Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) infection caused severe pneumonia and multiorgan dysfunction and had a higher crude fatality rate (around 50% vs 10%) than SARS coronavirus (SARS-CoV) infection. To understand the pathogenesis, we studied viral replication, cytokine/chemokine response, and antigen presentation in MERS-CoV–infected human monocyte–derived macrophages (MDMs) versus SARS-CoV–infected MDMs. Only MERS-CoV can replicate in MDMs. Both viruses were unable to significantly stimulate the expression of antiviral cytokines (interferon α [IFN-α] and IFN-β) but induced comparable levels of tumor necrosis factor α and interleukin 6. Notably, MERS-CoV induced significantly higher expression levels of interleukin 12, IFN-γ, and chemokines (IP-10/CXCL-10, MCP-1/CCL-2, MIP-1α/CCL-3, RANTES/CCL-5, and interleukin 8) than SARS-CoV. The expression of major histocompatibility complex class I and costimulatory molecules were significantly higher in MERS-CoV–infected MDMs than in SARS-CoV–infected cells. MERS-CoV replication was validated by immunostaining of infected MDMs and ex vivo lung tissue. We conclusively showed that MERS-CoV can establish a productive infection in human macrophages. The aberrant induction of inflammatory cytokines/chemokines could be important in the disease pathogenesis.
Keywords: MERS-CoV, SARS-CoV, viral replication, pathogenesis, cytokine and chemokine response
In September 2012, a novel human coronavirus, HCoV-EMC, later renamed Middle East respiratory syndrome coronavirus (MERS-CoV), was identified in 2 patients with severe pneumonia complicated with renal failure who once traveled to or resided in Saudi Arabia [1, 2]. Retrospective analysis of archived specimens showed that the first virologically confirmed cases could be traced back to early April 2012 [3]. As of 1 August 2013, World Health Organization has confirmed 94 cases of infection with 46 deaths, and therefore an appalling fatality rate of around 50% [4]. Coronaviruses are the largest of all RNA viruses, with positive single-stranded RNA genomes of 26–32 kb. Many of them are globally distributed and detectable in a wide range of animals and humans [5]. They are classified into 4 genera: alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacoronavirus [6, 7]. HCoV-OC43 (lineage A betacoronavirus) and HCoV-229E (alphacoronavirus) are known causative agents of the common cold and rarely cause severe respiratory diseases [8, 9]. In contrast, SARS coronavirus (SARS-CoV; lineage B betacoronavirus) caused the severe acute respiratory syndrome (SARS) outbreak during 2002–2003, affecting >8000 patients in >30 countries, with an overall fatality rate of 9.6% [10–13]. The discovery of SARS-CoV aroused a growing recognition that human coronaviruses are potentially highly pathogenic. The heightened awareness sparked an intense hunting for novel coronaviruses, which led to the subsequent identification of HCoV-NL63 (alphacoronavirus) and HCoV-HKU1 (lineage A betacoronavirus), that occasionally cause severe lower respiratory tract infections in young children, elderly individuals, and immunocompromised patients [14, 15]. Ten years after the SARS outbreak, another novel betacoronavirus of lineage C, MERS-CoV, might have jumped species barriers from bats to humans and caused an ongoing epidemic of severe human infections in the Middle East [6, 16–20]. Most patients with MERS presented with rapidly progressive pneumonia. Many of them developed multiorgan dysfunction, deranged coagulation profile, and hematological changes, including lymphopenia, neutrophilia, and thrombocytopenia [3]. Although the clinical syndromes of MERS resembled those described in severe SARS, MERS often had renal failure and substantially surpassed SARS in terms of crude fatality rate. Because of the increasing number of person-to-person transmission in both nosocomial and community settings [21, 22], there is a growing concern for another SARS-like pandemic.
Among all coronaviruses that cause human infections, SARS-CoV is the most extensively characterized. Human airway epithelial cells are the primary targets of SARS-CoV [23]. However, virus-infected macrophages contributed significantly to disease pathogenesis [23, 24]. Macrophages are the sentinel phagocytes of innate immune system, which functions to contain and eliminate pathogens, remove apoptotic cells, and present antigens to T cells. Macrophage-produced cytokines and chemokines modulate immune response, eradicate invading pathogens, and maintain tissue homeostasis [25]. Compared with other human primary macrophages, peripheral blood monocyte–derived macrophages (MDMs) are readily available and frequently used to recapitulate the initial innate immune responses in macrophages during viral infection.
Before the identification of MERS-CoV, SARS-CoV had been regarded as the most dangerous human coronavirus. However, a 5-fold higher fatality rate than that of SARS-CoV highlighted the possibly higher pathogenicity of MERS-CoV. Recent studies demonstrated that, similar to SARS-CoV, MERS-CoV was capable of infecting and productively replicating in primary human airway epithelial cells and ex vivo human lung tissues. Furthermore, MERS-CoV has a much broader tissue tropism than SARS-CoV and resembled SARS-CoV in its ability to suppress interferon production [26–29]. The high pathogenicity of MERS-CoV prompted us to explore the potential host-virus interaction in macrophages, the cells that were implicated in the pathogenesis of SARS [24]. Therefore, we studied viral infection/replication, cellular immune response, and antigen presentation in MERS-CoV–inoculated MDMs in comparison with SARS-CoV–inoculated MDMs. Further understanding of the pathogenesis of MERS will be important for optimizing treatment strategies for this highly fatal emerging infectious disease.
MATERIALS AND METHODS
Virus Culture and Virus Titration by a 50% Tissue Culture Infective Dose (TCID50) Assay
A clinical isolate of MERS-CoV was kindly provided by Fouchier et al [2]. The isolate was cultured in Vero cells (ATCC) with serum-free minimum Dulbecco's modified Eagle's medium (DMEM; Life Technologies), supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin, at 37°C with 5% CO2. SARS-CoV was cultured in FRhK-4 cells (ATCC) with the same medium. Two or 3 days after virus inoculation, culture supernatants were collected and stored in −80°C freezer in aliquots. For virus titration, aliquots of MERS-CoV and SARS-CoV were applied on confluent Vero cells in 96-well plates for TCID50 assay. Briefly, serial 10-fold dilutions of each virus were inoculated in a Vero cell monolayer in sextuplets and cultured in penicillin/streptomycin-supplemented DMEM. The plates were observed for cytopathic effect for 4–5 days. Viral titer was calculated with the Reed and Münch end point method. One TCID50 is interpreted as the amount of virus that causes cytopathic effect in 50% of inoculated wells.
MDM Culture and Virus Infection
Healthy adult blood samples were collected from Hong Kong Red Cross Blood Transfusion Service according to a protocol approved by the Institutional Review Board of the University of Hong Kong. Monocyte preparation and differentiation were performed according to a well-established protocol described previously [30]. The purity of the macrophages assessed by flow cytometry analysis of CD14 and CD68 was >93% on average [31]. For viral infection, MDMs in 24-well plates were inoculated with MERS-CoV or SARS-CoV at 2 TCID50/cell or were mock inoculated for 1 hour at 37°C. Supernatants and cell lysates were harvested 0, 5, 10, 24, 48, and 72 hour(s) after infection. TCID50 assay was performed on cell-free supernatants for viral titration. Cell lysates were extracted for RNA to detect viral RNA and cellular messenger RNA (mRNA). For immunostaining, MDMs were seeded in 24-well plates on glass coverslips. The cells were inoculated with MERS-CoV at 2 TCID50/cell. Twelve and 48 hours after infection, the cells were fixed with 4% paraformaldehyde and immunostained.
Quantification of Viral and Cellular RNA Transcript by Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Cellular RNA extraction and RT-qPCR were performed as described previously [32]. Viral RNA in the supernatant was extracted with the PureLink Viral RNA/DNA mini kit (Life Technologies). Virus-specific primer for MERS-CoV or SARS-CoV was used in reverse transcription to generate complementary DNAs (cDNAs) for the viruses and oligo(dT) for cellular cDNAs. Specific primers used in the qPCR analysis were listed in Supplementary Table 1. Cellular gene expression results were normalized to GAPDH and presented as the fold change in gene expression of virus-infected MDMs relative to that of mock-infected cells.
Ex Vivo Lung Tissue Culture and Virus Infection Experiment
The ex vivo lung tissue culture and virus infection experiment was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster. Fresh normal lung tissue was obtained from a patient undergoing resection of lung tumor. Lung tissue was cut into 2-mm3 cubes and subsequently infected by a MERS-CoV inoculum of 1 × 107 TCID50/mL or were mock-infected for 1 hour at 37°C. After inoculation, tissue cubes were maintained in DMEM/F12 medium supplemented with 10% human serum and penicillin/streptomycin before fixation and cryosectioning as described previously [32].
Immunofluorescence Staining
MERS-CoV was detected using our previously described guinea pig anti–nucleocapsid protein (NP) antibody [29] for 1 hour at room temperature, followed by fluorescein isothiocyanate–conjugated rabbit–anti-guinea pig immunoglobulin G (IgG; Life Technologies) or Alexa 594 goat–anti-guinea pig IgG (Abcam) as the secondary antibody. In tissue slides, macrophages were labeled with mouse anti-CD68 antibody (KP1, Abcam) followed by Alexa 488 goat–anti-mouse IgG (Abcam). All primary and secondary antibodies were diluted at 1:200. Finally, slides were mounted with ProLong Gold antifade reagent (Life Technologies) and examined with a Carl Zeiss LSM 710 microscope.
Statistical Analysis
Experimental results represented mean and standard errors of the mean from at least 3 different donors. Statistical comparison between the groups was performed by the Student t test, using GraphPad Prism 6. Differences were considered statistically significant when the P value was <.05.
RESULTS
Viral Infectivity and Replication of MERS-CoV in MDMs
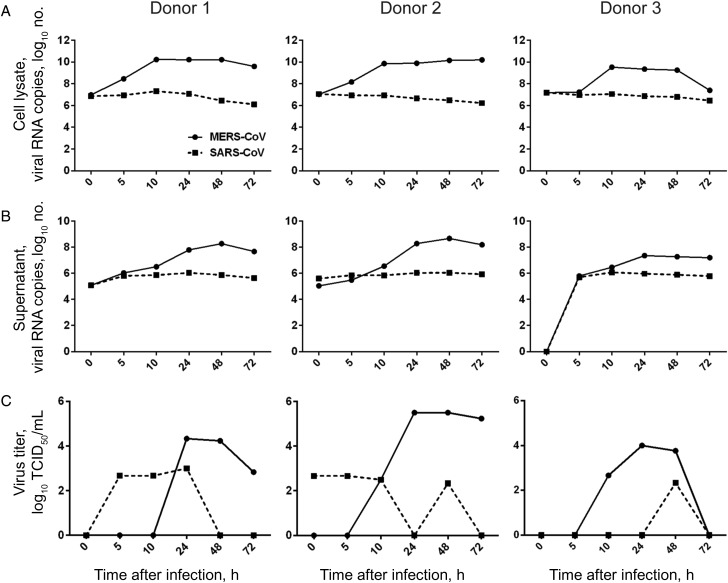
To understand the ability of MERS-CoV to infect human macrophages, we inoculated MDMs with MERS-CoV and SARS-CoV in parallel and observed the outcome of infection by examining levels of viral gene in the cell lysate and culture supernatant. As shown in Figure 1, the level of MERS-CoV RNA increased from 5 hours after infection in both cell lysate (Figure 1A) and supernatant (Figure 1B) of MDMs. Despite the variation among different donors, a 2–4-log increase in viral RNA level was consistently detected. MERS-CoV infection and replication in MDMs was confirmed with TCID50 assays, in which increasing titers of infectious virus were observed after inoculation. On the contrary, when the same dose of SARS-CoV was inoculated onto MDMs, no sign of viral replication were observed because the viral RNA levels gradually decreased in the cell lysate and culture supernatant. Infectious viruses detected in the supernatants of SARS-CoV–infected MDMs remained at low levels, indicating an abortive infection.
Figure 1.
Middle East respiratory syndrome coronavirus (MERS-CoV) replication kinetics in human monocyte–derived macrophages (MDMs), compared with SARS coronavirus (SARS-CoV). MERS-CoV or SARS-CoV was inoculated onto MDMs at 2 tissue culture infective doses (TCID50) per cell. At the indicated hours after infection, cell lysate (A) and culture supernatant (B) were collected to detect the levels of positive-strand viral RNA by reverse-transcription quantitative polymerase chain reaction. Viral titration of the culture supernatants was performed with a TCID50 assay (C). The results of 3 representative donors are demonstrated.
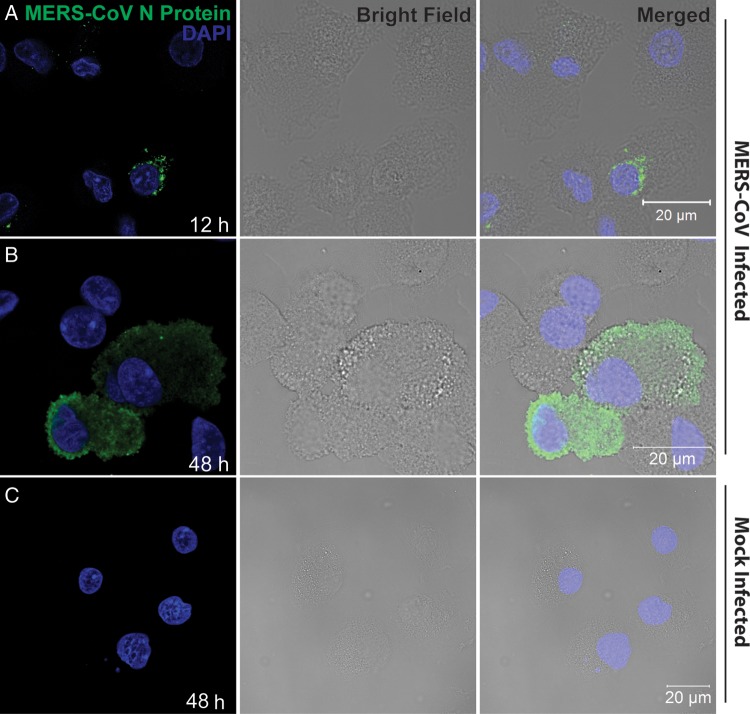
Replication of MERS-CoV in MDMs was further verified by immunofluorescence study. As shown in Figure 2, strong fluorescence signals for MERS-CoV NP were observed in the cytoplasm of virus-inoculated MDMs. Twelve hours after infection most of the infected cells displayed puncta-like positivity in the cytoplasm, whereas 48 hours after infection there were more cells displaying homogeneous immunoreactivity to NP throughout the cytoplasm. Moreover, there were more NP-positive cells 48 hours after infection than 12 hours after infection (data not shown), compatible with a productive infection and replication. Mock-infected MDMs did not display any immunoreactivity to NP. Moreover, replacing anti-NP sera with preimmune sera did not yield any positive signal in infected MDMs (data not shown). Collectively, we demonstrated the infection and replication capability of MERS-CoV in MDMs.
Figure 2.
Immunofluorescence staining of Middle East respiratory syndrome coronavirus (MERS-CoV) nucleocapsid protein (NP) in MERS-CoV–inoculated human monocyte–derived macrophages (MDMs). MDMs seeded on glass coverslips were inoculated with MERS-CoV at 2 tissue culture infective doses per cell or subjected to mock infection. Twelve hours (A) and 48 hours (B) after inoculation, cells were fixed, blocked, and permeabilized; incubated with anti-NP primary antibody for 1 hour; and stained with fluorescein isothiocyanate–conjugated secondary antibody for 1 hour. The slides were mounted with DAPI-containing mounting buffer and examined with a Carl Zeiss LSM 710 microscope. C, Findings for mock-inoculated MDMs that underwent the same procedure as that described for MERS-CoV–infected MDMs.
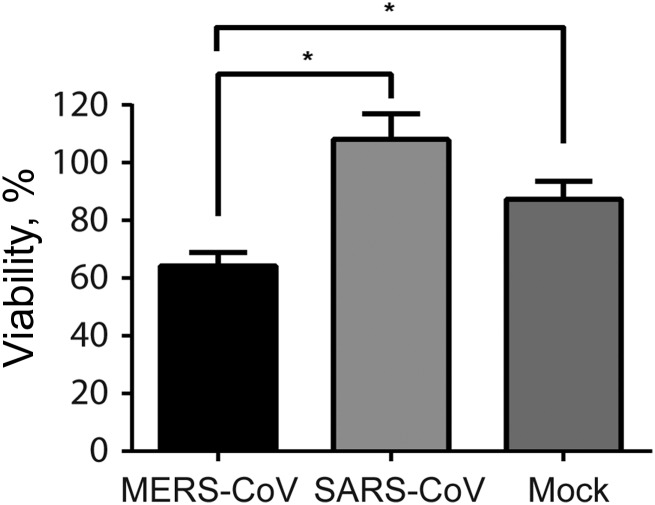
We next assessed the cell viability of MERS-CoV–infected, SARS-CoV–infected, and mock-infected MDMs by calculating the numbers of viable cells 48 hours after infection and comparing these data with values obtained right before infection. It was shown that approximately 87% of mock-infected MDMs remained viable 48 hours after infection, whereas the percentage of viable cells among MERS-CoV–infected MDMs was significantly reduced to 64% (Figure 3). In contrast, compared with mock infection, SARS-CoV appeared to have a protective effect from cell death. The difference, however, was not statistically significant. Our result suggested that MERS-CoV induced significantly higher cytotoxicity than SARS-CoV in infected MDMs.
Figure 3.
Cell viability of Middle East respiratory syndrome coronavirus (MERS-CoV)–infected and SARS coronavirus (SARS-CoV)–infected human monocyte–derived macrophages (MDMs). MDMs were seeded at 1.5 × 105/well in a 24-well plate and inoculated with MERS-CoV or SARS-CoV at 2 median tissue culture infective doses per cell or subjected to mock infection. Before infection and 48 hours after infection, MDMs were trypsinized, stained with trypan blue, and counted in duplicate with disposable hemocytometers. Viability represents the ratio of viable cell number of inoculated MDMs 48 hours after infection, compared with that before infection. Results are the mean and standard error of the mean of at least 3 representative donors. *P < .05, by the Student t test.
Innate Immune Response in MERS-CoV–Infected MDMs
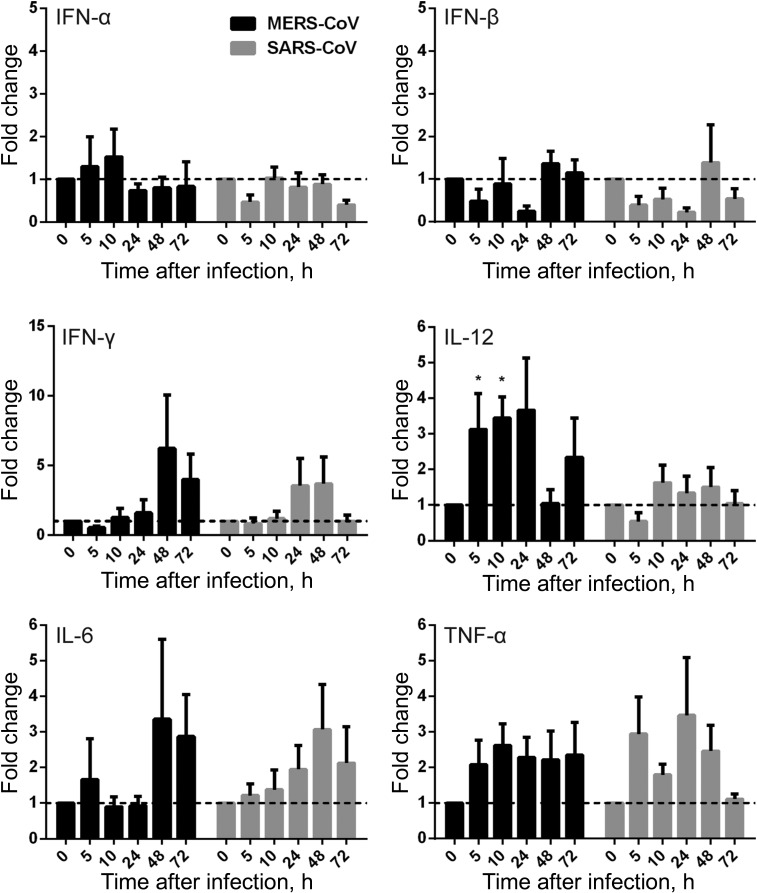
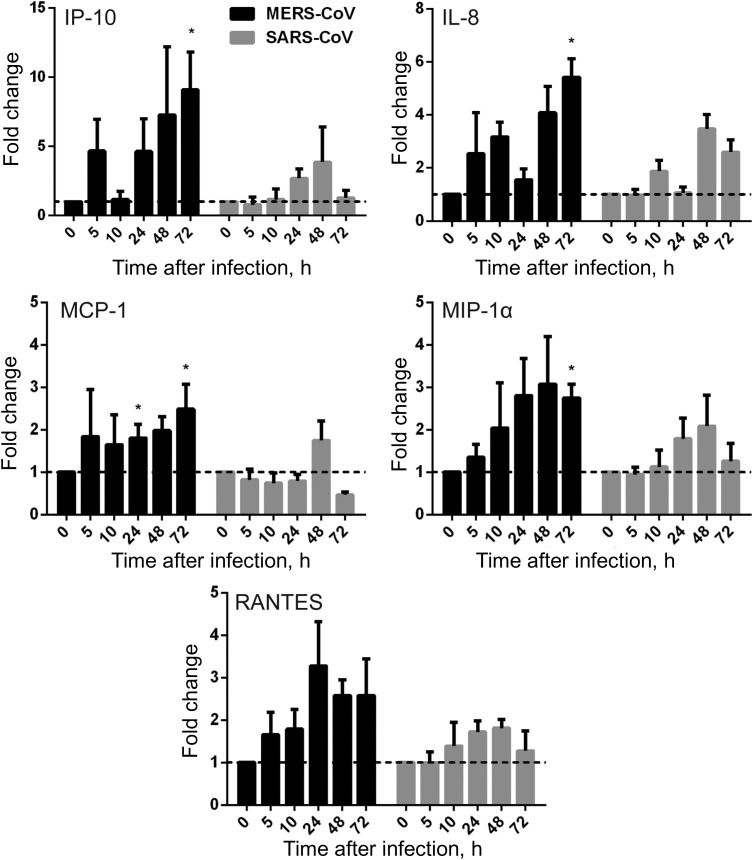
To evaluate the response of MDMs to active MERS-CoV replication, we assessed the mRNA expression levels of a series of antiviral cytokines, proinflammatory cytokines, and chemokines in MERS-CoV–infected, SARS-CoV–infected, and mock-infected MDMs. The expression of antiviral cytokines—type I interferon (interferon α [IFN-α] and IFN-β), type II interferon (IFN-γ), proinflammatory cytokines (tumor necrosis factor α [TNF-α], interleukin 6 [IL-6], and interleukin 12 [IL-12]), and chemokines (IP-10/CXCL-10, MCP-1/CCL-2, MIP-1α/CCL-3, RANTES/CCL-5, and interleukin 8 [IL-8])—was examined by RT-qPCR. As shown in Figure 4, both MERS-CoV–infected and SARS-CoV–infected MDMs displayed fairly weak and comparable expression levels of IFN-α and IFN-β, whereas these IFNs were highly induced in influenza A(H5N1)–infected MDMs (data not shown). However, IFN-γ expression was highly induced by both viruses. IFN-γ induction was more prominent in MERS-CoV–inoculated MDMs than in SARS-CoV–inoculated cells. TNF-α and IL-6 were comparably induced in both MERS-CoV–infected and SARS-CoV–infected cells. IL-12 was more significantly induced by MERS-CoV than SARS-CoV. Consistent with the high induction of IFN-γ, IP-10 (IFN-γ–inducible protein 10), which is secreted by a variety of cells in response to INF-γ, was highly upregulated by both viruses and was more significantly induced in MERS-CoV–infected cells (Figure 5).
Figure 4.
Cytokine response in Middle East respiratory syndrome coronavirus (MERS-CoV)–infected and SARS coronavirus (SARS-CoV)–infected human monocyte–derived macrophages (MDMs). MDMs were inoculated with MERS-CoV or SARS-CoV at 2 median tissue culture infective doses per cell or subjected to mock infection. At the indicated hours after infection, cells were lysed for RNA extraction, and reverse-transcription quantitative polymerase chain reaction was performed to detect the messenger RNA expression levels for antiviral and proinflammatory cytokines. Results were normalized to those for GAPDH and are presented as the fold change in gene expression of virus-infected MDMs in relation to that of the mock-infected MDMs. Data displayed in this figure represent mean and standard error of the mean from at least 3 different donors. Statistical analyses between MERS-CoV– and SARS-CoV–inoculated MDMs at different time points were performed using the Student t test. *P < .05. Abbreviations: IFN-α, interferon α; IFN-β, interferon β; IFN-γ, interferon γ; IL-6, interleukin 6; IL-12, interleukin 12; TNF-α, tumor necrosis factor α.
Figure 5.
Chemokine response in Middle East respiratory syndrome coronavirus (MERS-CoV)–infected and SARS coronavirus (SARS-CoV)–infected human monocyte–derived macrophages (MDMs). MDMs were inoculated with MERS-CoV or SARS-CoV at 2 median tissue culture infective doses per cell or subjected to mock infection. At the indicated hours after infection, cells were lysed for RNA extraction, and reverse-transcription quantitative polymerase chain reaction was performed to detect the messenger RNA expression levels for immunoregulatory chemokines. Results were normalized to those for GAPDH and are presented as the fold change in gene expression of virus-infected MDMs relative to that of the mock-infected MDMs. Statistical analyses between MERS-CoV– and SARS-CoV–infected MDMs at different time points were performed using the Student t test. *P < .05. Abbreviation: IL-8, interleukin 8.
Chemokines were extensively studied and implicated in the immunopathogenicity of SARS-CoV [33, 34]. Our data demonstrated that MERS-CoV induced significantly higher levels of MCP-1, MIP-1α, and IL-8 when compared with SARS-CoV (Figure 5). A trend of higher RANTES expression was documented in MERS-CoV–inoculated MDMs. Notably, all of these chemokines, as well as IP-10, exhibited a sustained induction curve from an early time point (5 hours after infection) to a late time point (72 hours after infection) in MERS-CoV–inoculated cells. The expression patterns of many of these cytokines/chemokines in SARS-CoV–infected MDMs in this study were in agreement with those in a previous report [35]. Collectively, we found that, similar to SARS-CoV, MERS-CoV was unable to induce effective antiviral IFN response (IFN-α and IFN-β). Both viruses similarly upregulated the expression of TNF-α and IL-6, but MERS-CoV induced more IFN-γ expression. Remarkably, MERS-CoV profoundly and sustainably induced immune cell–recruiting chemokines and cytokines, such as IP-10/CXCL-10, MCP-1/CCL-2, MIP-1α/CCL-3, RANTES/CCL-5, IL-8, and IL-12, in MDMs.
Antigen-Presenting Function of MERS-CoV–Infected MDMs
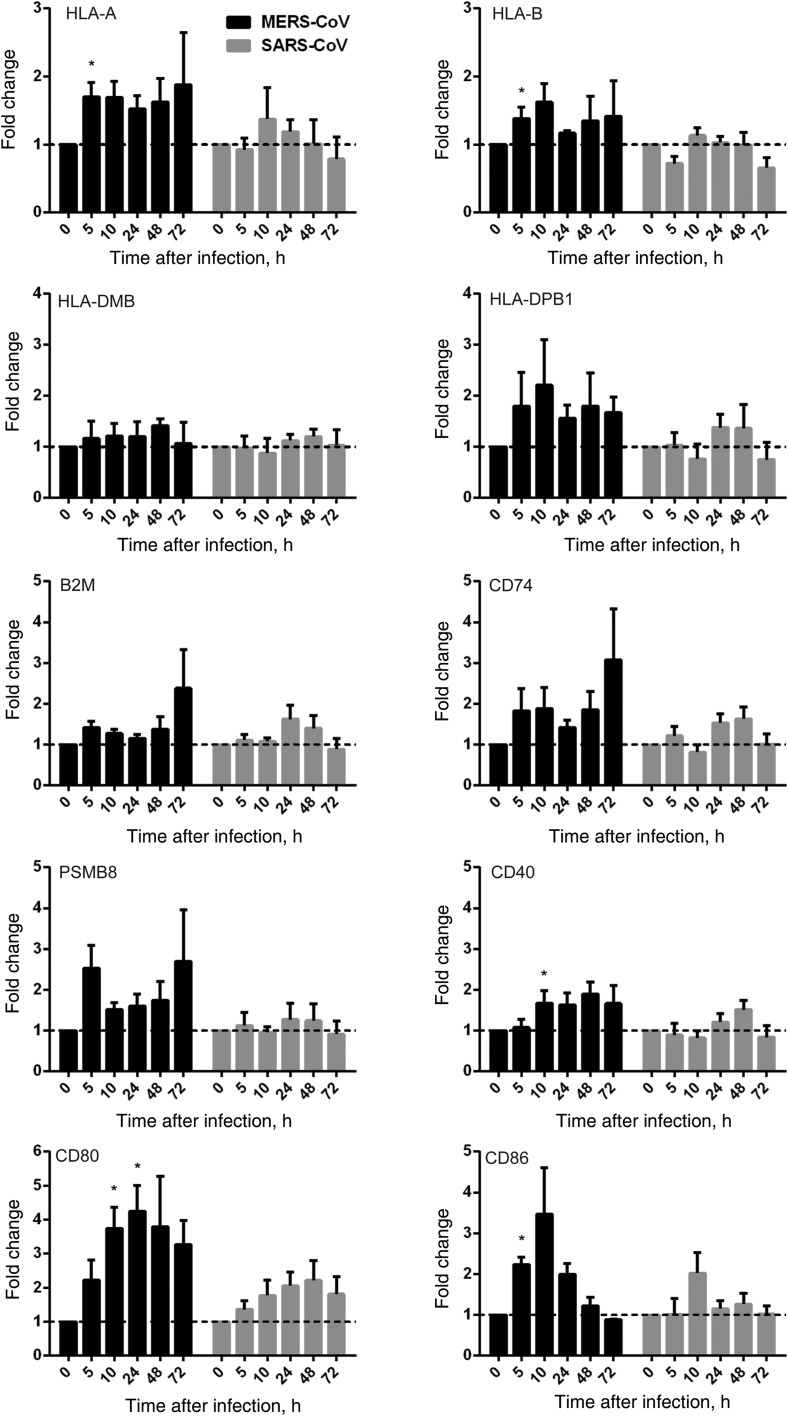
Apart from dendritic cells, macrophages are the foremost antigen-presenting cells in the host immune system. Viral antigens displayed on infected cells, via major histocompatibility complex (MHC) class I molecules, can be recognized by specific cytotoxic T cells that trigger the elimination of infected cells and thereby abrogate further viral replication and dissemination. On the other hand, macrophages present viral antigens, coupled with MHC class II molecules, to helper T cells and B cells to elicit the cellular and humoral immune responses. The effective host immune response is an important determinant for disease presentation and outcome of viral infection. A number of molecules for antigen processing (PSMB8 and CD74), antigen expression and stability in MHC class I complex (B2M), and costimulation and activation (CD40, CD80, and CD86), together with MHC class I (HLA-A and HLA-B) and MHC class II (HLA-DMB and HLA-DPB1) molecules, were examined upon inoculation with MERS-CoV, SARS-CoV, or mock control. We demonstrated that SARS-CoV largely failed to stimulate the expression of MHC class I and MHC class II molecules in inoculated MDMs. However, MERS-CoV modestly upregulated the expression of MHC class I–related molecules, HLA-A, HLA-B, B2M, and PSMB8. As for MHC class II–related genes, MERS-CoV failed to induce the expression of HLA-DMB but upregulated the expression of HLA-DPB1 and CD74. Additionally, MERS-CoV significantly induced higher levels of CD40, CD80, and CD86 expression than SARS-CoV (Figure 6). In summary, our data suggested that MERS-CoV triggered the higher expression of MHC class I–, MHC class II–, and costimulation-related genes in MDMs than SARS-CoV.
Figure 6.
Antigen presentation function of human monocyte–derived macrophages (MDMs) upon Middle East respiratory syndrome coronavirus (MERS-CoV) versus SARS coronavirus (SARS-CoV) infection. MDMs were inoculated with MERS-CoV or SARS-CoV at 2 median tissue culture infective doses per cell or subjected to mock infection. At the indicated hours after infection, cells lysates were collected for RNA extraction, and reverse-transcription quantitative polymerase chain reaction was performed to detect the messenger RNA expression levels for major histocompatibility complex (MHC) class I– and MHC class II–related molecules and costimulation molecules involved in antigen presentation. Results were normalized to those for GAPDH and are presented as the fold change in gene expression of virus-infected MDMs relative to that of the mock-infected MDMs.
MERS-CoV Infected Lung Macrophages in Ex Vivo Culture of Human Lung Tissues
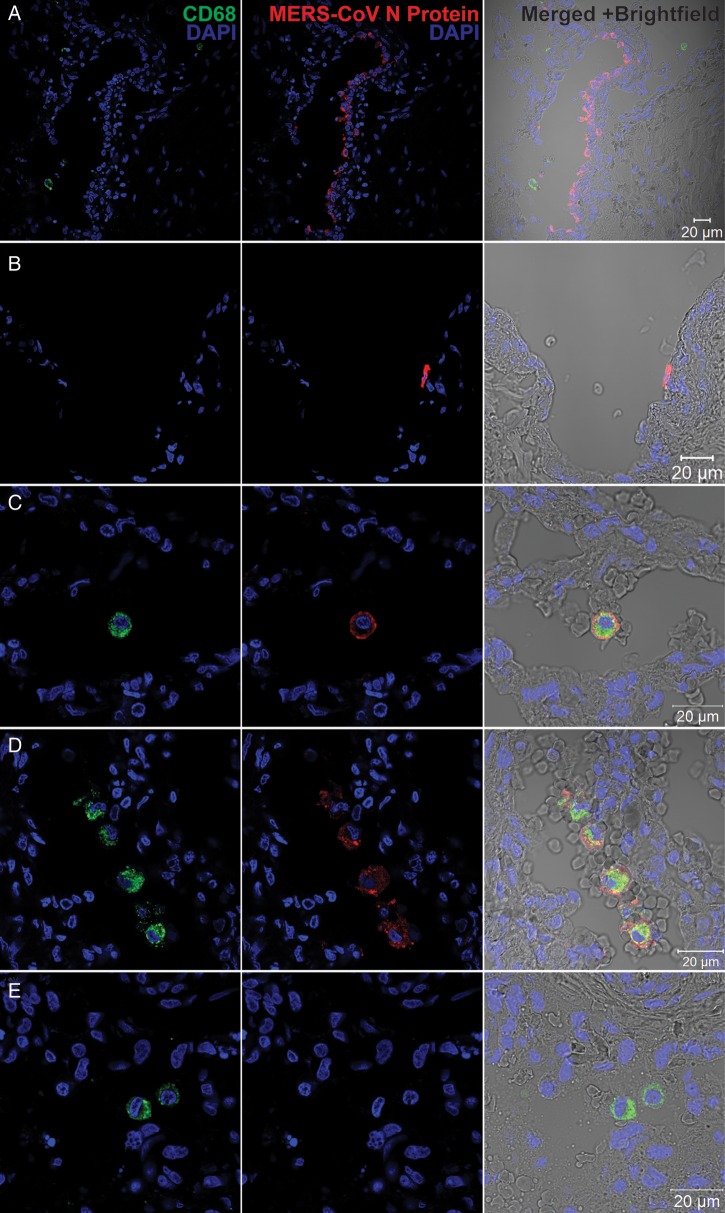
Although MDMs can be productively infected in vitro, they cannot fully represent the scenario of a real MERS-CoV infection. To this end, we performed the MERS-CoV inoculation in ex vivo culture of human lung tissue, where macrophages are abundant. We found that airway epithelial cells, predominantly pneumocytes and epithelial cells of terminal bronchioles, were infected, based on the cell and tissue morphology (Figure 7A). Moreover, intense immunoreactivity to MERS-CoV NP was observed in the endothelial cells of blood vessels in the interstitia (Figure 7B). We did not specifically define these NP-positive cells by costaining with epithelial cell– and endothelial cell–specific markers, because they were not the focus of this study. In stark contrast to a recent study [26], we found that lung macrophages can be infected by MERS-CoV, as shown by the colocalization of MERS-CoV NP with CD68 (Figure 7C and 7D), although the fluorescence intensity of NP was stronger in respiratory epithelial cells than in macrophages. There was no colocalization of NP and CD68 in mock-infected tissues (Figure 7E). On the basis of these observations, we demonstrated that human lung macrophages were susceptible to MERS-CoV infection ex vivo.
Figure 7.
Immunofluorescence double staining of Middle East respiratory syndrome coronavirus (MERS-CoV)–infected ex vivo lung tissue for MERS-CoV nucleocapsid protein (NP; red) and CD68 (green). Normal lung tissues were infected with MERS-CoV at 1 × 107 tissue culture infective doses per milliliter or subjected to mock infection for 1 hour at 37°C. A total of 48 hours after infection, tissues were fixed, cryoprotected, and cryosectioned. Slides were sequentially stained with guinea pig anti-NP sera, Alexa 694 goat–anti-guinea pig immunoglobulin G (IgG), mouse anti-CD68 antibody and Alexa 488 goat–anti-mouse IgG. Cell nuclei were counterstained by DAPI in the mounting buffer. Images were captured with a Carl Zeiss LSM 710 microscope. Immunoreactivity to NP was detected in epithelial cells of terminal bronchiole (A) and vascular endothelial (B) cells. C and D, Colocalization of MERS-CoV NP and CD68. E, A representative image of mock-infected lung tissue.
DISCUSSION
This study aimed to elucidate the pathogenesis of MERS-CoV. First, we found that the vascular endothelial cells in pulmonary interstitium and macrophages could be infected by MERS-CoV (Figure 7). This corroborated with the clinical observation that the virus was systemically disseminated in patients with MERS. A recent study described a patient with MERS who developed rapidly progressive pneumonia with respiratory failure, acute renal failure and septic shock, and eventually died [36]. The patient had high levels of viral RNA transcripts in bronchoalveolar lavage fluid. The patient's urine and stool samples also were positive for viral RNA. Despite a negative result for MERS-CoV in the blood sample of this patient, another study verified the presence of viral RNA transcripts in whole blood of a patient with MERS [21]. According to the gene expression profile in normal human tissue from public databases, DPP4, the MERS-CoV receptor [37], is abundantly expressed in a variety of human cells and tissues, which may facilitate the viral dissemination. Our previous study also provided strong evidence that cells derived from many human tissues can support MERS-CoV infection and replication [29]. Our present findings may provide the direct pathological basis for the systemic virus dissemination in MERS.
Second, immune cell–recruiting cytokines/chemokines were highly and endurably induced in MDMs upon MERS-CoV infection (Figures 4 and 5). Additionally, other proinflammatory cytokines also contributed to this chemoattractant cascade. This might lead to large number of immune cells infiltrating a patient's lower respiratory tract, causing severe inflammation and tissue damage. Clinically, most patients with MERS presented with rapidly progressive pneumonia with multilobar consolidations in radiographs and computed tomography scans. Cytological examination of bronchoalveolar lavage fluid often showed high number of neutrophils and macrophages [2, 21]. MERS-CoV–inoculated rhesus macaques displayed milder respiratory symptoms than patients with MERS. But histopathological examination of lung tissue showed acute pneumonia with infiltrating neutrophils and macrophages [38]. We believe cytokine/chemokine induction may have been responsible for the infiltration by immune cells and substantially contributed to the severe pneumonia and respiratory dysfunction in patients with MERS. Furthermore, most patients with MERS developed lymphopenia in the course of infection [2, 21, 22, 36]. This clinical manifestation was attributed to the immune cell recruitment and sequestration in the lower respiratory tract that was triggered by cytokine/chemokine induction. Additionally, we found that IP-10 and MCP-1, which can suppress proliferation of human myeloid progenitor cells [39], were highly induced upon MERS-CoV infection. The induction of these chemokines may inevitably aggravate the lymphopenia in patients with MERS. Taken together, our experimental findings are compatible with patient's clinical presentations and also elucidate the pathogenic mechanism of this life-threatening disease.
We compared MERS-CoV with SARS-CoV in 3 aspects: viral replication, cytokine/chemokine production, and antigen presentation in MDMs. It has been well recognized that SARS-CoV infection in MDMs was abortive [35, 40]. Unlike the abortive infection of SARS-CoV in MDMs, MERS-CoV can establish a productive infection in MDMs (Figures 1 and 2). The decreased viral titer in donors 1 and 3 at 72 hours after virus inoculation may have been due to excessive cell death over time. The infectivity of MERS-CoV to human macrophages was substantiated by the presence of viral NP in the macrophages of virus-inoculated ex vivo lung tissue (Figure 7). Macrophages are among the first line of host defense against viral invasion. Efficient viral replication in these cells implicates that the virus can overcome the host defense and is highly virulent. The productive viral replication in macrophages may turn these phagocytes into viral reservoirs and vehicles for further replication and dissemination, as shown for human immunodeficiency virus [41].
In response to viral infection, macrophages secrete proinflammatory cytokines/chemokines to activate various antiviral mechanisms. However, direct infection of these immune cells by virus may cause dysregulated induction of these inflammatory mediators, which could be damaging to neighboring tissues. Among these proinflammatory mediators, aberrant induction of IL-6 and TNF-α is almost a hallmark of severe respiratory viral infection, such as SARS and human infection by avian influenza viruses [34, 42]. The immunostimulatory effects of IFN-γ outweighed its antiviral effects in SARS [33]. IL-12 functions to stimulate the growth and activity of T cells and natural killer (NK) cells and induce the production of proinflammatory cytokines in these cells. MIP-1α, MCP-1, and IP-10 are notable for their strong chemoattraction of monocytes/macrophages, T cells, NK cells, and acute inflammatory cells to the site of infection. Apart from its chemotactic effect for granulocytes and T cells, RANTES can also promote T-cell and NK-cell activation. IL-8 shows strong chemotaxis for neutrophils and other granulocytes. It is also a potent inducer for other chemokines. A number of studies indicated that antiviral IFNs and cytokines were not induced in SARS-CoV–infected macrophages and dendritic cells [35, 43, 44], possibly because SARS-CoV evolved strategies to antagonize the innate responses [45]. In contrast, proinflammatory cytokines (IL-6 and TNF-α) and chemokines (IP-10, MIP-1α, and MCP-1) were upregulated [35, 43]. A plethora of clinical studies reported that levels of proinflammatory cytokines/chemokines, including IL-6, IL-8, IFN-γ, MCP-1, and IP-10, were highly elevated in patients with SARS, some of which were correlated with disease severity or mortality [33, 34]. Additionally, macrophages in the patient's lung tissue were believed to be the producer of cytokine storm and underlie the pathogenesis of SARS [24]. We compared and found that many of the cytokines/chemokines in MERS-CoV–infected MDMs shared a similar expression profile to SARS-CoV–infected cells. Notably, immune cell–recruiting chemokines and immunostimulating cytokines were induced at a significantly higher magnitude and prolonged interval by MERS-CoV, compared with SARS-CoV. Therefore, it is conceivable that immunopathogenesis, which is operative in SARS [46], may be aggravated in MERS, causing more severe disease and higher fatality. The detailed expression profile of the cytokine/chemokine responses in MERS-CoV–infected and SARS-CoV–infected MDMs should be investigated with microarray analysis.
The expression of MHC class I–, MHC class II–, and costimulation-related genes were modestly induced in MERS-CoV–infected MDMs and were marginally stronger than that in SARS-CoV–infected cells (Figure 6). This contrasts with a recent report showing a decreased antigen-presentation profile in a MERS-CoV–infected lung cancer cell line [47]. The disagreement is possibly attributable to differences in the properties of MDMs and the lung cancer cell line. The full scenario of antigen presentation in MERS definitely warrants further investigation. Nevertheless, based on our results, the increase in antigen presentation in MERS-CoV–infected MDMs may potentially contribute to exaggerate the immunopathology.
Taken together, we demonstrate that MERS-CoV can infect and effectively replicate in MDMs. MERS-CoV replication in MDMs causes extensive cytotoxicity and induces the expression of a number of proinflammatory cytokines/chemokines. The mixture of high viral replication and immune-mediated pathology could be important factors in the pathogenesis of MERS.
Supplementary Material
Notes
Acknowledgments. We thank the Core Imaging Facility, Li Ka Shing Faculty of Medicine, University of Hong Kong, for facilitation of the study.
Financial support. This work was supported by the Providence Foundation Limited in memory of the late Dr Lui Hac Minh and by the HKSAR Health and Medical Research Fund.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. The results shown in this paper have not been presented in any meetings.
References
- 1.Chan JF, Li KS, To KK, Cheng VC, Chen H, Yuen KY. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65:477–89. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Chan JF, Lau S, Woo PC. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns”. J Formos Med Assoc. 2013;112:372–81. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV)—update. 1 August 2013. http://www.who.int/csr/don/2013_08_01/en/index.html . Accessed 27 September 2013.
- 5.Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 2009;234:1117–27. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 6.Chan JF, To KK, Tse H, Jin DY, Yuen KY. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–55. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo PC, Wang M, Lau SK, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–85. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birch CJ, Clothier HJ, Seccull A, et al. Human coronavirus OC43 causes influenza-like illness in residents and staff of aged-care facilities in Melbourne, Australia. Epidemiol Infect. 2005;133:273–7. doi: 10.1017/s0950268804003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–41. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 12.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88–97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–94. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo PC, Lau SK, Chu CM, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Assiri A, McGeer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reusken CB, Haagmans BL, Muller MA, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annan A, Baldwin HJ, Corman VM, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–9. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau SK, Li KS, Tsang AK, et al. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol. 2013;87:8638–50. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan KH, Chan JF, Tse H, et al. Cross-reactive antibodies in convalescent SARS patients’ sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013;67:130–40. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guery B, Poissy J, el Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–72. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368:2487–94. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 23.Gu J, Gong E, Zhang B, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–24. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–8. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan RW, Chan MC, Agnihothram S, et al. Tropism and innate immune responses of the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87:6604–14. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kindler E, Jonsdottir HR, Muth D, et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4:e00611–12. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zielecki F, Weber M, Eickmann M, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87:5300–4. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan JF, Chan KH, Choi GK, et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis. 2013;207:1743–52. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu H, Wang JJ, Qi M, et al. The intracellular virus-containing compartments in primary human macrophages are largely inaccessible to antibodies and small molecules. PLoS One. 2012;7:e35297. doi: 10.1371/journal.pone.0035297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu H, Wang JJ, Qi M, et al. Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe. 2012;12:360–72. doi: 10.1016/j.chom.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Tan T, Tian Y, et al. Kruppel-like factor 15 activates hepatitis B virus gene expression and replication. Hepatology. 2011;54:109–21. doi: 10.1002/hep.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang KJ, Su IJ, Theron M, et al. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol. 2005;75:185–94. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Y, Xu J, Zhou C, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171:850–7. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 35.Cheung CY, Poon LL, Ng IH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–26. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N Engl J Med. 2013;368:1560–2. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broxmeyer HE, Sherry B, Cooper S, et al. Comparative analysis of the human macrophage inflammatory protein family of cytokines (chemokines) on proliferation of human myeloid progenitor cells. Interacting effects involving suppression, synergistic suppression, and blocking of suppression. J Immunol. 1993;150:3448–58. [PubMed] [Google Scholar]
- 40.Tseng CT, Perrone LA, Zhu H, Makino S, Peters CJ. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J Immunol. 2005;174:7977–85. doi: 10.4049/jimmunol.174.12.7977. [DOI] [PubMed] [Google Scholar]
- 41.Orenstein JM. The macrophage in HIV infection. Immunobiology. 2001;204:598–602. doi: 10.1078/0171-2985-00098. [DOI] [PubMed] [Google Scholar]
- 42.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law HK, Cheung CY, Ng HY, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–74. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziegler T, Matikainen S, Ronkko E, et al. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J Virol. 2005;79:13800–5. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–57. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5:917–27. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Josset L, Menachery VD, Gralinski LE, et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4:e00165–13. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.