ABSTRACT
A total of 50 poultry farms of commercial broilers (N = 39) and commercial layers (N = 11) suffered from respiratory problems and mortality during the period from January 2016 to December 2017 were investigated. Also, samples were collected from quail (N = 4), Bluebird (Sialis, N = 1), and Greenfinch (Chloris chloris, N = 1) for analysis. Respiratory viral pathogens were screened by PCR and positive samples were subjected to virus isolation and genetic identification. Antigenic relatedness of isolated avian influenza (AI) H5 subtype was evaluated using cross-hemagglutination inhibition. Results revealed that the incidence of single virus infections in commercial broilers was 64.1% (25/39), with the highest incidence for ND (33.3%) and H9N2 (20.5%), followed by H5N1 (7.7%) and H5N8 (2.7). Meanwhile, H9N2/ND mixed infection was the most observed case (7.7%). Other mixed infections H5N1/ND, H5N1/H9N2/ND, H5N1/H9N2/ND/IB, H9N2/IB, and H9N2/ILT were also observed (2.6% each). In commercial layers, H5N1 and ILT were the only detected single infections (18.1% each). Mixed H9N2/ND was the most predominant infection in layers (27.3%). Other mixed infections of H9N2/IB, H5N1/H5N8/H9N2, and H9N2/ND/IB were observed in 3 separate farms (9.1% each). The H5N8 virus was detected in one quail farm and 2 out of 3 wild bird's samples. Partial HA gene sequence analysis showed the clustering of the selected AI H5N8 within the 2.3.4.4 clade, while H5N1 clustered with the clade 2.2.1.2. Interestingly, the H5N8 isolated from chickens possessed 6 amino acids substitutions at HA1 compared to those isolated from wild birds with low antigenic relatedness to AI H5N1 clades 2.2.1 or 2.2.1.2. In conclusion, mixed viral infections were observed in both broiler and layer chickens in Egypt. The detected triple H5N1, H9N2, and H5N8 influenza co-infection raises the concern of potential AI epidemic strain emergence. The low genetic and antigenic relatedness between AI H5N1 and H5N8 viruses suggest the need for modification of vaccination strategies of avian influenza in Egypt along with strict biosecurity measures.
Keywords: AIV, H5N1, H5N8, co-infection, egypt
INTRODUCTION
Respiratory disease outbreaks in commercial chicken flocks are among the most serious challenges in the poultry industry in Egypt. The etiology of the respiratory disease is multifactorial, often associated with mixed infection by different pathogens, where an interaction between these pathogens and/or commensal bacteria may determine the severity of the disease (Samy and Naguib, 2018). There is a wide range of viruses that cause respiratory infections in Egyptian poultry farms, including avian influenza virus subtypes H5N1, H9N2, and H5N8, infectious bronchitis virus (IBV), Newcastle disease virus (NDV), and infectious laryngotracheitis (ILT).
The highly pathogenic avian influenza (HPAI) H5N1 was first reported in Egypt in 2006 and declared to be enzootic in 2008 (Aly et al., 2008; Peyre et al., 2009). During 2010 and 2011, the low pathogenic avian influenza (LPAI) H9N2 was also reported in chickens and quail, which added a new challenge for poultry industry (El-Zoghby et al., 2012). Recently, the HPAI H5N8 was isolated from common-coot in Egypt during December 2016 (Selim et al., 2017) and multiple introductions of reassorted H5N8 viruses in wild and domestic birds were reported (Anis et al., 2018; Salaheldin et al., 2018; Yehia et al., 2018).
Since ND was identified for the first time in 1948 in Egypt (Daubney and Mansy, 1948), ND genotypes II, VI, and VII have been frequently reported in Egypt in vaccinated and non-vaccinated birds resulting in severe economic losses due to severe respiratory manifestations and high mortality rates (Selim et al., 2018). On the hand, IBV has been reported in Egypt in the 1950s and outbreaks continued in Egyptian flocks despite vaccination with the Massachusetts (Mass) strains. This is possibly due to the emergence of new Egyptian variants, which are closely related to the Israeli variant 2 (Abdel-Moneim et al., 2006). More recently, Egyptian variant II, which is closely related to Israeli variant 1 was reported (Abdel-Moneim et al., 2012; Hosseini et al., 2015) and the continued evolution of IBV field strains lead to outbreaks in vaccinated chicken populations (Abdel-Sabour et al., 2017). Currently, IBV outbreaks are associated with respiratory distress and nephropathy with high mortalities due to the circulation of nephropathogenic IBV variant II strains (Ali et al., 2018).
The co-circulation of H5N1, H9N2, and H5N8 might play a potential role in the genesis of strains that could present a real threat for both poultry industry and human health. Co-infection with pathogens causing respiratory problems has been observed before in many natural cases, e.g., LPAI H9N2 with HPAIV H5N1 in China, Israel, Bangladesh, and Egypt (Arafa et al., 2012; Hassan et al., 2016), IBV co-infection with HPAIV and/or LPAIV (Roussan et al., 2008; Hassan et al., 2016), and avian influenza with Staphylococcus spp. (Tashiro et al., 1987). Mixed viral infections were studied under experimental conditions in chickens such as NDV and LPAI (Costa-Hurtado et al., 2014; Costa-Hurtado et al., 2016) and IBV and LPAI in chickens (Hassan et al., 2017) and found to aggravate the disease condition. Though avian influenza viruses H5N1 and H9N2 subtypes are circulating in different poultry production sectors in Egypt for almost 7 yr, no reassortant evidence has been reported. However, a recent study demonstrated the genetic compatibility of both viruses to produce new reassortant viruses that showed variable pathogenicity with no zoonotic potential in the ferret model (Naguib et al., 2017).
The aim of the present study was to investigate the viral etiologies of the respiratory syndrome in Egypt, namely, H5N1, H5N8, NDV, IBV, and ILT during the period from 2016 to 2017. Additionally, the genetic and antigenic relatedness of the isolated AI H5N8 viruses to their ancestors and the circulating H5N1 viruses were studied.
MATERIAL AND METHODS
Clinical Samples
Samples (trachea, tracheal swabs and oropharyngeal swabs) were collected during the period between January 2016 to December 2017 from commercial chicken farms (n = 50), and quail farms (n = 3). Samples from wild birds included tracheas from bluebird (n = 2) and Greenfinch (n = 1). Samples were allocated in 9 Egyptian Governorates, namely, Sharkia, Qalubia, Beni-Suef, Giza, Behaira, Damietta, Port Said, Mattroh, and Aswan. Available history of investigated farms, including bird type, number, mortality rates, and previous vaccination regimes were recorded (Supplementary Table S1). Postmortem lesions examination was performed on dead birds. Samples were processed for the isolation and identification of causative agents and aliquots were frozen at −80° C for further analysis.
Molecular Detection of Avian Respiratory Viruses
Viral genome was extracted using RNA/DNA extraction kit (Qiagen, Germany) according to the manufacturer's instructions. All samples were tested using specific oligonucleotide primers for avian influenza subtypes H5, H9, NDV, IBV, and ILT (Table 1). For avian influenza, samples were tested by RT- PCR using IAV-matrix (M) gene as described by (Zakia et al., 2013). Positive influenza A samples were subtyped using standard RT- PCR and H5, H9, and N8 gene-specific primers (Table 1). The PCR products were analyzed by electrophoresis using 1.5% w/v agarose gel and representative materials were purified and used for sequencing. The obtained HA gene-specific PCR products were sequenced at the Virus Gene Analysis Unit of the Reference Laboratory for Quality Control of Poultry Production “RLQP”, Dokki, Giza, Egypt, using BigDye Terminator v3.1 cycle sequencing kit. Sequences were edited using EditSeq (DNASTAR Inc., Madison, Wisconsin).
Table 1.
Oligonucleotide primers used for detection of avian respiratory viral pathogens.
| Virus | Targeting gene | Primer | Size bp | Reference |
|---|---|---|---|---|
| Avian influenza | Matrix | M-F TGA TCT TCT TGA AAA TTT GCA | 270 | (Zakia et al., 2013) |
| M-R TGT TGA CCA AAT GAC CCT CG | ||||
| H5 | H5-F GAT TGT AGT GTA GCY GGA TGG | 400 | (Hussein et al., 2015) | |
| H5-R CTT GTC TGC TCT KCM KCA TC | ||||
| H9 | H9-F CTY CAC ACA GAR CAC AAT GG | 500 | (Lee et al., 2014) | |
| H9-R GTC ACA CTT GTT GTT GTR TC | ||||
| N8 | N8F-ATA GTG ACC GTT GGC TCC ATT TCA | 600 | This study | |
| N8R- TGT CTA TTG GCT GGA GCG TCA GTC AT | ||||
| H5 HA1 | UNI-RG-5 TAT TCG TCT CAG GGA GCG AAA GCA GG | 1123 | (Lee and Suarez, 2008) | |
| EGY-H5-R: GCT CGT TGC TAT GGT GGT AC | (Shany, 2015) | |||
| NDV | HA | NDV-F ATG GGC TCC AAA CCT TCT ACC AGG | 1500 | This study |
| NDV-R ATG CTC TCG TGG TGG CTC TCA TCT GAT | ||||
| IBV | S1 | XCE1-CAC TGG TAA TTT TTC TGA TGG | 464 | (Cavanagh et al., 1999) |
| XCE1-CTC TAT AAA CAC CCT TAC A | ||||
| ILT | ICP4 | ICP4-2F- CTT CAG ACT CCA GCT CAT CTG | 636 | (Chacon and Ferreira, 2009) |
| ICP4-2R- AGT CAT GCG TCT ATG GCG TTG AC |
Virus Isolation and Identification
Tracheal swabs were processed and prepared as 10% suspension using sterile phosphate-buffered saline containing penicillin (1000 IU/ml) and streptomycin (100 μg/ml). Homogenates were clarified by centrifugation at 2000 rpm for 10 min at 4°C. Supernatants were filtered using a bacteriological filter (0.45 um, Millipore) and inoculated into the allantoic cavity of 10-day-old specific pathogens free embryonated chicken eggs “SPF-ECE” (Nile SPF company, Fayoum, Egypt). Inoculated eggs were candled daily for embryo viability. At 72 h post-inoculation, eggs were chilled and the allantoic fluid was collected and tested by the hemagglutination (HA) assay (OIE, 2015). HA positive samples were further identified by the hemagglutination inhibition (HI) assay using specific hyperimmune sera against ND, H9, and H5 (OIE, 2015).
Amplification and Sequencing of the HA1 of H5N8 Viruses
The UNI-RG-5 and EGY-H5-R primers (Table 1) were used for amplification of HA1 of the 3 isolated H5N8 viruses. The obtained gene-specific PCR products were sequenced at Macrogen Co (Seoul, Korea). Sequences were edited using EditSeq (DNASTAR Inc., Madison, Wisconsin). The sequences were aligned with additional virus sequences retrieved from the GenBank for phylogenetic analysis using the neighbor-joining method, the Tamura–Nei model, and 1000 bootstrap replicates implemented in MEGA7.0 (Tamura et al., 2013).
Cross-reactivity of the Isolated H5N8 Viruses with Different Clade H5N1 Viruses
Viruses
To evaluate the antigenic relatedness of the H5N8 viruses isolated in the current study to HP H5N1 viruses, 6 H5N1 viruses representing 2.2.1, 2.2.1.1, and 2.2.1.2 clades, namely, A/chicken/Egypt/083-NLQP/2008 (acc. no. JF746737), A/chicken/Egypt/086Q-NLQP/2008 (acc. no. EU496398), A/chicken/Egypt/Q1995D/2010 (acc. no. CY099579), A/duck/Egypt/M2583D/2010 (acc. no. CY099580), A/chicken/Egypt/M7217B/2013 (acc. no. KF881625), and A/chicken/Egypt/A10542E/2015 (acc. no. KU715949) were used for the preparation of specific antisera.
Preparation of Hyperimmune Sera
Selected viruses were propagated and titrated in specific pathogen free (SPF) embryonated chicken eggs then suspensions were inactivated using 0.02% formalin (Sigma Chemical Co., St. Louis, MO) for 36 h at 37°C. Complete inactivation was assured by passaging the inactivated suspensions in SPF-ECE for 3 successive passages via the allantoic sac route. The hemagglutination activities were calculated and 350 HAU/dose were then mixed with MontanideTM ISA 70 VG adjuvant (SEPPIC SA, Puteaux, France) at a ratio of 30:70 (w/w) at 3000 rpm using the Silverson L5M high-shear laboratory mixer (Silverson Machines, Inc., Buckinghamshire, United Kingdom).
For preparation of hyperimmune sera, White Leghorn SPF chickens were purchased from Nile SPF company (Kom–Osheim, Fayoum, Egypt). Experimental procedures were reviewed and approved by the Animal Care and Use Committee (ACUC#180215E014) of the Middle East for Veterinary Vaccines (ME VAC) Company, Egypt. A total of five 3-week-old SPF chickens/virus were inoculated with 350 HA unit/0.5 ml with adjuvant subcutaneously. The antisera were collected at 3 wk post-inoculation and tested for antibodies using HI test.
Hemagglutination Inhibition Test and Antigenic Relatedness
The cross-HI test was performed according to the standard protocol (OIE, 2015). The HI antibody titers were expressed as a reciprocal of the highest serum dilution that had complete inhibition of hemagglutination. The antigenic relatedness between H5N8 and H5N1 viruses was expressed as R-value using cross-HI titers based on the Archetti and Horsfall formula (R = 100 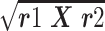 , where, r1 = titer of strain A with antiserum B/titer of strain A with antiserum A, r2 = titer of strain B with antiserum A/titer of strain B with antiserum B) (Archetti and Horsfall, 1950).
, where, r1 = titer of strain A with antiserum B/titer of strain A with antiserum A, r2 = titer of strain B with antiserum A/titer of strain B with antiserum B) (Archetti and Horsfall, 1950).
RESULTS
Clinical Findings of the Examined Flocks
In the present study, investigated chicken farm's size ranged between 5000 and 180 000 bird/farm. The vaccination schedule consisted of inactivated H9N2 & H5N1 vaccines at the age of 5 and 9 d, respectively. For NDV, live attenuated vaccines were administered at day-old then every 10 to 12 d. Live attenuated IBV vaccines were applied at day-old. Clinical manifestations in broiler farms (n = 39) involved respiratory distress in the form of coughing, sneezing, rales, and nasal discharge. The most common postmortem lesions were hemorrhagic tracheitis; tracheal caseation and generalized congestion of visceral organs. In some cases, nervous manifestations were observed in the form of tremors of the neck and torticollis. Mortality rates in commercial broiler and cross-breeds “Sasso breed” farms varied from 2 to 40% at the age of 16 to 36 d old. In commercial layers (n = 11), the clinical disease was more severe and mortality rates ranged between 2.4 and 30%, and were mainly observed at the young age (18 to 70 d old) (Supplementary Table S1).
Detection of Avian Respiratory Viruses
As shown in Table 2, out of 39 commercial broiler flocks, 25 (64.1%) flocks suffered from single viral respiratory infection while the mixed infection was reported in 8 flocks (20.5%). None of the tested viruses was detected in 6 flocks (15.4%). The incidence of single infections in commercial broilers with ND, H9N2, H5N1, and H5N8 were 13/39 (33.3%), 8/39 (20.5%), 3/39 (7.7%), and 1/39 (2.6%), respectively. Notably, none of the tested farms showed a single IB infection. Meanwhile, H9N2 and ND mixed infections were the most observed cases [3/39 (7.7%)]. Other mixed infections reported in commercial broilers included H5N1/ND, H5N1/H9N2/ND, H5N1/H9N2/ND/IB, H9N2/IB, H9N2/ILT, [1/39 (2.6%) each]. H5N8 was not detected in mixed infections in any of the tested commercial broiler farms.
Table 2.
Prevalence of avian respiratory viruses in commercial broilers and layer chicken farms in Egypt (2016–2017).
| No. of positive flocks (%) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single infection | Mixed | ||||||||||||||
| Governorate | Production type | H5N1 | H5N8 | H9N2 | ND | ILT | H5N1-ND | H9N2-IB | H9N2-ILT | H9N2-ND | H5N1/H9N2- ND | H5N1/H9N-H5N8 | H9N2-ND-IB | H5N1-H9N2 –ND-IB | Negative |
| Sharkia | Broiler N = 17 | 1 | 0 | 4 | 5 | 0 | 1 | 1 | 0 | 3 | 1 | 0 | 0 | 0 | 1 |
| Layers N = 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | |
| Beni-Suef | Broiler N = 5 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Layers N = 3 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Giza | Broiler N = 6 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| Layers N = 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Behaira | Broiler N = 10 | 1 | 0 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Layers N = 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| Qalubia | Broiler N = 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Layers N = 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | Broilers N = 39 (%) | 3 (7.7) | 1 (2.6) | 8 (20.5) | 13 (33.3) | 0 | 1 (2.6) | 1 (2.6) | 1 (2.6) | 3 (7.7) | 1 (2.6) | 0 | 0 | 1 (2.6) | 6 |
| Layers N = 11 (%) | 2 (18.1) | 0 | 0 | 0 | 2 (18.1) | 0 | 1 (9.1) | 0 | 3 (27.3) | 0 | 1 (9.1) | 1 (9.1) | 0 | 1 | |
In commercial layers, out of 11 commercial layer flocks, 4 (36.4%) flocks were suffered from single viral respiratory infection while mixed infections were predominant in most of the tested farms [6 flocks (54.5%)]. H5N1 and ILT were the only detected single infections in commercial layers [1/11 (18.1%) each]. Double mixed infections with H9N2/ND and H9N2/IB were reported in (3/11) and 1/11 farms, respectively. Triple infection was observed also in 2 separate layer farms (H5N1/H5N8/H9N2 and H9N2/ND/IB infection), 9.1% each. Of samples collected during 2016, H5N8 virus was detected in wild birds only [quail (1/4), bluebird (1/1) and Greenfinch (1/2)], but in 2017 the virus was detected in both broiler and layer farms.
Virus Isolation and Identification from Single Infection Samples
Virus isolation was only attempted from single infection samples and resulted in the recovery of the following viruses; H5N1 (N = 3), H5N8 (N = 4), H9N2 (N = 8), ILTV (N = 2), and NDV (N = 13). The identity and purity of the isolated viruses were confirmed using PCR. The sequence of 3 selected H9N2 viruses exhibited an ancestor relationship to the low pathogenic H9N2 Quail/HK/G1/1997 prototype without significant genetic changes (Data not shown). Also, partial genetic analysis of NDV isolates revealed that all isolates (N = 13) belonged to genotype VIId (data not shown).
Molecular Characterization of HA Genes of Isolated H5N1 and H5N8 Strains
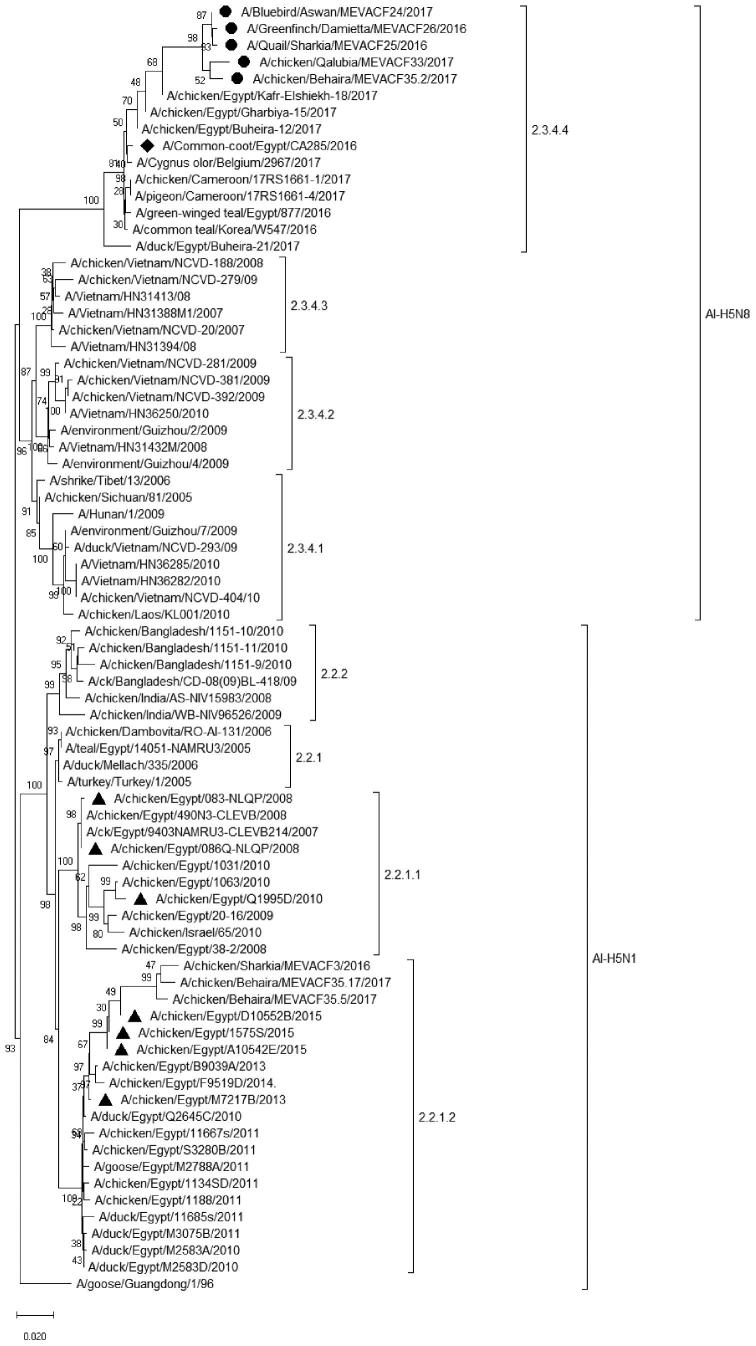
Avian influenza viruses H5N1 (N = 3) and H9N2 (N = 3) were further characterized by partial HA gene sequence analysis. The isolated H5N1 viruses; A/chicken/Sharkia/MEVACF3/2016, A/chicken/Behaira/MEVACF35.5/2017, and A/chicken/Behaira/MEVACF35.17/2017 (acc. No. MH349009- MH349011) belon-ged to the predominant 2.2.1.2 clade in Egypt (Figure 1). Sequence analysis of H5N8 viruses revealed that the A/chicken/Behaira/MEVACF35.2/2017 and A/Greenfinch/Damietta/MEVACF26/2016 A/chicken/Qalubia/MEVACF33/2017, A/Quail/Sharkia/MEVACF25/2016, A/Bluebird/Aswan/MEVACF24/2017, (access. no. MH349012- MH349016) had clustered within the 2.3.4.4 clade.
Figure 1.
Phylogenetic analysis of isolated H5N1 and H5N8. The tree was generated using neighbor-joining method, the Tamura-Nei model, and 1000 bootstrap replicates implemented in MEGA7.0. H5N8 (circle) were clustered with 2.3.4.4 viruses however, new H5N1 isolates (square) clustered with 2.2.1.2 clades. The viruses used for antigenic relatedness studies are indicated (triangle). The first isolated H5N8 virus is indicated (Diamond).
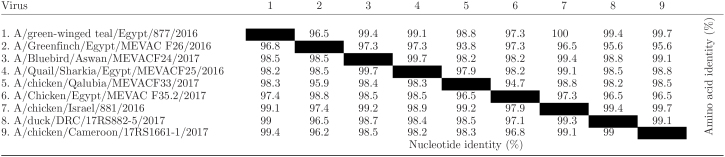
Further sequence analysis of HA1 of A/Bluebird/Aswan/MEVACF24/2017, A/chicken/Qalubia/MEVACF33/2017, and A/Quail/Sharkia/MEVACF25/2016 strains revealed that these viruses had the typical multi-basic PLREKRRK-R*GLF cleavage site (aa 321–332, H5 numbering). The receptor binding pocket of the HA1 molecule had amino acid residues 222Q and 224 G, according to H5 numbering, (Table 4). Analysis of potential glycosylation sites (N-XT/S motif in which X can be any amino acid except proline) of HA1 of current H5N8 viruses revealed the presence of 4 potential N-linked glycosylation sites [positions 10 (NNST), 23 (NVTV), 165 (NNTN), and 286 (NSSM)]. The sequences of antigenic sites of H5N8 viruses were analyzed, site A contains A133, T140, and 141P, site B contains N154, A156, A184, site E contains I71, A83, and A86. Other antigenic sites are S94, 120S, 162Y, 227 D, and 252Y. The A/chicken/Qalubia/MEVACF33/2017 isolated from chickens, possessed S56I, D88G, A134P, N146K, M175L, and G342W different amino acids substitutions compared to the first isolated H5N8 viruses from wild birds (Table 4). When predicted amino acid sequences (aa 1–342) of H5N8 viruses were aligned with H5N1, amino acid sequences of H5N8 were 79.6 to 83.3% identical to the analyzed H5N1 viruses. On the other hand, amino acid identities among isolated H5N8 strains ranged between 97.9–99.1% (Table 5).
Table 4.
Predicted amino acids of the HA1 gene of the isolated H5N8 isolates compared to other H5N8 viruses.
| RBS | HA amino acid substitutions | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus | Cleavage site | 222 | 224 | 72 | 104 | 150 | 162 | 180 | 191 | 293 | 285 | 358 |
| A/Bluebird/Aswan/MEVACF24/2017 | PLREKRRKRGLF | Q | G | S | D | A | N | Y | M | K | V | G |
| A/Quail/Sharkia/MEVACF25/2016 | PLREKRRKRGLF | Q | G | S | D | A | N | Y | M | K | M | G |
| A/chicken/Qalubia/MEVACF33/2017* | PLREKRRKRGLF | Q | G | I | G | P | K | Y | L | K | M | W |
| A/Greenfinch/Egypt/MEVAC F26/2016* | – | – | – | – | D | A | N | F | M | K | – | – |
| A/Chicken/Egypt/MEVAC F35.2/2017* | – | – | – | – | D | A | N | Y | M | K | – | – |
| A/green-winged teal/Egypt/877/2016 | PLREKRRKRGLF | Q | G | S | D | A | N | Y | M | K | V | G |
| A/Common-coot/Egypt/CA285/2016** | PLREKRRKRGLF | Q | G | S | D | A | N | Y | M | K | V | G |
| A/duck/DRC/17RS882-5/2017 | PLREKRRKRGLF | Q | G | S | D | A | N | Y | M | R | V | G |
| A/chicken/Israel/881/2016 | PLREKRRKRGLF | Q | G | S | D | A | N | Y | M | K | V | G |
*Partial sequence analysis of HA1 (aa 92 to 204 according to H5 numbering).
**First isolated H5N8 from wild birds in Egypt.
Table 5.
Nucleotide and amino acid identities of the HA1 gene of the isolated H5N8 isolates compared to the first isolated H5N8 from Egypt and other isolates.
|
Antigenic Relatedness of the Isolated H5N8 to Different H5N1 Virus Clades
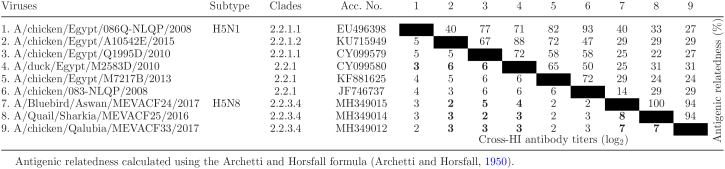
Six Egyptian H5N1 viruses representing different clusters 2.2.1.1, 2.2.1.2, 2.2.1, and 3 H5N8 viruses (clade 2.3.4.4) were used in the antigenic analysis. Chicken sera raised against inactivated H5N1 showed very low HI titers with H5N8 viruses. While the antigenic relatedness among H5N1 strains ranged between 40 and 100%, the antigenic relatedness between H5N1 and H5N8 strains varied from 22 to 40% (Table 3).
Table 3.
Cross-reactivity (hemagglutination inhibition titers) and antigenic relatedness (%) between H5N8 and H5N1 of different clades.
|
DISCUSSION
The epidemiology of avian respiratory pathogens in Egypt has undergone substantial changes since the emergence of HP H5N1 in 2006, vNDV and LP H9N2 in 2011, and recently the HP H5N8 in late 2016 (Slomka et al., 2007; Hassan et al., 2016; Arafa et al., 2016a; Kayali et al., 2016a). The current study aimed to monitor the incidence of different respiratory viruses and mortality rates in sick commercial broiler and layer chickens. Few samples were also collected from wild migratory birds from northern Egypt in 2016.
Variable mixed infections that mainly involved H9N2 virus were observed in commercial broilers and layers. Therefore, farms that exhibited mixed infections mainly showed higher mortality rates compared to those with single infections. Respiratory co-infections, especially those involving H9N2 and IBV, were reported to be more common during the last decade in Egypt and showed significant alterations in clinical signs, severity, and mortality rates (Hassan et al., 2016; Young et al., 2016; Hassan et al., 2017; Samy and Naguib, 2018). Notably, a triple H5N1, H9N2, and H5N8 avian influenza co-infection was detected in a commercial layer flock from Sharkia governorate. This unique epidemiological situation in Egypt raises a concern about the potential emergence of a new influenza A virus epidemic strains (Kim, 2018).
Genetic analysis of the isolated H9N2 viruses showed that they belong to the G1 lineage. It has been claimed that co-infection of H9N2 with other respiratory viruses complicates the clinical picture (Hassan et al., 2017), possibly via the IBV trypsin-like proteases activate the HA cleavage of H9N2 (Ng and Liu, 2000). The NDV was detected in 13 commercial broiler flocks despite intensive vaccination regimes. The widespread of the genotype VII NDV infection in commercial broilers might be attributed to the short lifespan of broiler chickens that does not allow full protective immunity by inactivated or classical live vaccines (Ji et al. 2018). Additionally, the retarded biosecurity, mixed infections, and/or the difference between the circulating and vaccine strains may count for such evidence (Hassan et al., 2016). All isolated ND viruses confirmed that they belonged to genotype VII, which is in consent with previous studies (Mohamed et al., 2011; Radwan et al., 2013; Saad et al., 2017).
Due to the continuous evolution of H5N1 virus under vaccination pressure, subclades 2.2.1 and 2.2.1.1 emerged between 2008 and 2011 in association with backyard and commercial poultry breeding (Grund et al., 2011; Kayali et al., 2016b). In this study, single and/or mixed HP H5N1 virus infections were common in both poultry production sectors. All isolated viruses belonged to clade 2.2.1.2. This clade has been reported to represent the most predominant H5N1 viruses in Egypt compared to clades 2.2.1 and 2.2.1.1 (El-Shesheny et al., 2012; Arafa et al., 2016b).
In the current study, the HPAI H5N8 virus was only detected in samples collected from bluebird, quail, and greenfinch during 2016. However, by early 2017 the H5N8 virus was detected in both commercial broiler and layer chickens (Salaheldin et al., 2018; Yehia et al., 2018). Sequence analysis of the isolated H5N8 revealed that it belongs to the 2.2.3.4b clade. Though partial HA gene sequences were recovered, different amino acid differences were noticed among the isolated H5N8 viruses from domestic chickens compared to their counterparts from wild birds (93.8 to 98.2% amino acids identities). For instance, the strain A/chicken/Qalubia/MEVACF33/2017 H5N8 possessed 6 amino acids substitutions (S56I, D88G, A134P, N146K, M175L, G342W) compared to the first detected H5N8 in Egypt (A/Common coot/Egypt/CA285/2016). None of these amino acid changes is in the previously reported important genetic sites, however, their biological properties need to be elucidated. Meanwhile, all the isolated H5N8 viruses retain the preferential binding to α-2–3-Gal avian-like receptors as indicated by aa 222Q and 224 G at the receptor binding pocket of the HA1 molecule (Kandeil et al., 2017). The differences between domestic and wild birds H5N8 isolates further support previous reports of separate introductions of HPAI H5N8 viruses in domestic poultry in Egypt (Salaheldin et al., 2018; Yehia et al., 2018).
In Egypt, licensed avian influenza commercial vaccines represent HPAI H5N1 of 2.2.1, 2.2.1.1, and 2.2.1.2 clades. Though the efficacy of some commercial vaccines against HP H5N8 viruses ranged between 80 to 90%, all these trials were under experimental conditions (Kapczynski et al., 2017; Santos et al., 2017; Kandeil et al., 2018). Nevertheless, field observations indicate that both commercial broiler and layer chickens are not protected by H5N1 vaccination. Thus, the cross-reactivities of different H5N1 clades with the isolated H5N8 viruses were analyzed. Antisera raised against inactivated H5N1 viruses belonging to clades 2.2.1 and 2.2.1.2 showed minimal reactivity with H5N8 viruses suggesting poor cross-protection. Though the heterologous protection of H5N1 vaccines against H5N8 challenge was attributed to the antibody-dependent cell-mediated cytotoxicity (ADCC), it was not clear if the increase of ADCC response would be adequate to protect vaccinated mice against H5N8 virus infection (Park et al., 2016).
Though not included in the current study, the role of other factors including immunosuppressive diseases, potential viral interference conditions during mixed infections (Aouini et al., 2018), and secondary bacterial infections (Chu et al., 2017) in increased pathogenicity of different viruses need to be taken in consideration. To conclude, the high rate of mixed infections and poor biosecurity measures are contributing to the increased vulnerability to infection. Hence, modification of vaccination strategies using H5N1 vaccines (e.g., double dose regimes in broilers) accompanied with strict biosecurity measures and/or the development of H5N8 specific vaccine were suggested.
SUPPLEMENTARY DATA
Table (1): Prevalence of respiratory viruses in commercial broiler farms.
ACKNOWLEDGEMENTS
The authors would like to thank Middle East for Veterinary Vaccines (ME VAC) Company laboratory animal team for their technical support during hyperimmune sera preparation.
Conflict of interest
The authors declare that there is no conflict of interest.
REFERENCES
- Abdel-Moneim A. S., Afifi M. A., El-Kady M. F.. 2012. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch. Virol. 157:2453–2457. [DOI] [PubMed] [Google Scholar]
- Abdel-Moneim A. S., El-Kady M. F., Ladman B. S., Gelb J. Jr. 2006. S1 gene sequence analysis of a nephropathogenic strain of avian infectious bronchitis virus in Egypt. Virol. J. 3:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Sabour M. A., Al-Ebshahy E. M., Khaliel S. A., Abdel-Wanis N. A., Yanai T.. 2017. Isolation and molecular characterization of novel infectious bronchitis virus variants from vaccinated broiler flocks in Egypt. Avian Dis. 61:307–310. [DOI] [PubMed] [Google Scholar]
- Ali A., Kilany W. H., Zain El-Abideen M. A., Sayed M. E., Elkady M.. 2018. Safety and efficacy of attenuated classic and variant 2 infectious bronchitis virus candidate vaccines. Poult. Sci. 97:4238–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly M. M., Arafa A., Hassan M. K.. 2008. Epidemiological findings of outbreaks of disease caused by highly pathogenic H5N1 avian influenza virus in poultry in Egypt during 2006. Avian Dis. 52:269–277. [DOI] [PubMed] [Google Scholar]
- Anis A., AboElkhair M., Ibrahim M.. 2018. Characterization of highly pathogenic avian influenza H5N8 virus from Egyptian domestic waterfowl in 2017. Avian Pathol. 47:400–409. [DOI] [PubMed] [Google Scholar]
- Aouini R., Laamiri N., Ghram A.. 2018. Viral interference between low pathogenic avian influenza H9N2 and avian infectious bronchitis viruses in vitro and in ovo. J. Virol. Methods. 259:92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafa A., El-Masry I., Kholosy S., Hassan M. K., Dauphin G., Lubroth J., Makonnen Y. J.. 2016. Phylodynamics of avian influenza clade 2.2.1 H5N1 viruses in Egypt. Virol. J. 13:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafa A., El-Masry I., Khoulosy S., Hassan M. K., Soliman M., Fasanmi O. G., Fasina F. O., Dauphin G., Lubroth J., Jobre Y. M.. 2016b. Predominance and geo-mapping of avian influenza H5N1 in poultry sectors in Egypt. Geospat. Health. 11:492. [DOI] [PubMed] [Google Scholar]
- Arafa A. S., Hagag N., Erfan A., Mady W., El-Husseiny M., Adel A., Nasef S.. 2012. Complete genome characterization of avian influenza virus subtype H9N2 from a commercial quail flock in Egypt. Virus Genes. 45:283–294. [DOI] [PubMed] [Google Scholar]
- Archetti I., Horsfall F. L.. 1950. Persistent antigenic variation in Influenza A viruses after incomplete neutralization in ovo with heterologous immune serurn. J. Exp. Med. 92:441–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Britton P., Naylor C. J.. 1999. Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specific polymerase chain reactions. Avian Pathol. 28:593–605. [DOI] [PubMed] [Google Scholar]
- Chacon J. L., Ferreira A. J.. 2009. Differentiation of field isolates and vaccine strains of infectious laryngotracheitis virus by DNA sequencing. Vaccine. 27:6731–6738. [DOI] [PubMed] [Google Scholar]
- Chu J., Zhang Q., Zuo Z., El-Ashram S., Guo Y., Zhao P., Huang S., He C., Khan A.. 2017. Co-infection of Chlamydia psittaci with H9N2, ORT and Aspergillus fumigatus contributes to severe pneumonia and high mortality in SPF chickens. Sci. Rep. 7:13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Hurtado M., Afonso C. L., Miller P. J., Shepherd E., DeJesus E., Smith D., Pantin-Jackwood M. J.. 2016. Effect of infection with a mesogenic strain of newcastle disease virus on infection with highly pathogenic avian influenza virus in chickens. Avian Dis. 60:269–278. [DOI] [PubMed] [Google Scholar]
- Costa-Hurtado M., Afonso C. L., Miller P. J., Spackman E., Kapczynski D. R., Swayne D. E., Shepherd E., Smith D., Zsak A., Pantin-Jackwood M.. 2014. Virus interference between H7N2 low pathogenic avian influenza virus and lentogenic Newcastle disease virus in experimental co-infections in chickens and turkeys. Vet. Res. 45:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubney R., Mansy W.. 1948. The occurrence of Newcastle disease in Egypt. J. Comp. Pathol. Ther. 58:189–200. [DOI] [PubMed] [Google Scholar]
- El-Shesheny R., Kayali G., Kandeil A., Cai Z., Barakat A. B., Ghanim H., Ali M. A.. 2012. Antigenic diversity and cross-reactivity of avian influenza H5N1 viruses in Egypt between 2006 and 2011. J. Gen. Virol. 93:2564–2574. [DOI] [PubMed] [Google Scholar]
- El-Zoghby E. F., Arafa A. S., Kilany W. H., Aly M. M., Abdelwhab E. M., Hafez H. M.. 2012. Isolation of avian influenza H5N1 virus from vaccinated commercial layer flock in Egypt. Virol J. 9:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund C., Abdelwhab el S. M., Arafa A. S., Ziller M., Hassan M. K., Aly M. M., Hafez H. M., Harder T. C., Beer M.. 2011. Highly pathogenic avian influenza virus H5N1 from Egypt escapes vaccine-induced immunity but confers clinical protection against a heterologous clade 2.2.1 Egyptian isolate. Vaccine. 29:5567–5573. [DOI] [PubMed] [Google Scholar]
- Hassan K. E., Ali A., Shany S. A. S., El-Kady M. F.. 2017. Experimental co-infection of infectious bronchitis and low pathogenic avian influenza H9N2 viruses in commercial broiler chickens. Res. Vet. Sci. 115:356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan K. E., Shany S. A., Ali A., Dahshan A. H., El-Sawah A. A., El-Kady M. F.. 2016. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult. Sci. 95:1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini H., Fard M. H., Charkhkar S., Morshed R.. 2015. Epidemiology of avian infectious bronchitis virus genotypes in Iran (2010-2014). Avian Dis. 59:431–435. [DOI] [PubMed] [Google Scholar]
- Hussein A. H., Emara M. M., Rohaim M. A.. 2015. Genetic diversity of avian influenza H5N1 Subclade 2.2.1/C in commercial poultry in Egypt during 2013. Global Vet. 1:14 8. [Google Scholar]
- Ji Y., Liu T., Du Y., Cui X., Yu Q., Wang Z., Zhang J., Li Y., Zhu Q.. 2018. A novel genotype VII Newcastle disease virus vaccine candidate generated by mutation in the L and F genes confers improved protection in chickens. Vet Microbiol. 216:99–106. [DOI] [PubMed] [Google Scholar]
- Kandeil A., Kayed A., Moatasim Y., Webby R. J., McKenzie P. P., Kayali G., Ali M. A.. 2017. Genetic characterization of highly pathogenic avian influenza A H5N8 viruses isolated from wild birds in Egypt. J. Gen. Virol. 98:1573–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeil A., Sabir J. S. M., Abdelaal A., Mattar E. H., El-Taweel A. N., Sabir M. J., Khalil A. A., Webby R., Kayali G., Ali M. A.. 2018. Efficacy of commercial vaccines against newly emerging avian influenza H5N8 virus in Egypt. Sci. Rep. 8:9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapczynski D. R., Pantin-Jackwood M. J., Spackman E., Chrzastek K., Suarez D. L., Swayne D. E.. 2017. Homologous and heterologous antigenic matched vaccines containing different H5 hemagglutinins provide variable protection of chickens from the 2014 U.S. H5N8 and H5N2 clade 2.3.4.4 highly pathogenic avian influenza viruses. Vaccine. 35:6345–6353. [DOI] [PubMed] [Google Scholar]
- Kayali G., Kandeil A., El-Shesheny R., Kayed A. S., Maatouq A. M., Cai Z., Ali M. A.. 2016. Avian influenza A(H5N1) virus in Egypt. Emerg. Infect. Dis. 22:379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayali G., Kandeil A., El-Shesheny R., Kayed A. S., Maatouq A. M., Cai Z., McKenzie P. P., Webby R. J., El Refaey S., Kandeel A., Ali M. A.. 2016. Avian iknfluenza A(H5N1) virus in Egypt. Emerg. Infect. Dis. 22:379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H. 2018. Challenge for one health: co-circulation of zoonotic H5N1 and H9N2 avian influenza viruses in Egypt. Viruses 10:pii: E121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. W., Suarez D. L.. 2008. Reverse genetics of the avian influenza virus. Methods Mol. Biol. 436:99–111. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Park J. K., Yuk S. S., Erdene-Ochir T. O., Kwon J. H., Lee J. B., Park S. Y., Choi I. S., Lee S. W., Song C. S.. 2014. Complete genome sequence of a natural reassortant H9N2 avian influenza virus found in bean goose (Anser fabalis): direct evidence for virus exchange between Korea and China via wild birds. Infect. Genet. Evol. 26:250–254. [DOI] [PubMed] [Google Scholar]
- Mohamed M. H., Kumar S., Paldurai A., Samal S. K.. 2011. Sequence analysis of fusion protein gene of Newcastle disease virus isolated from outbreaks in Egypt during 2006. Virol. J. 8:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib M. M., Ulrich R., Kasbohm E., Eng C. L. P., Hoffmann D., Grund C., Beer M., Harder T. C.. 2017. Natural reassortants of potentially zoonotic avian influenza viruses H5N1 and H9N2 from Egypt display distinct pathogenic phenotypes in experimentally infected chickens and ferrets. J. Virol. 91:pii: e01300–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L. F., Liu D. X.. 2000. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology. 272:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE 2015. Terresterial Manual 2015. Avian Influenza; Accessed Jan. 2018. http://www.oie.int/fileadmin/Home/fr/Health_standards/tahm/2.03.04_AI.pdf. [Google Scholar]
- Park S. J., Si Y. J., Kim J., Song M. S., Kim S. M., Kim E. H., Kwon H. I., Kim Y. I., Lee O. J., Shin O. S., Kim C. J., Shin E. C., Choi Y. K.. 2016. Cross-protective efficacies of highly-pathogenic avian influenza H5N1 vaccines against a recent H5N8 virus. Virology. 498:36–43. [DOI] [PubMed] [Google Scholar]
- Peyre M., Samaha H., Makonnen Y. J., Saad A., Abd-Elnabi A., Galal S., Ettel T., Dauphin G., Lubroth J., Roger F., Domenech J.. 2009. Avian influenza vaccination in Egypt: limitations of the current strategy. J. Mol. Genet. Med. 3:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan M. M., Darwish S. F., El-Sabagh I. M., El-Sanousi A. A., Shalaby M. A.. 2013. Isolation and molecular characterization of Newcastle disease virus genotypes II and VIId in Egypt between 2011 and 2012. Virus Genes. 47:311–316. [DOI] [PubMed] [Google Scholar]
- Roussan D. A., Totanji W. S., Khawaldeh G. Y.. 2008. Molecular subtype of infectious bronchitis virus in broiler flocks in Jordan. Poult. Sci. 87:661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad A. M., Samy A., Soliman M. A., Arafa A., Zanaty A., Hassan M. K., Sultan A. H., Bazid A. I., Hussein A. H.. 2017. Genotypic and pathogenic characterization of genotype VII Newcastle disease viruses isolated from commercial farms in Egypt and evaluation of heterologous antibody responses. Arch. Virol. 162:1985–1994. [DOI] [PubMed] [Google Scholar]
- Salaheldin A. H., El-Hamid H. S., Elbestawy A. R., Veits J., Hafez H. M., Mettenleiter T. C., Abdelwhab E. M.. 2018. Multiple Introductions of Influenza A(H5N8) Virus into Poultry, Egypt, 2017. Emerg. Infect. Dis. 24 doi 10.3201/eid2405.171935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samy A., Naguib M. M.. 2018. Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet. Sci. 5:943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. J. S., Obadan A. O., Garcia S. C., Carnaccini S., Kapczynski D. R., Pantin-Jackwood M., Suarez D. L., Perez D. R.. 2017. Short- and long-term protective efficacy against clade 2.3.4.4 H5N2 highly pathogenic avian influenza virus following prime-boost vaccination in turkeys. Vaccine. 35:5637–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim A. A., Erfan A. M., Hagag N., Zanaty A., Samir A. H., Samy M., Abdelhalim A., Arafa A. A., Soliman M. A., Shaheen M., Ibraheem E. M., Mahrous I., Hassan M. K., Naguib M. M.. 2017. Highly pathogenic avian influenza virus (H5N8) clade 2.3.4.4 infection in migratory birds, Egypt. Emerg. Infect. Dis. 23:1048–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim K. M., Selim A., Arafa A., Hussein H. A., Elsanousi A. A.. 2018. Molecular characterization of full fusion protein (F) of Newcastle disease virus genotype VIId isolated from Egypt during 2012-2016. Vet World. 11:930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shany S. A. S. 2015. Further studies on the current situation of avian influenza in Egypt. PhD Beni-Suef University, Beni-Suef, Egypt. [Google Scholar]
- Slomka M. J., Pavlidis T., Banks J., Shell W., McNally A., Essen S., Brown I. H.. 2007. Validated H5 Eurasian real-time reverse transcriptase-polymerase chain reaction and its application in H5N1 outbreaks in 2005–2006. Avian Dis. 51:373–377. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M., Reinacher M., Rott R.. 1987. Aggravation of pathogenicity of an avian influenza virus by adaptation to quails. Arch. Virol 93:81–95. [DOI] [PubMed] [Google Scholar]
- Yehia N., Naguib M. M., Li R., Hagag N., El-Husseiny M., Mosaad Z., Nour A., Rabea N., Hasan W. M., Hassan M. K., Harder T., Arafa A. A.. 2018. Multiple introductions of reassorted highly pathogenic avian influenza viruses (H5N8) clade 2.3.4.4b causing outbreaks in wild birds and poultry in Egypt. Infect. Genet. Evol. 58:56–65. [DOI] [PubMed] [Google Scholar]
- Young S. G., Carrel M., Malanson G. P., Ali M. A., Kayali G.. 2016. Predicting Avian Influenza Co-Infection with H5N1 and H9N2 in Northern Egypt. IJERPH 13:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakia A. M., Ahmed H. A., Hussein, Rohaim M. A.. 2013. Efficacy of composting poultry mortality and farms wastes with mixed respiratory infection viruses H9N2 and NDV in Egypt. Global Vet. 11:640–648. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.