure 3). The grouped sequences from other countries, which contained the sequences from India that belonged to the subgroup Bi of CAstV, were grouped according to capsid sequence diversity. No other sequences from other countries grouped with the Brazilian sequences obtained here.
DISCUSSION
Enteric diseases were reported in mammals, and several outbreaks of diarrhea, were associated with enteric viruses, such as astrovirus (Dai et al., 2010; De Benedictis et al., 2011; Zsak et al., 2013), rotavirus (Rajendran and Kang, 2014; Wu et al., 2014), and coronavirus (Costa et al., 2014; Pinto et al., 2014). Chickens and turkeys are also affected by enteric diseases, and outbreaks of enteric diseases have been described in several parts of the world (McNulty et al., 1980a; Pantin-Jackwood et al., 2008; Guy et al., 2011; Day and Zsak, 2013; Nuñez and Piantino Ferreira, 2013a). Viruses that are associated with mammalian enteric diseases, including ARtV (Spackman et al., 2010; Moura-Alvarez et al., 2013), AReoV (Davis et al., 2013), ChPV (Zsak et al., 2013; Nuñez et al., 2015b), and CAstV (Smyth et al., 2009). CAstV (Baxendale and Mebatsion, 2004), TAstV (Saif et al., 1985), and ANV (Imada et al., 1979; Imada et al., 2000) were identified in poultry, and all of these viruses are associated mainly with enteric problems. CAstV was reported in the United States (Pantin-Jackwood et al., 2008; Smyth et al., 2013), the United Kingdom (Todd et al., 2009a), Korea (Koo et al., 2013), Italy (Canelli et al., 2012), India (Bulbule et al., 2013), and Brazil (Mettifogo et al., 2014; Nuñez et al., 2015c).
The present work reported the presence of CAstV in chicks with white plumage and weakness that died within a few h of life (i.e., white chicks). The presence of this condition was recently reported by (Smyth et al., 2013), who showed that CAstV was involved in the presentation of this alteration in recently hatched chickens. The first report of white chicks showed that several countries in Europe and North America where experiencing this condition in chicken flocks (Smyth et al., 2013). Over the past several y, there have been many reports of the presence of white chicks in Brazilian hatcheries, and these chicks showed an increase in mortality and hatchability problems. The present study showed that CAstV is involved with this condition in chickens in Brazil.
The present study also suggests that CAstV is vertically transmitted and that the emergence of white chicks may not be a unique condition related with CAstV, but could also be associated with the occurrence of cases of enteric disease (diarrhea) in young chickens where CAstV has already been detected at one d of age (Mettifogo et al., 2014), which confirms the vertical transmission. The present work demonstrates that CAstV was distributed in several organs, principally in the digestive tract, gizzard, and intestine. However, there were fewer proventriculus positive samples. CAstV also was detected in the yolk sac remnant, which suggests that the virus originated there and began to develop in the intestine with continuous propagation in the gizzard. The virus may subsequently move to the proventriculus. However, difficulty in the isolation of CAstV hinders our understanding of the path that the virus uses for propagation in the embryonic organism (Nuñez et al., 2015c). Further, CAstV was described in the accessory glands, such as the pancreas and liver, and in the lymphoid organs, such as the thymus, bursa, and spleen. However, few liver samples were positive for CAstV, which contrasts a Smyth et al. (2013) report in which the liver exhibited more prevalence for CAstV. Moreover, in the present work, CAstV was detected in the brain of analyzed chicks. To the authors’ knowledge, there are no reports of CAstV in the brains of chickens. These results suggest that the viremia may reach several organs, including the brain.
Astrovirus in chickens was detected using transmission electronic microscopy in feces, where it was visualized as astrovirus-like particles (McNulty et al., 1980b; McNulty et al., 1990). Molecular tests were developed subsequently to detect CAstV with more accuracy (Pantin-Jackwood et al., 2006; Day et al., 2007; Smyth et al., 2009; Pantin-Jackwood et al., 2011). RT-PCR amplified a specific region of the ORF 1b gene of the CAstV genome in the present work (Day et al., 2007). The sequences obtained here from our chickens with the condition of white chicks showed a high similarity of nucleotides and amino acids with sequences from several parts of the world, which demonstrates that the CAstV related with white chicks in Brazilian flocks is molecularly related with other CAstV. However, the Brazilian CAstV was grouped into a separate cluster. Different molecular analyses showed that CAstV was classified into several groups: group A principally included sequences of the United Kingdom, and group B (Smyth et al., 2012) formed two subgroups, Bi and Bii. The sequences obtained in the present work were grouped into a unique group, and the other sequences were grouped with isolates from India that belonged to Subgroup Bi (Bulbule et al., 2013). However, a high similarity of nt and aa were shared with the Brazilian sequences, which suggests that the sequences obtained here may also belong to the Subgroup Bi. Moreover, the sequences obtained here were compared with sequences from groups A and Bii according to Bulbule et al. (2013), and the results showed a low similarity of nt and aa (data not shown). More studies must be performed to understand the molecular features of CAstV that are related to white chicks and determine whether the complete genome of this virus is completely related to the viruses detected in chickens with enteric diseases to determine its importance in the occurrence of enteric diseases.
CAstV is related principally with enteric disorders, such as RSS (Koci and Schultz-Cherry, 2002; Day et al., 2007; Pantin-Jackwood et al., 2007; Pantin-Jackwood et al., 2011). CAstV was also involved with an outbreak of gout (Bulbule et al., 2013), in which CAstV was related to chicken embryos with discolored feathers and mortality, which are features that are very similar to the characteristics of the chicks analyzed in the present work. Experimental infection with CAstV associated with gout in specific pathogen-free (SPF) chicken embryos also resulted in the stunting of embryos, necrosis of the liver, and pale and swollen kidneys (Bulbule et al., 2013). CAstV in the present work was detected in the kidneys and lungs, which demonstrates that these organs are another target for virus replication.
CAstV is circulating in Brazilian chicken flocks, and the actual data show the presence of CAstV in the kidneys of analyzed chicks. These results suggest that CAstV, in addition to causing enteric diseases, may be causing renal alterations, but this hypothesis requires further investigation to determine whether these factors are related also to gout.
Normally, broiler chicks hatch with yellow plumage, and the color is due to a genetic effect (Park et al., 2013) and carotenoid pigments (Perez-Vendrell et al., 2001). Carotenoid pigments are common colorants of egg yolk, feathers, and bare parts (such as the beak and legs) in birds. Carotenoids are supplied in the diet, and they are usually absorbed in different sections of the intestine, principally the ileum. The absorption of lutein occurs in the duodenum and jejunum (Tyczkowski and Hamilton, 1986). White chicks exhibit a white plumage at hatching, from which the name was derived. Hypothetically, this unusual white plumage pigmentation may be due to a lack of carotenoid accumulation and a decrease in carotenoid (lutein) absorption provoked by CAstV replication in the intestinal mucosa.
Enteric diseases are a very significant health problem in the poultry industry. Over the last several years, several outbreaks of enteric diseases were reported in some states of Brazil, including primary agents, such as CAstV, ChPV, FAdV-1, ARtV, and AReoV (Mettifogo et al., 2014). A compromised enteric tract is a concern for the poultry industry because of the low performance, high production costs, and increase in the use of therapeutic drugs that are required to reduce the impact of associated pathogens.
Detection of CAstV in chicks with clinical signs of white chicks in the present study confirmed that the virus is circulating in Brazilian chicken flocks and that it may be vertically transmitted and causing hatchability problems. The molecular analyses showed that the Brazilian CAstV is similar to CAstV around the world. Further studies must be performed to understand the role of this virus in chicken health.
Acknowledgments
The authors would like to thank the Poultry companies, in Brazil that gently sent the samples for the development of this study and for diagnosis of enteric viruses. This work was supported by grants from CNPq (Conselho Nacional de Pesquisa e Desenvolvimento Tecnológico – grant no. 453920/2014–4) and FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo – grant no 2013/08560–5). Also, A. J. Piantino Ferreira is a recipient of CNPq fellowships.
REFERENCES
- Baxendale W., Mebatsion T. The isolation and characterization of astroviruses from chickens. Avian Pathol. 2004;33:364–370. doi: 10.1080/0307945042000220426. [DOI] [PubMed] [Google Scholar]
- Bulbule N.R., Mandakhalikar K.D., Kapgate S.S., Deshmukh V.V., Schat K.A., Chawak M.M. Role of chicken astrovirus as a causative agent of gout in commercial broilers in India. Avian Pathol. 2013;42:464–473. doi: 10.1080/03079457.2013.828194. [DOI] [PubMed] [Google Scholar]
- Canelli E., Cordioli P., Barbieri I., Catella A., Pennelli D., Ceruti R., Moreno A., Lavazza A. Astroviruses as causative agents of poultry enteritis: genetic characterization and longitudinal studies on field conditions. Avian Dis. 2012;56:173–182. doi: 10.1637/9831-061311-Reg.1. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Welchman D.D.B., Britton P., Gough R.E. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 2002;31:81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- Costa E.M., de Castro T.X., Bottino F.O., Garcia R.C. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet. Microbiol. 2014;168:8–15. doi: 10.1016/j.vetmic.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Xu Q., Wu X., Hu G., Tang Y., Li J., Chen Q., Nie J. Development of real-time and nested RT-PCR to detect astrovirus and one-year survey of astrovirus in Jiangmen City, China. Arch. Virol. 2010;155:977–982. doi: 10.1007/s00705-010-0664-6. [DOI] [PubMed] [Google Scholar]
- Davis J.F., Kulkarni A., Fletcher O. Reovirus Infections in Young Broiler Chickens. Avian Dis. 2013;57:321–325. doi: 10.1637/10515-021313-Case.1. [DOI] [PubMed] [Google Scholar]
- Day J.M., Spackman E., Pantin-Jackwood M.J. A multiplex RT-PCR test for the differential identification of turkey astrovirus type 1, turkey astrovirus type 2, chicken astrovirus, avian nephritis virus, and avian rotavirus. Avian Dis. 2007;51:681–684. doi: 10.1637/0005-2086(2007)51[681:AMRTFT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Day J.M., Zsak L. Recent progress in the characterization of avian enteric viruses. Avian Dis. 2013;57:573–780. doi: 10.1637/10390-092712-Review.1. [DOI] [PubMed] [Google Scholar]
- De Benedictis P., Schultz-Cherry S., Burnham A., Cattoli G. Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit J.J., ten Dam G.B.M., van de Laar J.M.A., Biermann Y., Verstegen I., Edens F., Schrier C.C. Detection and characterization of a new astrovirus in chicken and turkeys with enteric and locomotion disorders. Avian Pathol. 2011;40:453–561. doi: 10.1080/03079457.2011.596813. [DOI] [PubMed] [Google Scholar]
- Goodwin M.A., Davis J.F., McNulty M.S., Brown J., Player E.C. Enteritis (so-called runting stunting syndrome) in Georgia broiler chicks. Avian Dis. 1993;37:451–458. [PubMed] [Google Scholar]
- Guy J.S., Barnes H.J., Smith L.G., Breslin J. Antigenic characterization of a turkey coronavirus identified in poult enteritis- and mortality syndrome-affected turkeys. Avian Dis. 2011;41:583–590. [PubMed] [Google Scholar]
- Imada T., Yamaguchi S., Kawamura H. Pathogenicity for baby chicks of the G-4260 strain of the picornavirus “avian nephritis virus”. Avian Dis. 1979;23:582–588. [PubMed] [Google Scholar]
- Imada T., Yamaguchi S., Mase M., Tsukamoto K., Kubo M., Morooka A. Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA. J. Virol. 2000;74:8487–8493. doi: 10.1128/jvi.74.18.8487-8493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal N., Chander Y., Patnayak D.P., Mor S.K., Ziegler A.F., Goyal S.M. A multiplex RT-PCR for the detection of astrovirus, rotavirus, and reovirus in turkeys. Avian Dis. 2012;56:592–596. doi: 10.1637/9958-100911-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Kang K.I., Icard A.L., Linnemann E., Sellers H.S., Mundt E. Determination of the full length sequence of a chicken astrovirus suggests a different replication mechanism. Virus Genes. 2012;44:45–50. doi: 10.1007/s11262-011-0663-z. [DOI] [PubMed] [Google Scholar]
- Kang K.I., El-Gazzar M., Sellers H.S., Dorea F., Williams S.M., Kim T., Collet S., Egbert M. Investigation into the aetiology of runting and stunting syndrome in chickens. Avian Pathol. 2012;41:41–50. doi: 10.1080/03079457.2011.632402. [DOI] [PubMed] [Google Scholar]
- Koci M.D., Schultz-Cherry S. Avian astroviruses. Avian Pathol. 2002;31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- Koo B.S., Lee H.R., Jeon E.O., Jang H.S., Han M.S., Mo I.P. An unusual case of concomitant infection with chicken astrovirus and group A avian rotavirus in broilers with a history of severe clinical signs. J. Vet. Sci. 2013;14:231–233. doi: 10.4142/jvs.2013.14.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty M.S., Curran W., McFerran J.B. Detection of astroviruses in turkey faeces by direct electron microscopy. Vet. Rec. 1980;106:561. doi: 10.1136/vr.106.26.561. [DOI] [PubMed] [Google Scholar]
- McNulty M.S., Allan G.M., Todd D., McFerran J.B., McKillop E.R., Collins S.D., McCracken R.M. Isolation of rotaviruses from turkeys and chickens: demonstration of distinct serotypes and RNA electropherotypes. Avian Pathol. 1980;9:363–375. doi: 10.1080/03079458008418420. [DOI] [PubMed] [Google Scholar]
- McNulty M.S., Connor T.J., McNeilly F., McFerran J.B. Biological characterization of avian enteroviruses and enterovirus-like viruses. Avian Pathol. 1990;19:75–87. doi: 10.1080/03079459008418658. [DOI] [PubMed] [Google Scholar]
- Mettifogo E., Nuñez L.F., Chacón J.L., Santader-Parra S.H., Astolfi-Ferreira C.S., Jerez J.A., Jones R.C., Ferreira A.J.P. Emergence of enteric viruses in production chickens is a concern for avian health. ScientificWorldJournal. 2014;2014:450423. doi: 10.1155/2014/450423. doi: 10.1155/2014/450423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans G., Boschmans M., Berg V.D., Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 2001;30:655–660. doi: 10.1080/03079450120092143. [DOI] [PubMed] [Google Scholar]
- Moser L.A., Schultz-Cherry S. Pathogenesis of astrovirus infection. Viral Immunol. 2005;18:4–10. doi: 10.1089/vim.2005.18.4. [DOI] [PubMed] [Google Scholar]
- Moura-Alvarez J., Chacon J.L., Scanavini L.S., Nuñez L.F., Astolfi-Ferreira C.A., Jones R.C., Piantino Ferreira A.J. Enteric viruses in Brazilian turkey flocks: Single and multiple virus infection frequency according to age and clinical signs of intestinal disease. Poult. Sci. 2013;92:945–955. doi: 10.3382/ps.2012-02849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez L.F., Piantino Ferreira A.J. Viral agents related to enteric disease in commercial chicken flocks, with special reference to Latin America. World's Poult. Sci. J. 2013;69:853–864. [Google Scholar]
- Nuñez L.F., Santander-Parra S.H., Mettifogo E., Astolfi-Ferreira C.S., Ferreira A.J.P. Isolation and molecular characterization of chicken parvovirus from Brazilian flocks with enteric disorders. Brit. Poult. Sci. 2015;56:39–47. doi: 10.1080/00071668.2014.981797. [DOI] [PubMed] [Google Scholar]
- Nuñez L.F., Santander-Parra S.H., Mettifogo E., Astolfi-Ferreira C.S., Ferreira A.J.P. Isolation of Chicken Astrovirus from specific páthogen-free chicken embryonated eggs. Poult. Sci. 2015;56:455–462. doi: 10.3382/ps/pev086. [DOI] [PubMed] [Google Scholar]
- Olsen D. E. Isolation of a reovirus-like agent from broiler chicks with diarrhea and stunting Proc. 26th Western Poult. Dis. Conf. 1977 131 139
- Otto P., Liebler-Tenorio E.M., Elschner M., Reetz J., Löhren U., Diller R. Detection of rotaviruses and intestinal lesions in broiler chicks from flocks with runting and stunting syndrome (RSS) Avian Dis. 2006;50:411–418. doi: 10.1637/7511-020106R.1. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Day J.M., Jackwood M.W., Spackman E. Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis. 2008;52:235–244. doi: 10.1637/8174-111507-Reg.1. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Spackman E., Day J.M., Rives D. Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms. Avian Dis. 2007;51:674–680. doi: 10.1637/0005-2086(2007)51[674:PMOCTF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Spackman E., Woolcock P.R. Molecular characterization and typing of chicken and turkey astroviruses circulating in the United States: implications for diagnostics. Avian Dis. 2006;50:397–404. doi: 10.1637/7512-020606R.1. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Strother K.O., Mundt E., Zsak L., Day J.M., Spackman E. Molecular characterization of avian astroviruses. Arch. Virol. 2011;156:235–244. doi: 10.1007/s00705-010-0849-z. [DOI] [PubMed] [Google Scholar]
- Park M.N., Choi J.A., Lee K., Lee H., Choi B., Kim H., Kim T., Cho S., Lee T. Genome-wide Association Study of Chicken Plumage Pigmentation. Asian-Australas. J. Anim. Sci. 2013;26:1523–1528. doi: 10.5713/ajas.2013.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vendrell A.M., Hernandez J.M., Llaurado L., Schierle J., Brufau J. Influence of source and ratio of xanthophyll pigments on broiler chicken pigmentation and performance. Poult. Sci. 2001;80:320–326. doi: 10.1093/ps/80.3.320. [DOI] [PubMed] [Google Scholar]
- Pinto L.D., Barros I.N., Budaszewski R.F., Weber M.N., Mata H., Antunes J.R., Boabaid F.M., Wouters A.T.B., Driemeier D., Brandao P.E., Canal C.W. Characterization of pantropic canine coronavirus from Brazil. Vet. J. 2014;202:659–662. doi: 10.1016/j.tvjl.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran P., Kang G. Molecular epidemiology of rotavirus in children and animals and characterization of an unusual G10P[15] strain associated with bovine diarrhea in south India. Vaccine. 2014;32(Suppl 1):A89–94. doi: 10.1016/j.vaccine.2014.03.026. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Saif Y.M., Theil K.W. Enteric viruses in diarrheic turkey poults. Avian Dis. 1985;29:798–811. [PubMed] [Google Scholar]
- Smyth V.J., Jewhurst H.L., Adair B.M., Todd D. Detection of chicken astrovirus by reverse transcriptase-polymerase chain reaction. Avian Pathol. 2009;38:293–299. doi: 10.1080/03079450903055397. [DOI] [PubMed] [Google Scholar]
- Smyth V.J., Jewhurst H.L., Wilkinson D.S., Adair B.M., Gordon A.W., Todd D. Development and evaluation of real-time TaqMan® RT-PCR assays for the detection of avian nephritis virus and chicken astrovirus in chickens. Avian Pathol. 2010;39:467–474. doi: 10.1080/03079457.2010.516387. [DOI] [PubMed] [Google Scholar]
- Smyth V.J., Todd D., Trudgett J., Lee A., Welsh M.D. Capsid protein sequence diversity of chicken astrovirus. Avian Pathol. 2012;41:151–159. doi: 10.1080/03079457.2011.652938. [DOI] [PubMed] [Google Scholar]
- Smyth V.J., Trudgett J., Wylie M. Chicken astrovirus detected in hatchability problems associated with “white chicks”. Vet. Rec. 2013;173:403–404. doi: 10.1136/vr.f6393. [DOI] [PubMed] [Google Scholar]
- Spackman E., Day J.M., Pantin-Jackwood M.J. Astrovirus, reovirus, and rotavirus concomitant infection causes decreased weight gain in broad-breasted white poults. Avian Dis. 2010;54:16–21. doi: 10.1637/8986-070909-Reg.1. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D., Smyth V.J., Ball N.M., Donnelly B.M., Wylie M., Knowles N.J., Adair B.M. Identification of chicken enterovirus-like viruses, duck hepatitis virus type 2 and duck hepatitis virus type 3 as astroviruses. Avian Pathol. 2009;38:21–29. doi: 10.1080/03079450802632056. [DOI] [PubMed] [Google Scholar]
- Todd D., Wilkinson D.S., Jewhurst H.L., Wylie M., Gordon A.W., Adair B.M. A seroprevalence investigation of chicken astrovirus infections. Avian Pathol. 2009;38:301–309. doi: 10.1080/03079450903055421. [DOI] [PubMed] [Google Scholar]
- Tyczkowski J., Hamilton P.B. Absorption, transport, and deposition in chickens of lutein diester, a carotenoid extracted from marigold (Tagetes erecta) petals. Poult. Sci. 1986;65:1526–1531. doi: 10.3382/ps.0651526. [DOI] [PubMed] [Google Scholar]
- Wu F.T., Bányai K., Jiang B., Wu C.Y., Chen H.C., Fehér E., Huang Y.C., Lin J.S., Huang F.C., Hsing C.A., Huang J.C., Wu H.S. Molecular epidemiology of human G2P[4] rotaviruses in Taiwan, 2004–2011. Infect. Genet. Evol. 2014;28:530–536. doi: 10.1016/j.meegid.2014.09.033. [DOI] [PubMed] [Google Scholar]
- Zsak L., Cha R.M., Day J.M., Cha A.R.M. Chicken parvovirus-induced runting-stunting syndrome in young broilers. Avian Dis. 2013;57:123–127. doi: 10.1637/10371-091212-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Zsak L., Strother K.O., Day J.M., Strother A.K.O. Development of a polymerase chain reaction procedure for detection of chicken and turkey parvoviruses. Avian Dis. 2009;53:83–88. doi: 10.1637/8464-090308-Reg.1. [DOI] [PubMed] [Google Scholar]


