Abstract
In 2012, a novel coronavirus (CoV) associated with severe respiratory disease, Middle East respiratory syndrome (MERS-CoV; previously known as human coronavirus–Erasmus Medical Center or hCoV-EMC), emerged in the Arabian Peninsula. To date, 114 human cases of MERS-CoV have been reported, with 54 fatalities. Animal models for MERS-CoV infection of humans are needed to elucidate MERS pathogenesis and to develop vaccines and antivirals. In this study, we developed rhesus macaques as a model for MERS-CoV using intratracheal inoculation. The infected monkeys showed clinical signs of disease, virus replication, histological lesions, and neutralizing antibody production, indicating that this monkey model is suitable for studies of MERS-CoV infection.
Keywords: animal model, coronavirus, MERS, pathology, rhesus monkey
(See the brief report by Aburizaiza et al on pages 243–6, and the editorial commentary by Hui and Zumla on pages 173–6.)
Coronaviruses (CoVs) can infect humans and a wide variety of animals, causing respiratory, enteric, hepatic, and neurological diseases of varying clinical severity. Based on their genotypic and serological characteristics, they have been classified into 3 genera: Alphacoronavirus, Betacoronavirus, and Gammacoronavirus [1]. Coronaviruses are well known for their high frequency of recombination and high mutation rates, which may allow them to adapt to new hosts and ecological niches. This is best exemplified by the severe acute respiratory syndrome (SARS) epidemic, which was caused by SARS-CoV [2]. SARS-CoV was shown to have originated from animals, with horseshoe bats (Rhinolophus sinicus) as the natural reservoir and the palm civet (Paguma larvata) as the intermediate host, allowing animal-to-human transmission [3]. Since the SARS epidemic, many other novel CoVs have been discovered in both humans and animals. A novel Betacoronavirus lineage C, including the Tylonycteris bat CoV HKU4 (Ty-BatCoV HKU4) and Pipistrellus bat CoV HKU5 (Pi-BatCoV HKU5), was discovered in the lesser bamboo bat (T. pachypus) and the Japanese pipistrelle (P. abramus) in 2007 in Hong Kong, China. These viruses were found to be closely related to a novel strain of human CoV, referred to as Middle East respiratory syndrome CoV (MERS-CoV), which was identified in Saudi Arabia [4–6]. This novel human virus is now classified as a lineage C Betacoronavirus that differs from other CoVs previously found in humans, including SARS [7]. To date, 114 human cases of MERS-CoV have been reported, with 54 fatalities [8].
MERS-CoV is associated with severe respiratory-tract infection, renal failure, and fatalities. Similar to SARS-CoV, MERS-CoV is closely related to bat CoVs, suggesting that bats may be the natural reservoir of this family of viruses [9, 10]. Specifically, a virus from Egyptian tomb bats showed 100% nucleotide identity to virus from the human index case-patient [11]. It is currently unclear whether the human cases were a result of direct zoonotic transmission from bats to humans or whether an intermediate host was involved. However, a recent study showed MERS-CoV reactive antibodies in retired racing camels in Oman, which neighbors Saudi Arabia [12]. In Middle Eastern nations, large amounts of camel meat were consumed every year, much of which was imported from African countries like Egypt. This epidemiological evidence may suggest a bat-camel-human viral linkage, while more investigations are needed to define the source of MERS-CoV. Currently, there is no evidence of efficient transmission between humans by MERS-CoV. However, the occurrence of some clusters of cases suggests that human-to-human transmission is possible and has raised concerns regarding the potential of this virus to cause a pandemic similar to that caused by SARS-CoV in 2002/2003 [13, 14].
The pathogenic mechanism of MERS-CoV is not clear, and neither an effective vaccine nor therapeutic drugs are available for prevention and treatment. The development of animal models for MERS-CoV infection of humans is of utmost importance to study the pathogenesis of this virus and to test the efficacy of potential therapeutic or prophylactic intervention strategies. At present, there are few reports of animal model for MERS-CoV infection, thus limiting further study [15, 16]. Nonhuman primates have played an essential role in our understanding of the various forms of the pathogen, which could reflect variable clinical symptoms and pathology in humans. Previous studies have reported nonhuman primate disease models for influenza, SARS, and other viruses [17–19]. Therefore, in the present study, we explored the suitability of the rhesus monkey as an animal model for MERS-CoV isolate human coronavirus–Erasmus Medical Center (hCoV-EMC) infection or disease.
METHODS
Ethics Statement
The research on MERS-CoV virus was discussed among the staff members of the Department of Pathogen Biology at the Institute of Laboratory Animal Science (ILAS) of the Chinese Academy of Medical Sciences and Peking Union Medical College (PUMC). The MERS-CoV animal model experiments and protocols were discussed explicitly and extensively among the staff members of the Department of Pathogen Biology. These discussions were followed by additional discussions with biosafety officers and facility managers at the ILAS of PUMC, as well as with numerous specialists from the SARS-CoV and general infectious disease fields throughout China. All research procedures were approved by the ILAS Institutional Animal Care and Use Committee and Laboratory Safety Committee (LSC). The committee recommended that the number of animals be reduced to comply with the 3R (reduction, replacement, refinement) principles; we therefore designed the experiments to include 6 animals to test the animal model of MERS-CoV, 4 monkeys infected with virus and 2 uninfected monkeys as controls. The approved registration number is ILAS-PC-2013-004.
All experiments were conducted within the animal biosafety level 3 (ABSL-3) facility, which was constructed and accredited based on National Standard GB19489 at the ILAS of PUMC, Beijing, China.
Virus and Cell Culture
MERS-CoV strain hCoV-EMC was a kind gift from Professor Fouchier [4]. Seed stocks of hCoV-EMC were propagated in Vero cells. The seed stocks were diluted to the designated titer and used for determining the hCoV-EMC 50% tissue culture infection dose (TCID50) and performing the neutralizing antibody assays.
Vero cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 international units (IU)/mL penicillin, and 100 µg/mL streptomycin and cultured at 37°C in 5% CO2.
MERS-CoV Monkey Model Study Design
Four monkeys, aged 2–3 years, were inoculated intratracheally with hCoV-EMC (6.5 × 107 TCID50/1 mL) diluted in DMEM. The monkeys were anesthetized, and 1 mL of the inoculum was administered intratracheally. Mock-infected monkeys (2 monkeys) intratracheally inoculated with DMEM were included as controls. The monkeys were observed twice daily, with detailed recording of clinical signs, symptoms, morbidity, and mortality, including the nature, onset, severity, and duration of all gross or visible changes. Chest X-rays were performed before inoculation and 3 and 5 days postinoculation (d.p.i.) with MERS-CoV. Two infected monkeys and a control monkey were sacrificed at 3 d.p.i. Tissue specimens, including lung, trachea, heart, spleen, kidney, brain, liver, and colon tissue, were collected for various pathological, virological, and immunological tests. For the virological and immunological tests, swab samples of the oropharyngeal, nasal turbinates, and cloacal regions were collected at 1, 3, 5, 7, 9, 11, 14, 21, and 28 d.p.i., and blood was collected at 7, 14, 21, and 28 d.p.i.
RNA Extraction and Detection of Viral RNA by Reverse Transcription Polymerase Chain Reaction
Total RNA was isolated from individual samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Reverse transcription (RT) reactions were performed using the Superscript III First Strand Synthesis Kit (Invitrogen) according to the manufacturer's instructions. All complementary DNA (cDNA) samples were stored at −20°C until use in the polymerase chain reaction (PCR). For most experimental samples and the positive control (hCoV-EMC), duplicate cDNA samples were obtained from each total RNA sample. The primers used for hCoV-EMC were described previously [20] and are specific for the RNA-dependent RNA polymerase (RdRp) gene. The sequences of the primers used for PCR detection were as follows: RdRpSeq-Fwd (TGCTATWAGTGCTAAGAATAGRGC; R = A/G, W = A/T) and RdRpSeq-Rev (GCATWGCNCWGTCACACTTAGG; W = A/T, N = A/C/T/G). The amplification protocol was as follows: 95°C for 3 minutes, followed by 45 cycles of 95°C for 15 seconds, 56°C for 15 seconds, and 72°C for 30 seconds, with a terminal elongation step of 72°C for 2 minutes. For cases in which no amplification products were obtained with the PCR assay, a 50-µL second-round reaction was set up containing 2 µL of the reaction mixture from the first round, the primer RdRpSeq-Fwd (the same as that used in the first round), and RdRpSeq-Rnest (CACTTAGGRTARTCCCAWCCCA). Thermal cycling was performed as follows: 95°C for 3 minutes, followed by 45 cycles of 95°C for 15 seconds, 56°C for 15 seconds, and 72°C for 30 seconds, followed by a 2-minute extension step at 72°C.
Determination of Viral Burden in Tissue and Swab Samples
Tissue samples were homogenized to a final 10% (weight per volume) suspension in DMEM and clarified by low-speed centrifugation at 4500 g for 30 minutes at 4°C. Swab samples were immersed in 1 mL DMEM, vortexed, and clarified by low-speed centrifugation at 5000 g for 10 minutes at 4°C. Virus titers were determined in Vero cells monolayers grown in 96-well plates. Vero cells were seeded (1.5 × 104/well) in a 96-well plate and incubated overnight at 37°C in a CO2 incubator. Then, 100 µL of 10-fold serially diluted suspension was added to each well in quadruplicate. The virus was allowed to adsorb to the cells at 37°C for 1 hour. After adsorption, the viral inocula were removed, and 0.1 mL medium (DMEM, 2% FBS) was added to each well. The plates were incubated in a CO2 incubator at 37°C for 3 days, after which the cytopathic effects (CPEs) were observed microscopically at 40× magnification. The virus titer of each specimen, expressed as the TCID50, was calculated by the Reed–Muench method.
Virus Neutralization Assay
For the neutralization test, serum samples were diluted 2-fold from an initial 1:10 dilution to a final dilution of 1:1280, mixed with 50 µL of 2000 TCID50/mL virus, and incubated at 37°C for 1 hour; this step was performed in quadruplicate. Thereafter, 100 µL virus-serum mixture was added to Vero cells previously seeded at 1.5 × 104/well. The inoculated plates were incubated in a CO2 incubator at 37°C for 3 days, after which the CPEs of the virus were observed microscopically at 40× magnification.
Histopathology and Immunohistochemistry
Animal necropsies were performed according to a standard protocol. Samples for histological examination were stored in 10% neutral-buffered formalin, embedded in paraffin, sectioned at 4 µm, and stained with hematoxylin and eosin prior to examination by light microscopy. Alternatively, the sections were immunohistochemically stained using a polyclonal antibody against the nucleoprotein of hCoV-EMC.
RESULTS
Clinical Observations
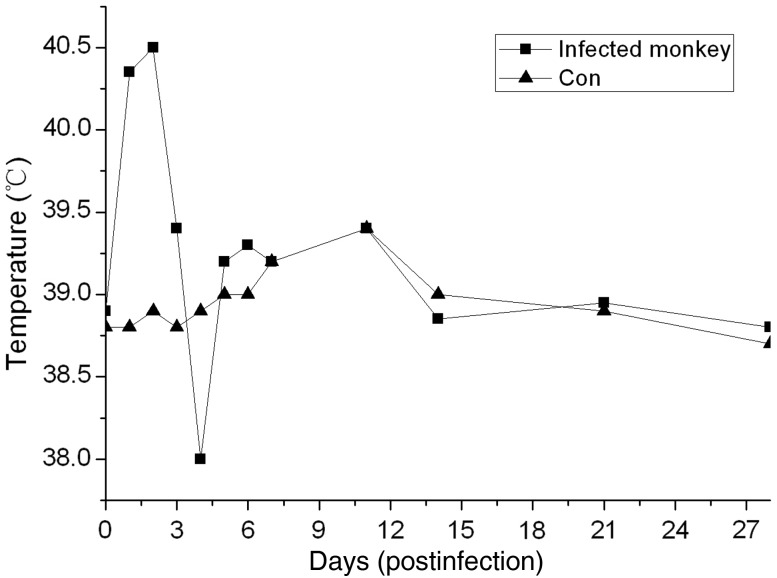
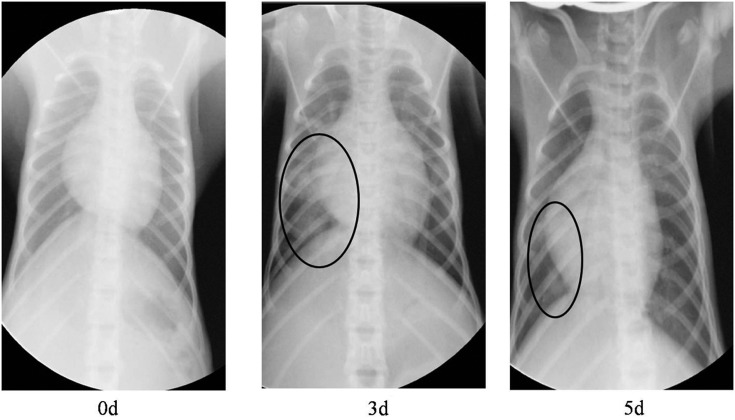
The monkeys were observed daily for clinical signs of disease for 28 d.p.i., and body weight and temperature were recorded. The rectal temperature was obtained daily in the first week after challenge, after which the temperature was measured at specific time points; graphs of the temperature profiles (Figure 1) depict the mean temperatures for the mock-infected and MERS-CoV–infected monkeys. The temperature of the infected monkeys increased at 1–2 d.p.i, and returned to normal thereafter. Moreover, within the first few days, the infected monkeys drastically decreased their water intake. None of the infected monkeys showed overt weight loss when compared with the mock-infected monkeys. Radiographic imaging revealed varying degrees of localized infiltration and interstitial markings (Figure 2). None of the infected monkeys died during the experiment.
Figure 1.
Temperature fluctuation in mock-infected and MERS-CoV–infected monkeys. Rectal temperature was obtained daily in the first week after challenge, and then the temperature was measured in specific time. Abbreviations: Con, control; MERS-CoV, Middle East respiratory syndrome coronavirus.
Figure 2.
Radiographs of the chest of monkey before inoculation and 3 and 5 days after inoculation with MERS-CoV. The circled areas are regions of interstitial infiltrates indicative of viral pneumonia. Abbreviation: MERS-CoV, Middle East respiratory syndrome coronavirus.
Detection of Viral RNA by RT-PCR
Total RNA was isolated from tissue homogenates and oropharyngeal, nasal turbinates, and cloacal swab samples and analyzed using primers specific for the hCoV-EMC RdRp gene. Viral RNA was detected in lung homogenates obtained from the infected monkey at 3 d.p.i.; however, the other organs remained RT-PCR negative (Table 1). No viral RNA was detected in any of the swab samples (Table 2).
Table 1.
MERS-CoV (hCoV-EMC) vRNA Detected in Monkey Organsa
| Group | Lung | Heart | Liver | Spleen | Kidney | Brain |
|---|---|---|---|---|---|---|
| Infected 1 | +b | …c | … | … | … | … |
| Infected 2 | + | … | … | … | … | … |
| Control | … | … | … | … | … | … |
Abbreviations: hCoV-EMC, human coronavirus–Erasmus Medical Center; MERS-CoV, Middle East respiratory syndrome coronavirus; TCID50, 50% tissue culture infection dose; vRNA, viral RNA.
a The monkeys were infected intratracheal with the viruses (6.5 × 107 TCID50/1 mL). Organs were collected on day 3 after infection, and clarified homogenates were titrated for virus infectivity in Vero cell.
b “+” Virus RNA was detected in the samples.
c “−” Virus RNA was not detected in the samples.
Table 2.
MERS-CoV (hCoV-EMC) vRNA Detected in Monkey Swabsa
| Number | Sites | 1d | 3d | 5d | 7d | 11d | 14d | 21d | 28d |
|---|---|---|---|---|---|---|---|---|---|
| Infected 1 | Nasal turbinate | …b | … | ND | ND | ND | ND | ND | ND |
| Oropharyngeal | … | … | ND | ND | ND | ND | ND | ND | |
| Cloacal | … | … | ND | ND | ND | ND | ND | ND | |
| Infected 2 | Nasal turbinate | … | … | ND | ND | ND | ND | ND | ND |
| Oropharyngeal | … | … | ND | ND | ND | ND | ND | ND | |
| Cloacal | … | … | ND | ND | ND | ND | ND | ND | |
| Infected 3 | Nasal turbinate | … | … | … | … | … | … | … | … |
| Oropharyngeal | … | … | … | … | … | … | … | … | |
| Cloacal | … | … | … | … | … | … | … | … | |
| Infected 4 | Nasal turbinate | … | … | … | … | … | … | … | … |
| Oropharyngeal | … | … | … | … | … | … | … | … | |
| Cloacal | … | … | … | … | … | … | … | … | |
| Con1 | Nasal turbinate | … | … | ND | ND | ND | ND | ND | ND |
| Oropharyngeal | … | … | ND | ND | ND | ND | ND | ND | |
| Cloacal | … | … | ND | ND | ND | ND | ND | ND | |
| Nasal turbinate | … | … | … | … | … | … | … | … | |
| Con2 | Oropharyngeal | … | … | … | … | … | … | … | … |
| Cloacal | … | … | … | … | … | … | … | … |
Abbreviations: Con, control; hCoV-EMC, human coronavirus–Erasmus Medical Center; MERS-CoV, Middle East respiratory syndrome coronavirus; ND, not done; TCID50, 50% tissue culture infection dose; vRNA, viral RNA.
a The monkeys were inoculated with 6.5 × 107 TCID50 of the virus in 1 mL volume intratracheally, and observed for 4 weeks after infection.
b “−“ Virus RNA was not detected in the samples.
MERS-CoV Titer in Tissue and Swab Samples
Nasal turbinates, oropharyngeal, and cloacal swabs were collected from inoculated animals at 1, 3, 5, 7, 11, 14, 21, and 28 d.p.i., and virus titers were determined by end-point titration in Vero cells. The oropharyngeal, nasal turbinates, and cloacal swabs contained no detectable virus titers during the experiment, which was in agreement with the RT-PCR results. Samples of the tissues described above were homogenized, and virus titers were determined. For the animals inoculated with MERS-CoV, virus could be detected in the lung (101.67 TCID50/mL); however, samples from the kidney, trachea, brain, heart, liver, spleen, and intestine had no detectable virus titers.
Neutralizing Antibody to hCoV-EMC
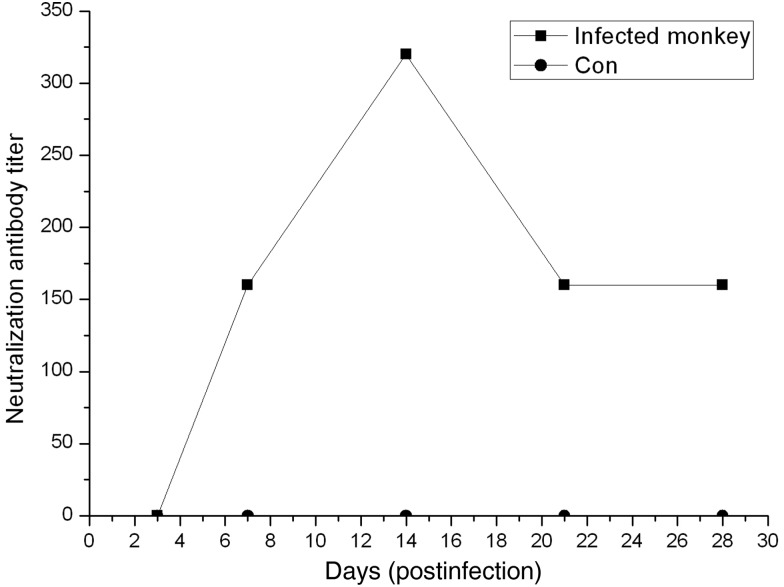
The levels of hCoV-EMC–neutralizing antibody were evaluated. No neutralizing antibodies were detected in mock-infected monkeys. By contrast, in the infected monkeys, neutralizing antibodies were detected beginning at 7 d.p.i., reaching a peak titer of 1:320 at 14 d.p.i., and decreasing slightly to 1:160 at 28 d.p.i. (Figure 3).
Figure 3.
Reciprocal neutralizing antibody titers from mock-infected or MERS-CoV–infected monkeys. The blood was collected on 7, 14, 21, and 28 d.p.i. for neutralizing antibody tests. Abbreviations: Con, control; d.p.i., days postinfection; MERS-CoV, Middle East respiratory syndrome coronavirus.
Histopathology and Immunohistochemistry
Two MERS-CoV–infected animals were euthanized at 3 d.p.i., and the trachea, lung, brain, heart, liver, spleen, kidney, and intestine were histologically and virologically examined. Lung lesions were present in the inoculated monkeys. Grossly, there was congestion, and palpable nodules, which showed as being scattered in distribution, were particularly evident in the posterior regions of the lobes (Figure 4). Microscopically, lung tissue showed multifocal mild-to-moderate interstitial pneumonia and exudative pathological changes (Figure 5).
Figure 4.
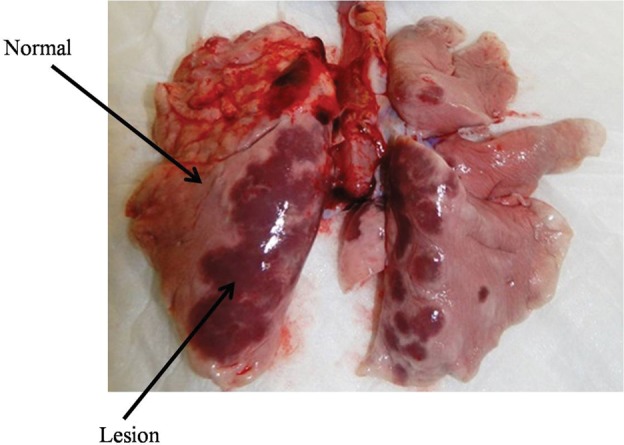
Histopathological findings in lungs of rhesus macaques inoculated with MERS-CoV. The infected monkeys were killed at 3 d.p.i. The arrows show normal and affected tissue. Abbreviations: d.p.i., days postinfection; MERS-CoV, Middle East respiratory syndrome coronavirus.
Figure 5.
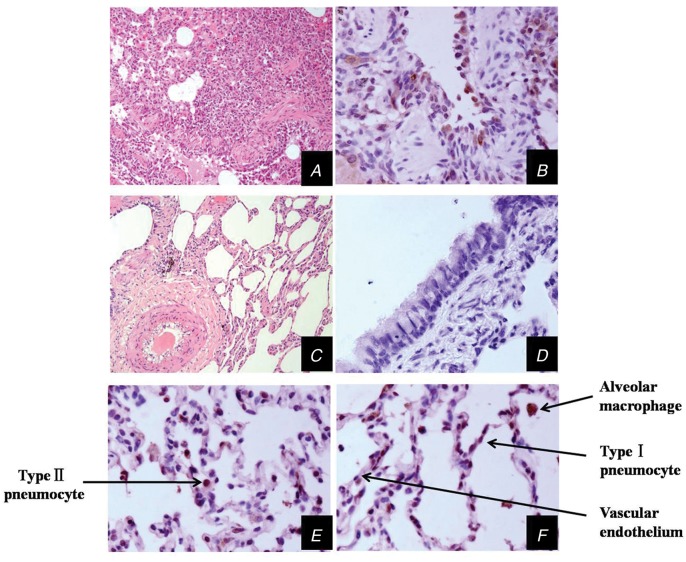
Histopathological and immunohistochemical (IHC) analyses of lung tissues of monkeys inoculated with MERS-CoV virus, collected at 3 d.p.i. Lung tissues of monkeys inoculated with MERS-CoV viruses, collected at 3 d.p.i. (A, HE ×100; B, IHC ×400; E, IHC ×400; F, IHC ×400) and DMEM (C, HE ×100; D, IHC ×400). The arrows show different cell types in the lung. At 3 d.p.i., lung tissues showed mild or moderate interstitial pneumonia. Abbreviations: d.p.i., days postinfection; DMEM, Dulbecco's Modified Eagle Medium; HE, hematoxylin-eosin; MERS-CoV, Middle East respiratory syndrome coronavirus.
Immunohistochemistry was also performed to assess the presence of MERS-CoV–infected cells in tissues collected from the infected animals and control animals (Figure 5). In monkeys inoculated with the MERS-CoV, infected cells were identified in the lung and in the same regional area as the pathological changes. Types I and II pneumocytes and alveolar macrophages were found to contain viral antigen. No vascular endothelium was observed (Figure 5). In addition, infected cells were not observed in other organs. In the control group, no positive areas were identified in any organs.
DISCUSSION
Currently, humans infected with MERS-CoV have been reported to develop a severe acute respiratory illness with symptoms of fever, cough, shortness of breath, and often, renal failure. More than half of the reported patients have died. Some laboratory-confirmed cases reportedly experience a mild respiratory illness. The development of animal models that reflect these variations in the human population is challenging. In this study, we sought to develop an animal model that displays these symptoms. Our studies have tested mouse, ferret, and guinea pig models and found these animals are not susceptible to MERS-CoV (unpublished data).
Nonhuman primates have been useful for evaluating vaccines and studying disease pathogenesis for several respiratory viruses, including influenza, SARS-CoV, respiratory syncytial virus, and human parainfluenza viruses. In this study, the rhesus monkey was explored as a model for MERS-CoV. Not only did this monkey model support viral growth, it also manifested respiratory and generalized illness along with tissue pathology.
The MERS-CoV dose used in this study was 6.5 × 107 TCID50 by intratracheal infection. After infection, we observed a temperature increase in the infected animals. Fever is one of the markers of MERS-CoV infection in humans [21], and in the included monkeys, the temperature increase was significant from 1–2 d.p.i. The body weight of each monkey was also recorded prior to viral infection, but no significant change in mean weight was observed. Virus replication was detected in the lungs of the infected animals. However, we did not detect virus RNA or measurable titers in the nasal turbinate or oropharyngeal samples, suggesting that the monkeys did not shed virus in the upper-respiratory tract. In humans, MERS-CoV was detected in lower-respiratory-tract specimens with a high viral load, whereas nasopharyngeal samples were weakly positive or inconclusive [22]. This monkey model showed similar results.
The primary MERS-CoV infection–related pathology in humans was acute pneumonia accompanied by multiorgan dysfunction and acute renal failure, which is compatible with a broad range of tissue tropisms in cell culture susceptibility testing [23]. The pathological presentation of pneumonia was also observed in the infected monkeys (Figure 3); however, no extrapulmonary lesions were found. These findings are consistent with the report by Vincent et al, who reported similar findings of acute localized-to-widespread pneumonia that resulted in mild-to-moderate clinical disease [15]. In humans, renal failure is a major symptom of MERS; however, we observed no pathological changes in the kidneys. The reasons for this discrepancy are not clear and deserve further study. It is possible that dissemination of the virus may not yet have occurred and the virus was limited to the lung, as we did not detect the virus in the blood.
To determine whether the animals were infected, we developed a neutralizing antibody assay against MERS-CoV. There was evidence of seroconversion in all the inoculated animals. MERS-CoV–specific antibody was detected in the monkey serum samples beginning at 7 d.p.i., and the titer increased gradually. Neutralizing antibody protection assays showed that the infected monkeys produced neutralizing antibodies that could prevent infection due to rechallenge by MERS-CoV. The results also indicate the potent immunogenicity of MERS-CoV, suggesting the possibility of using the virus as an inactivated or attenuated vaccine.
In summary, the viral detection assays, immunological response, and pathological changes in the lungs of the infected animals suggest that MERS-CoV was able to establish an infection in the inoculated monkeys. Additionally, the rhesus macaque appears to be a suitable model for studies of MERS-CoV pathogenesis or potential prophylactic or therapeutic intervention strategies.
Up to this stage, pathogenesis of MERS-CoV in human is still not clear. Rhesus macaques can be infected by MERS-CoV in the laboratory, but we recognize that it is necessary to search for more suitable animal model that will have similar disease presentations as that observed among those laboratory-confirmed human cases. However, before another animal model is defined, Rhesus macaques may be used as an infection model for the evaluation of vaccine and antiviral drugs as stated in a recent paper [24].
Notes
Acknowledgments. The authors would like to thank Professor Fouchier et al for providing MERS-CoV isolate hCoV-EMC/2012.
Financial support. This work was supported by grants from the National Science and Technology Major Projects of Infectious Disease (2012ZX10004501-004-003, 2012ZX10004501-004-004, and 2012ZX10004301-8), the Fundamental Research Funds for the Central Universities (2012Y02, 2012D15), and Open Fund of the Key Laboratory of Human Diseases Comparative Medicine, Ministry of Health (DWS201214).
Potential conflict of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Chan JF, Li KS, To KK, Cheng VC, Chen H, Yuen KY. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65:477–89. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SK, Woo PC, Li KS, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–5. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Woo PC, Wang M, Lau SK, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–85. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau SK, Li KS, Tsang AK, et al. Genetic characterization of vetacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel middle east respiratory syndrome coronavirus. J Virol. 2013;87:8638–50. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Boheemen S, de Graaf M, Lauber C, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. Global alert and response: Novel Coronavirus infection-update. Geneva: WHO; http://www.who.int/csr/don/2013_09_07/en/index.html. Accessed 7 September 2013. [Google Scholar]
- 9.Annan A, Baldwin HJ, Corman VM, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–9. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthony SJ, Ojeda-Flores R, Rico-Chavez O, et al. Coronaviruses in bats from Mexico. J Gen Virol. 2013;94:1028–38. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–23. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reusken CB, Haagmans BL, Muller MA, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–66. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 2013;18:20427. doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- 14.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368:2487–94. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 15.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N Engl J Med. 2013;368:1560–2. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wit E, Prescott J, Baseler L, et al. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLOS One. 2013;8:e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinfurter JT, Brunner K, Capuano SV, III, et al. Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates. PLOS Pathog. 2011;7:e1002381. doi: 10.1371/journal.ppat.1002381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin C, Wang J, Wei Q, et al. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J Pathol. 2005;206:251–9. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shipley TW, Kling HM, Morris A, et al. Persistent pneumocystis colonization leads to the development of chronic obstructive pulmonary disease in a nonhuman primate model of AIDS. J Infect Dis. 2010;202:302–12. doi: 10.1086/653485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman VM, Muller MA, Costabel U, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:1–9. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 21.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–51. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guery B, Poissy J, el Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–72. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan JF, Chan KH, Choi GK, et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis. 2013;207:1743–52. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falzarano D, de Wit E, Rasmussen AL, et al. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–17. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]