Abstract
The emerging novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) was recently isolated from patients with severe pneumonia and renal failure and was associated with an unexplained high crude fatality rate of 56%. We performed a cell line susceptibility study with 28 cell lines. HCoV-EMC was found to infect the human respiratory tract (polarized airway epithelium cell line Calu-3, embryonic fibroblast cell line HFL, and lung adenocarcinoma cell line A549), kidney (embryonic kidney cell line HEK), intestinal tract (colorectal adenocarcinoma cell line Caco-2), liver cells (hepatocellular carcinoma cell line Huh-7), and histiocytes (malignant histiocytoma cell line His-1), as evident by detection of high or increasing viral load in culture supernatants, detection of viral nucleoprotein expression by immunostaining, and/or detection of cytopathic effects. Although an infected human neuronal cell line (NT2) and infected monocyte and T lymphocyte cell lines (THP-1, U937, and H9) had increased viral loads, their relatively lower viral production corroborated with absent nucleoprotein expression and cytopathic effects. This range of human tissue tropism is broader than that for all other HCoVs, including SARS coronavirus, HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63, which may explain the high mortality associated with this disease. A recent cell line susceptibility study showed that HCoV-EMC can infect primate, porcine, and bat cells and therefore may jump interspecies barriers. We found that HCoV-EMC can also infect civet lung fibroblast and rabbit kidney cell lines. These findings have important implications for the diagnosis, pathogenesis, and transmission of HCoV-EMC.
Keywords: coronavirus, novel, HCoV-EMC, cell line, susceptibility
(See the editorial commentary by McIntosh on pages 1630–2.)
Coronaviruses (CoVs) were first identified as causative agents of respiratory tract infections, gastroenteritis, hepatitis, and encephalomyelitis in birds and mammals [1]. The first 2 identified human CoVs (HCoVs), HCoV-229E and HCoV-OC43, cause self-limiting respiratory infections, such as the common cold, in humans [2, 3]. The potential of CoVs in causing severe human infections was not recognized until the emergence of SARS-CoV, which produced an epidemic in 2003, leading to 774 deaths in >30 countries [4–9]. In addition to a high crude fatality rate of around 11%, protean clinical manifestations not limited to the upper respiratory tract were observed during severe acute respiratory syndrome (SARS). These included severe acute pneumonia, pulmonary vasculitis and thrombosis, gastroenteritis, hepatitis, acute renal impairment, and impaired coagulation [9, 10]. Following the SARS epidemic, 2 more HCoVs, HCoV-NL63 [11, 12] and HCoV-HKU1 [13], were identified as causes of respiratory tract infections that were usually mild in clinical severity. On 23 September 2012, the World Health Organization (WHO) reported 2 cases of severe community-acquired pneumonia, which were subsequently confirmed to be caused by a novel HCoV, human betacoronavirus 2c EMC/2012 (HCoV-EMC) [14–16]. Phylogenetic analysis showed that HCoV-EMC was closely related to the bat CoVs Tylonycteris-bat-CoV-HKU4 (Ty-BatCoV-HKU4) and Pipistrellus-bat-CoV-HKU5 (Pi-BatCoV-HKU5), found in the lesser bamboo bat (Tylonycteris pachypus) and Japanese pipistrelle bat (Pipistrellus abramus), respectively, of Hong Kong, China [17–19]. As of 30 November 2012, the total number of laboratory-confirmed cases of HCoV-EMC infection reported to the WHO has increased to 9, with 5 deaths [20]. Because none of the 2400 residents in the Kingdom of Saudi Arabia had serum antibody against HCoV-EMC [21], a crude fatality rate of 56% is alarmingly similar to that of human influenza A(H5N1) pneumonia [22, 23]. The severe clinical manifestations of acute respiratory and renal failure and the high fatality fate were exceptional among all of the known HCoVs. Although these laboratory-confirmed cases were concentrated in the Middle East, with 5 cases in the Kingdom of Saudi Arabia and 2 cases each in Qatar and Jordan, research preparedness against another SARS-like epidemic is urgently needed, as deaths of exposed healthcare workers were suspected in Jordan [20]. Critical information on the pathogenesis of the unusual clinical severity of HCoV-EMC infection, the viral load and antigen expression patterns, and the likelihood of interspecies jumping of the virus from animals to humans must be ascertained to optimize laboratory testing protocols, treatment options, and infection control strategies. In this study, we provide insight into these key questions by correlating the differential cell line susceptibility, species tropism, viral replication efficiency, and antigen expression patterns with the clinical and epidemiologic characteristics of HCoV-EMC.
METHODS
Viral Isolate
A clinical isolate of HCoV-EMC described elsewhere [15] was kindly provided by R. Fouchier, A. Zaki, and colleagues. The isolate was amplified by 1 additional passage in Vero cell lines to make working stocks of the virus. All experimental protocols involving the live HCoV-EMC isolate followed the standard operating procedures of the approved biosafety level 3 facility, as we previously described [24].
Viral Culture
Twenty-eight cell lines derived from different tissues or organs and host species (Table 1) were prepared in 24-well plates and inoculated with 1 multiplicity of infection of HCoV-EMC for 1 hour. Nonattached virus was removed by washing the cells twice in serum-free minimum essential medium (MEM; Gibco, NY). The monolayer cells and suspension cells were maintained in MEM with 1% fetal calf serum or in Roswell Park Memorial Institute medium (Gibco) with 2% fetal calf serum. All infected cell lines were incubated at 37°C for 5 days. Cytopathic effects (CPEs) were examined on days 1, 3, and 5 by inverted light microscopy. For monolayer cell lines, the effect of trypsin on HCoV-EMC was examined by additional inoculation in serum-free medium supplemented with tosyl phenylalanyl chloromethyl ketone (TPCK)–treated trypsin (2 µg/mL) (Sigma–Aldrich, St. Louis, MO).
Table 1.
Human and Nonhuman Cell Lines Used in the Present Study
| Organism, Anatomic Site, Cell Line | Abbreviation | Source |
|---|---|---|
| Human | ||
| Upper and lower respiratory tract | ||
| Laryngeal epidermoid carcinoma | Hep-2 | ATCC no. CCL-23 |
| Lung adenocarcinoma | A549 | ATCC no. CCL-185 |
| Lung adenocarcinoma | Calu-3 | ATCC no. HTB-55 |
| Embryonic lung fibroblasts | HFL | In-house development |
| Gastrointestinal tract | ||
| Colorectal adenocarcinoma | Caco-2 | ATCC no. HTB-37 |
| Liver | ||
| Hepatocellular carcinoma | Huh-7 | JCRB0403, JCRB cell bank of Okayama University |
| Genitourinary tract | ||
| Cervical adenocarcinoma | HeLa | ATCC no. CCL-2.2 |
| Fetal kidney | HEK | In-house development |
| Neuromuscular cells | ||
| Neuron-committed teratocarcinoma | NT2 | ATCC no. CRL-1973 |
| Rhabdomyosarcoma | RD | ATCC no. CCL-136 |
| Immune cells | ||
| Peripheral blood monocytes from acute monocytic leukemia | THP-1 | ATCC no. TIB-202 |
| Monocytes from histiocytic lymphoma | U937 | ATCC no. CRL-1593.2 |
| B lymphocytes from Burkitt lymphoma | Raji | ATCC no. CCL-86 |
| T lymphocytes from T-cell leukemia | H9 | ATCC no. HTB-176 |
| Malignant histiocytoma | His-1 | In-house development |
| Nonhuman | ||
| Mammals | ||
| Rhesus monkey kidney | LLC-MK2 | ATCC no. CCL-7 |
| African green monkey kidney | Vero | ATCC no. CCL-81 |
| African green monkey kidney (clone of Vero-76) | Vero-E6 | ATCC no. CRL-1586 |
| Madin-Darby canine kidney | MDCK | ATCC no. CCL-34 |
| Feline kidney | CRFK | ATCC no. CCL-94 |
| Porcine kidney | PK-15 | ATCC no. CCL-33 |
| Rabbit kidney | RK-13 | ATCC no. CCL-37 |
| Civet lung fibroblasts | CL-1 | In-house development |
| Primary mouse embryonic fibroblasts | 3T3 | ATCC no. CCL-92 |
| Rattus norvegicus kidney | RK3E | ATCC no. CRL-1895 |
| Rattus norvegicus kidney | RMC | ATCC no. CRL-2573 |
| Chicken | ||
| Chicken fibroblasts | DF-1 | ATCC no. CRL-12203 |
| Insect | ||
| Aedes albopictus | C6-36 | ATCC no. CRL-1660 |
RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total nucleic acid was extracted from culture supernatants of the 28 cell lines infected by HCoV-EMC on days 1, 3 and 5, using the NucliSens easyMAG instrument (bioMerieux, the Netherlands) as we previously described [25]. Briefly, 250 µL of supernatant was added to 2 mL of lysis buffer and incubated for 10 minutes at room temperature. Nucleic acid was eluted after automatic magnetic separation. A real-time 1-step quantitative RT-PCR assay was used for the detection of HCoV-EMC, using the Invitrogen SuperScript III Platinum One-Step Quantitative Kit in a 7500 Sequence Detection System (Applied Biosystems, Foster City, CA) [26]. Briefly, 5 µL of purified total nucleic acid was amplified in a 25-µL reaction containing 0.5 µL of Superscript III Reverse Transcriptase/Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.05 µL of ROX reference dye (25 mM), 12.5 µL of 2X reaction buffer, 1.5 µM forward primer (5′-CAAAACCTTCCCTAAGAAGGAAAAG-3′; corresponding to nucleotides 29639–29663), 1.5 µM reverse primer (5′-GCTCCTTTGGAGGTTCAGACAT-3′; corresponding to nucleotides 29720–29700), and probe 100 nM (5′ FAM-ACAAAAGGCACCAAAAGAAGAATCAACAGACC BHQ1-3; corresponding to nucleotides 29666–29697) and designed by multiple alignment of the N protein gene sequences of HCoV-EMC and other betacoronaviruses available in GenBank [accession no. JX869059.1]. Reactions were incubated at 50°C for 30 minutes followed by 95°C for 2 minutes and then underwent thermal cycling for 50 cycles (at 95°C for 15 seconds and at 55°C for 30 seconds). A series of 6 log10 dilutions equivalent to 1 × 101 to 1 × 106 copies per reaction mixture were prepared to generate calibration curves and were run in parallel with the test samples.
Cloning and Purification of His6-Tagged Recombinant N Protein of HCoV-EMC
Primers (5′-GGAATTCCATATGATGGCATCCCCTGCTGCACCTC-3′ and 5′-ATAAGAATGCGGCCGCATCAGTGTTAACATCAATCATT-3′) were used to amplify the gene encoding the N protein of HCoV-EMC by RT-PCR [27]. The sequence encoding amino acid residues 1–413 of the N protein was amplified and cloned into the NdeI and NotI sites of expression vector pETH in frame and upstream of the series of 6 histidine residues. The recombinant N protein was expressed in Escherichia coli and purified by using Ni-nitrilotriacetic acid affinity chromatography (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Expression of the recombinant N protein was confirmed by Western blot analysis using mouse anti-His monoclonal antibody (Sigma–Aldrich).
Preparation of Specific Antibody Against the N Protein of HCoV-EMC
One hundred micrograms of purified recombinant N protein was mixed with an equal volume of complete Freund adjuvant (Sigma–Aldrich) and injected subcutaneously into Dunkin-Hartley guinea pigs. Incomplete Freund adjuvant (Sigma–Aldrich) was used in subsequent injections at 14-day intervals. Serum samples were taken after the fourth injection [27].
Antigen Detection of Infected Cell Lines by Immunofluorescence (IF)
Cell smears on days 1, 3, and 5 were prepared and fixed in chilled acetone at −20°C for 10 minutes. The fixed cells were incubated with guinea pig antiserum against the HCoV-EMC N protein, followed by fluorescein isothiocyanate–conjugated rabbit anti-guinea pig immunoglobulin G (Invitrogen). Cells were then examined under a fluorescence microscope. Uninoculated cells were used as negative control. The percentages of positive cells were recorded.
Statistical Analysis
The Student t test was used to compare the mean viral load of the different cell lines on days 1, 3, and 5 with the mean baseline viral load on day 0. All calculations were based on log-transformed viral load. A P value of < .05 was considered statistically significant. Computation was performed using Predictive Analytics Software, version 18.0 for Windows.
RESULTS
Human Cell Lines
A total of 15 human cell lines were tested (Table 1). Eleven of the 15 cells lines showed significantly increased mean viral loads consistently, as compared to the baseline mean viral load, by quantitative RT-PCR (Table 2). These included lower airway (A549, Calu-3, and HFL), intestinal tract (Caco-2), liver (Huh-7), kidney (HEK), neuronal (NT2), monocyte (THP-1 and U937), T lymphocyte (H9), and histiocyte (His-1) cell lines. Seven of these 11 cell lines, namely A549, Calu-3, HFL, Caco-2, Huh-7, HEK, and His-1, showed viral N protein expression by IF in addition to a high viral load (Table 3). Most of these 7 cell lines had peak N protein expression detected by IF on days 3–5 after infection (Figure 1). Although the Huh-7 cell line had a high mean viral load detected by quantitative RT-PCR, N protein expression detected by IF in this cell line was consistently lower than that in other cell lines with similar or even lower mean viral loads detected by quantitative RT-PCR. The viral N protein expression in the A549 cell line was less prominent than that in the other 6 cell lines, and a CPE was not observed. In the other lower airway cell lines (Calu-3 and HFL) and the HEK cell line, CPE was observed beginning on day 3 after infection. In the Caco-2, Huh-7, and His-1 cell lines, CPE was observed as early as day 1 after infection (Figure 2). The rapid onset of CPE in some of these cell lines on day 1 after infection correlated with the subsequent lowering of viral load on days 3 and 5 because most of the cells were unable to support further viral replication. Syncytia formation was prominent in human Calu-3 and Caco-2 cell lines. Marked shrinkage of infected Huh-7 cells, which gave the appearance of cellular aggregates, was observed on day 3 (Figure 2). Although the mean viral loads in the monocyte cell lines (THP-1 and U937), H9 cell line and NT2 cell line were statistically significantly higher than at baseline, none of these cell lines had N protein expression detected by IF or CPE. Laryngeal (Hep-2), cervical (HeLa), muscle (RD), and B lymphocyte (Raji) cell lines had the lowest mean viral load detected by quantitative RT-PCR and did not have N protein expression detected by IF or CPE. No significant difference in mean viral load was noted in monolayer cell lines after additional inoculation in serum-free medium supplemented with TPCK-treated trypsin.
Table 2.
Differential Cell Line Susceptibility to Human Betacoronavirus 2c EMC/2012, as Defined by Viral Load, on Days 1, 3, and 5 After Infection
| Viral Load, log10 copies/mL, mean ± SDa,b |
|||||||
|---|---|---|---|---|---|---|---|
| Cell Line | Day 0 | Day 1 | P | Day 3 | P | Day 5 | P |
| Human | |||||||
| Respiratory tract | |||||||
| Hep-2 | 5.87 ± 0.10 | 5.84 ± 0.04 | .784 | 5.85 ± 0.02 | .823 | 5.85 ± 0.10 | .859 |
| A549 | 5.90 ± 0.16 | 6.97 ± 0.11 | .025 | 7.18 ± 0.04 | .050 | 7.45 ± 0.31 | .045 |
| Calu-3 | 5.62 ± 0.04 | 9.63 ± 0.04 | <.001 | 10.42 ± 0.22 | .016 | 10.25 ± 0.06 | .001 |
| HFL | 5.72 ± 0.02 | 9.54 ± 0.35 | .041 | 10.73 ± 0.03 | .002 | 10.45 ± 0.04 | .003 |
| Gastrointestinal tract | |||||||
| Caco-2 | 5.82 ± 0.04 | 10.19 ± 0.61 | .060 | 10.49 ± 0.17 | .009 | 10.26 ± 0.16 | .018 |
| Liver | |||||||
| Huh-7 | 5.87 ± 0.07 | 10.47 ± 0.22 | .011 | 9.56 ± 0.11 | .002 | 9.63 ± 0.02 | .006 |
| Genitourinary tract | |||||||
| HeLa | 5.75 ± 0.06 | 5.94 ± 0.08 | .124 | 5.86 ± 0.17 | .522 | 5.99 ± 0.08 | .079 |
| HEK | 5.82 ± 0.05 | 10.45 ± 0.25 | .015 | 9.97 ± 0.17 | .012 | 9.90 ± 0.03 | .001 |
| Neuromuscular cells | |||||||
| NT2 | 5.72 ± 0.04 | 7.83 ± 0.02 | .002 | 8.22 ± 0.04 | <.001 | 8.40 ± 0.14 | .015 |
| RD | 5.66 ± 0.04 | 6.68 ± 0.24 | .098 | 6.90 ± 0.02 | .003 | 6.78 ± 0.01 | .013 |
| Immune cells | |||||||
| THP-1 | 6.75 ± 0.01 | 7.03 ± 0.02 | .020 | 7.28 ± 0.06 | .044 | 7.40 ± 0.06 | .036 |
| U937 | 6.70 ± 0.06 | 7.10 ± 0.07 | .028 | 7.19 ± 0.04 | .016 | 7.25 ± 0.07 | .015 |
| Raji | 6.71 ± 0.05 | 6.85 ± 0.02 | .106 | 6.76 ± 0.08 | .528 | 7.05 ± 0.04 | .019 |
| H9 | 6.52 ± 0.03 | 6.55 ± 0.57 | .592 | 6.73 ± 0.04 | .038 | 6.71 ± 0.03 | .020 |
| His-1 | 5.55 ± 0.04 | 9.44 ± 0.08 | .001 | 9.27 ± 0.05 | <.001 | 9.12 ± 0.01 | .004 |
| Nonhuman | |||||||
| Mammals | |||||||
| LLC-MK2 | 5.94 ± 0.04 | 10.73 ± 0.20 | .017 | 10.61 ± 0.10 | .009 | 10.34 ± 0.05 | .001 |
| Vero | 5.73 ± 0.04 | 9.61 ± 0.57 | .064 | 10.15 ± 0.36 | .034 | 10.93 ± 0.18 | .014 |
| Vero-E6 | 5.57 ± 0.08 | 9.34 ± 0.12 | .002 | 10.79 ± 0.09 | .002 | 10.55 ± 0.04 | <.001 |
| MDCK | 5.65 ± 0.04 | 5.78 ± 0.21 | .522 | 5.65 ± 0.00 | .772 | 5.72 ± 0.01 | .190 |
| CRFK | 5.43 ± 0.08 | 5.91 ± 0.01 | .072 | 5.85 ± 0.07 | .035 | 5.79 ± 0.03 | .080 |
| PK-15 | 5.39 ± 0.06 | 8.30 ± 0.10 | .003 | 10.16 ± 0.30 | .025 | 10.12 ± 0.08 | <.001 |
| RK-13 | 5.37 ± 0.05 | 6.12 ± 0.08 | .013 | 7.40 ± 0.05 | .036 | 7.75 ± 0.02 | .003 |
| CL-1 | 5.56 ± 0.05 | 7.17 ± 0.01 | .008 | 7.52 ± 0.03 | .001 | 8.19 ± 0.86 | .144 |
| 3T3 | 5.40 ± 0.01 | 5.47 ± 0.15 | .669 | 5.18 ± 0.03 | .044 | 5.22 ± 0.10 | .229 |
| RK3E | 5.31 ± 0.07 | 5.40 ± 0.05 | .302 | 5.34 ± 0.02 | .639 | 5.24 ± 0.01 | .443 |
| RMC | 5.29 ± 0.08 | 5.80 ± 0.21 | .154 | 5.32 ± 0.07 | .713 | 5.36 ± 0.09 | .516 |
| Chicken | |||||||
| DF-1 | 5.92 ± 0.02 | 6.32 ± 0.14 | .115 | 6.24 ± 0.04 | .041 | 6.20 ± 0.00 | .011 |
| Insect | |||||||
| C6-36 | 5.93 ± 0.01 | 6.50 ± 0.06 | .049 | 6.37 ± 0.11 | .110 | 6.36 ± 0.11 | .111 |
a All experiments were done in duplicate. The mean viral loads on days 1, 3, and 5 were compared with the mean baseline viral load on day 0.
b No significant difference in mean viral load was noted in monolayer cell lines after additional inoculation in serum-free medium supplemented with tosyl phenylalanyl chloromethyl ketone–treated trypsin (2 µg/mL).
Table 3.
Differential Cell Line Susceptibility to Human Betacoronavirus 2c EMC/2012, as Defined by Cytopathic Effect (CPE) and Immunofluorescent Antigen Staining, on Days 1, 3, and 5 After Infection
| CPE, Gradea |
Immunofluorescenceb |
|||||
|---|---|---|---|---|---|---|
| Cell lines | Day 1 | Day 3 | Day 5 | Day 1 | Day 3 | Day 5 |
| Human | ||||||
| Respiratory tract | ||||||
| Hep-2 | N | N | N | N | N | N |
| A549 | N | N | N | N | 1 | 5 |
| Calu-3 | N | 4+ | 4+ | 20 | 40 | 20 |
| HFL | N | 3+ | 4+ | N | 90 | 100 |
| Gastrointestinal tract | ||||||
| Caco-2 | 2+ | 3+ | 3+ | 5 | 80 | 100 |
| Liver | ||||||
| Huh-7 | 2+ | 4+ | 4+ | 30 | 30 | 50 |
| Genitourinary tract | ||||||
| HeLa | N | N | N | N | N | N |
| HEK | N | 4+ | 4+ | 1 | 100 | 100 |
| Neuromuscular cells | ||||||
| NT2 | N | N | N | N | <1 | <1 |
| RD | N | N | N | N | N | N |
| Immune cells | ||||||
| THP-1 | N | N | N | N | N | N |
| U937 | N | N | N | N | N | N |
| Raji | N | N | N | N | N | N |
| H9 | N | N | N | N | N | N |
| His-1 | 1+ | 3+ | 4+ | 30 | 70 | 60 |
| Nonhuman | ||||||
| Mammals | ||||||
| LLC-MK2 | N | 3+ | 4+ | 90 | 100 | 100 |
| Vero | N | 4+ | 4+ | 90 | 90 | 100 |
| Vero-E6 | N | 4+ | 4+ | 1 | 90 | 100 |
| MDCK | N | N | N | N | N | N |
| CRFK | N | N | N | N | N | N |
| PK-15 | N | 2+ | 4+ | N | 10 | 100 |
| RK-13 | N | N | N | N | N | <1 |
| CL-1 | N | N | 1+ | <1 | 40 | 60 |
| 3T3 | N | N | N | N | N | N |
| RK3E | N | N | N | N | N | N |
| RMC | N | N | N | N | N | N |
| Chicken | ||||||
| DF-1 | N | N | N | N | N | N |
| Insect | ||||||
| C6-36 | N | N | N | N | N | N |
a N is defined as negative, 1 is defined as 1%–25% involvement, 2 is defined as >25% to 50% involvement, 3 is defined as >50% to 75% involvement, and 4 is defined as >75% involvement.
b N is defined as negative, and numerals denote the percentage of positive cells.
Figure 1.
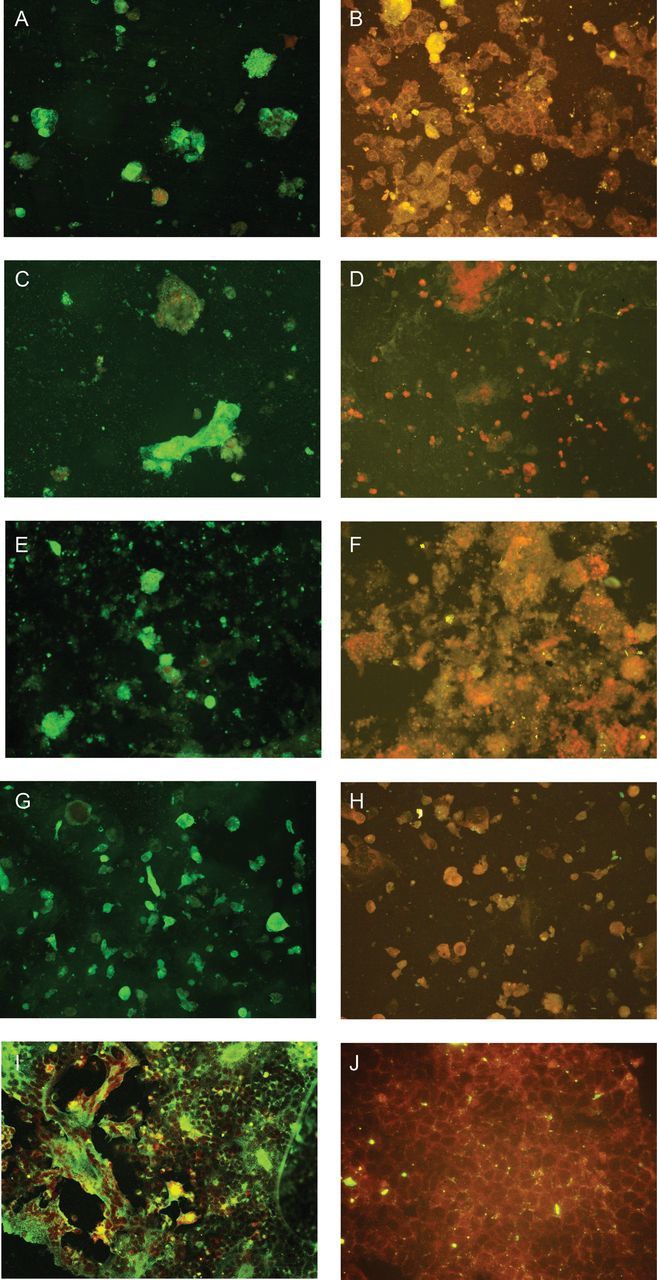
Expression of human betacoronavirus 2c EMC/2012 (HCoV-EMC) nucleoprotein as intense apple green cytoplasmic fluorescence in different cell lines on day 3 after infection stained by monospecific polyclonal serum obtained from a guinea pig infected with His6-tagged HCoV-EMC nucleoprotein (original magnification ×200). A, Infected lower airway (Calu-3) cells. B, Uninfected Calu-3 control. C, Infected intestinal (Caco-2) cells. D, Uninfected Caco-2 control. E, Infected liver (Huh-7) cells. F, Uninfected Huh-7 control. G, Infected histiocytes (His-1). H, Uninfected His-1 control. I, Infected African green monkey kidney (Vero) cells. J, Uninfected Vero control.
Figure 2.
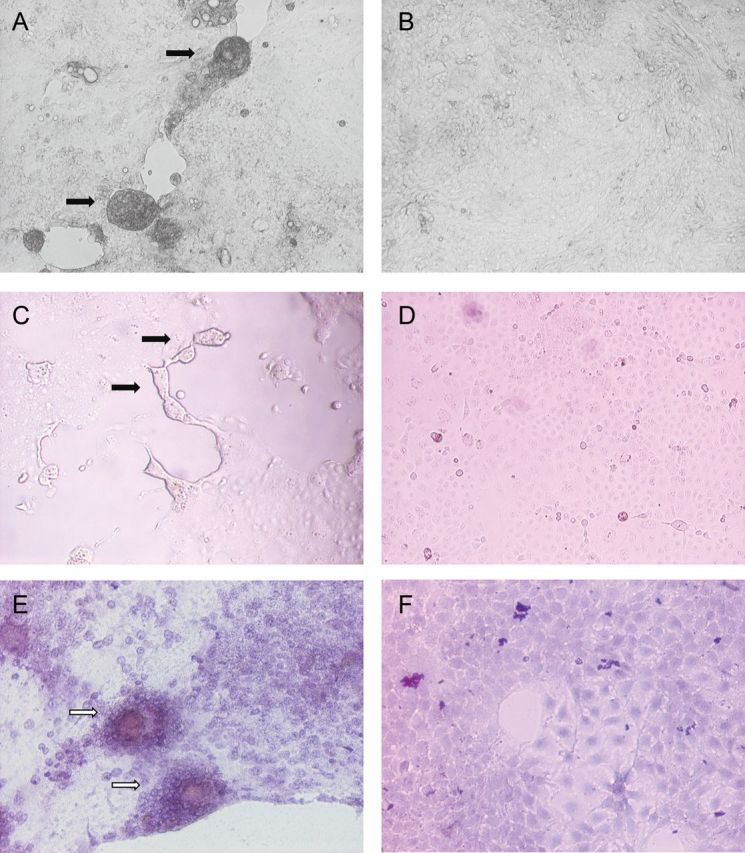
Cytopathic effects of cell rounding, detachment, and syncytia formation (black arrows) in the Calu-3 and Caco-2 cell lines and aggregates of infected cells with marked shrinkage (white arrows) in the Huh-7 cell line (hematoxylin-eosin stain) on day 3 after infection by human betacoronavirus 2c EMC/2012, observed under inverted microscopy (original magnification ×40–100). A, Infected lower airway (Calu-3) cells. B, Uninfected Calu-3 control. C, Infected intestinal (Caco-2) cells. D, Uninfected Caco-2 control. E, Infected liver (Huh-7) cells. F, Uninfected Huh-7 control.
Nonhuman Cell Lines
A total of 13 nonhuman cell lines were tested (Table 1). Six of the 13 cell lines had significantly increased mean viral loads (Table 2). The highest mean viral load was observed in primate (LLC-MK2, Vero, and Vero-E6), porcine (PK-15), civet (CL-1), and rabbit (RK-13) cell lines. The primate, porcine, and civet cell lines had viral N protein expression detected by IF and CPE, whereas the rabbit cell line did not (Table 3). The primate cell lines showed N protein expression by IF on as early as day 1 after infection, while the porcine and civet cell lines showed N protein expression since day 3 after infection. These 5 cell lines showed CPE since day 3–5 after infection. The remaining 7 cell lines, namely the canine (MDCK), feline (CRFK), rodent (3T3, RK3E, RMC), chicken (DF-1), and insect (C6-36) cell lines, did not have N protein expression detected by IF or CPE. Their mean viral loads were at least 2 logs lower than those in the primate, porcine, and civet cell lines.
DISCUSSION
CoVs are notorious for their resistance to culture by in vitro culture systems, and therefore routine viral culture service for CoVs is not available in most clinical laboratories. The isolation of the first HCoVs, HCoV-229E and HCoV-OC43, took many weeks and required the use of labor-intensive embryonic organ culture before subsequent adaptation to a limited number of cell lines. Even the more recently discovered HCoV-NL63 took 7–8 days before CPE was detected on tertiary monkey kidney cells [11, 12, 28]. HCoV-HKU1 produced significant viral load and antigen expression without CPE by 96 hours only in primary, well-differentiated polarized human airway epithelium cell line cultures generated by provision of an air-liquid interface for 4–6 weeks, but it still cannot be stably adapted to commonly used cell lines at this stage [29]. However, primary isolation of SARS-CoV was achieved by direct inoculation of patients’ specimens into embryonic monkey kidney cell lines, such as FRhK-4 or Vero E6, which produced CPE within 5–14 days. Moreover, SARS-CoV was able to grow in the polarized lung cancer cell line Calu-3 (2B4), human lung stem cells, hepatocytes, and intestinal cells after at least 3 days of infection [30–32]. This relatively rapid onset of CPE and the susceptibility of different tissue cells to SARS-CoV might explain the unusually severe clinical manifestations seen in SARS. This novel HCoV-EMC produced CPE on day 5 after inoculation during primary isolation and on subsequent passage by as early as day 1 in human Calu-3, Caco-2, Huh-7, and His-1 cells. Infected Calu-3 and Caco-2 cells showed frequent syncytia formation, in addition to cell rounding and detachment (Figure 2). In the Caco-2 cell line, approximately 10%–15% of cells showed syncytia formation 24–30 hours after infection. Additional incubation after the initial 24–30 hours resulted in cell detachment, with syncytia formation being less obvious. Although multinucleated giant cells could be found during autopsy of patients with SARS, syncytia formation was not found in SARS-CoV–infected cell cultures. None of the other HCoVs produced syncytia in cell lines. Furthermore, the ability of a virus to grow in cells from different host species might provide insight into its ability to cross interspecies barriers. We therefore performed a systematic study on the differential susceptibility to HCoV-EMC among 28 cell lines derived from different tissues, organs, and host species.
Viral replication in different cell lines, as demonstrated by quantitative RT-PCR, may reflect the breadth of tissue tropism of HCoV-EMC. Seventeen of the 28 cell lines supported replication of HCoV-EMC. Notably, the mean viral load in monolayer cell lines did not change significantly after additional inoculation in serum-free medium supplemented with TPCK-treated trypsin. Although many betacoronaviruses, including SARS-CoV, have a proteolytic cleavage site between the globular head S1, for receptor binding, and the stalk S2, for membrane fusion, the corresponding cleavage site in HCoV-EMC could not be found on bioinformatics analysis. In terms of viral N protein expression and CPE, 7 of the 15 human cell lines and 5 of the 13 nonhuman cell lines had viral N protein expression detected by IF and/or CPE. Among the human cell lines, the infected lower airway cell lines (A549, Calu-3, and HFL) produced mean viral loads that were statistically significantly higher than those at the baseline, with more N protein expression detected by IF and/or CPE. Calu-3 cells, which are polarized with tight junctions, resemble the normal human pneumocytes more closely than A549 cells, owing to their greater secretion of MUC5 mucin [33], showed greater susceptibility to HCoV-EMC than A549 cells. The upper airway cell line (Hep-2) did not support the growth of this novel virus. These findings are quite expected because all 9 laboratory-confirmed cases of HCoV-EMC infection had severe acute pneumonia, but they are markedly different from those of the other non-SARS HCoVs, which have very limited cell line tropism and usually cause self-limiting upper respiratory tract infections, although fatal cases of pneumonia have been occasionally reported in patients with multiple underlying illnesses and in elderly individuals [34, 35]. In the case of HCoV-229E, it infected only hepatocytes, primary embryonic lung fibroblasts, neural cells, monocytes, dendritic cells, and macrophages after adaptation, not human pneumocyte cell lines [36–39]. HCoV-OC43 infected only the human intestinal cell line HRT18 and neural tissue cell lines after adaptation, not human pneumocyte cell lines [40]. HCoV-NL63 could only infect the human intestinal cell line (Caco-2) after adaptation [28]. Even SARS-CoV could not infect human A549 cells or human embryonic lung fibroblasts but could selectively infect subclones of Calu-3 [32] and pulmonary stem cells [30, 31, 41]. Notably, HCoV-EMC induced CPE in susceptible human cell lines as early as day 1 in the intestinal and liver cell lines and on day 3 in the lower respiratory tract cell lines. These in vitro changes occurred even faster than those induced by SARS-CoV and could partly explain the apparently more severe clinical presentations and higher fatality rate in infections caused by HCoV-EMC. Specific testing for HCoV-EMC should therefore be performed preferably in lower instead of upper respiratory tract specimens from patients with unexplained severe pneumonia, as reported in the first case of HCoV-EMC infection. Although upper respiratory tract epithelial cells did not appear to support the growth of this novel virus, this finding did not preclude the collection of upper respiratory tract specimens from those with significant epidemiologic linkage and in whom lower respiratory tract specimens could not be obtained. More virologic data on HCoV-EMC–infected patients need to be collected before we know whether the current situation is similar to SARS, wherein virus was also shed in the upper respiratory tract, leading to human-to-human transmission [42].
Two clusters of laboratory-confirmed HCoV-EMC infections were reported, but human-to-human transmission could not be confirmed because these patients might have been exposed to the same animal or environmental source. In the absence of highly effective replication in the upper respiratory tract, human-to-human transmission is likely to be limited, as in the case of influenza A(H5N1) infection, except in the setting of marked aerosol generation during intubation, airway suction, and resuscitation [22, 23]. As in the case of influenza A(H5N1) infection, infection control measures involving segregation and immunization of animal hosts may be important in the control of viral transmission in the community if the animal hosts of HCoV-EMC are identified.
Acute renal failure occurred in 5 of the 9 laboratory-confirmed cases of HCoV-EMC infection [20]. This clinical presentation corroborated with our laboratory finding of effective lytic infection of the HEK cell line and another human kidney cell line, 769P [43]. About 6.7% of patients with SARS developed acute renal impairment, which occurred a median duration of 20 days after the onset of symptoms [44]. Histologically, these patients had predominantly acute tubular necrosis with no evidence of glomerular pathology. The acute renal impairment was likely the combined effect of prerenal and renal factors instead of direct viral CPE. In kidneys of broiler chickens infected with avian nephropathogenic infectious bronchitis virus, lymphoplasmacytic interstitial nephritis with characteristic tubular epithelial degeneration and sloughing were found [45]. The histopathologic changes in kidneys of patients infected with HCoV-EMC should be determined. If viral cytopathic changes are dominant, it might explain the much higher incidence of acute renal failure among individuals with HCoV-EMC infection. The viral load in urine specimens from HCoV-EMC–infected patients should also be investigated, because acute renal failure was an important risk factor for mortality in patients with SARS and may also apply to patients with HCoV-EMC infection. Among the other human cell lines, Huh-7 and Caco-2 also showed significant lytic infection by HCoV-EMC. Interestingly, no hepatitis or enterocolitis has been reported in patients with HCoV-EMC infection so far. This is in contrast to SARS-CoV, which induced lytic infection in both the Huh-7 and Caco-2 cell lines and manifested clinically as hepatitis without liver failure in 49.4% of infected patients [46] and as watery diarrhea without enterocolitis in 48.6% [42]. However, detailed clinical information for most of the 9 cases of HCoV-EMC infection is lacking. Except for the first case, in which elevated levels of hepatic parenchymal enzymes were detected, the apparent absence of hepatitis and enterocolitis may be due to underreporting. Additional data obtained from animal experiments and human cases are needed to fully characterize the complete spectrum of disease manifestations of HCoV-EMC infection, and collection of extrapulmonary specimens such as feces, urine, and blood should be considered for specific viral testing to characterize the viral load and shedding patterns of the virus.
Marked proinflammatory cytokine production was associated with fatal SARS cases with acute respiratory distress syndrome and multiorgan failure. Although a significant increase in viral load in monocytic cell lines (THP-1 and U937) was noted, no N protein expression detected by IF or CPE was noted. A possible interpretation of this finding was that the inoculated virus was phagocytosed by the monocytes, remained in intracellular compartments without viral replication, and was subsequently released into the culture supernatant. On the other hand, the histiocytic cell line (His-1) had a substantially higher mean viral load of 109–1010 copies/mL, evidence of lytic infection with N protein expression detected by IF, and CPE. This cell line would be important for further studies involving in vitro cytokine induction by HCoV-EMC, because the other immune cell lines are either ineffectively infected with this virus or are associated with suboptimal cytokine production. This may help in defining roles other than viral cytolysis of cytokine dysregulation in the pathogenesis of HCoV-EMC.
Our finding of HCoV-EMC's ability to infect primate and porcine cells concurs with another recent report, which demonstrated that cells of Rousettus, Rhinolophus, Pipistrellus, Myotis, and Carollia bats were also susceptible [43]. In addition, we showed that HCoV-EMC could infect cells from other animal species, including civets and rabbits. The virus’ broad species tropism and its close phylogenic relatedness with the bat CoVs Ty-BatCoV-HKU4 and Pi-BatCoV-HKU5 [17, 47] support the hypothesis that HCoV-EMC has emerged from animal hosts. In fact, we have previously discovered numerous CoVs not only in bats but also in other mammalian and avian species [48–50]. The presence of a receptor that is used by the virus and common in bats, primates, pigs, civets, rabbits, and humans might imply a broad species tropism that is unique among all the currently known HCoVs [43]. The definitive and amplifying animal hosts of HCoV-EMC should be defined to prevent further interspecies jumping among animals and humans [19, 47]. In the case of SARS-CoV, the virus was traced from humans to civets as amplifying hosts and to Chinese horseshoe bats as a natural reservoir [47]. Further studies should provide more information on the potential of HCoV-EMC to cause a SARS-like epidemic in the future [14].
Notes
Financial support. This work was supported by Mr Larry Chi-Kin Yung; the Hui Hoy and Chow Sin Lan Charity Fund Limited; the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease of the Department of Health, Hong Kong Special Administrative Region; and the University Development Fund and the Committee for Research and Conference Grant, University of Hong Kong.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 2009;234:1117–27. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 2.Tyrrell DA, Bynoe ML. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966;1:76–7. doi: 10.1016/s0140-6736(66)92364-6. [DOI] [PubMed] [Google Scholar]
- 3.Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–3. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 4.Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–41. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 5.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier RA, Kuiken T, Schutten M, et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 8.Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;347:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 9.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–94. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo PC, Lau SK, Tsoi HW, et al. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–5. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouchier RA, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo PC, Lau SK, Chu CM, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JF, Li KS, To KK, Cheng VC, Chen H, Yuen KY. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65:477–89. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 16.Bermingham A, Chand MA, Brown CS, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17 pii=20290 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290. Accessed 4 October 2012. [PubMed] [Google Scholar]
- 17.Woo PC, Lau SK, Li KS, Tsang AK, Yuen KY. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg Microbes Infect. 2012;1:e35. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Boheemen S, de Graaf M, Lauber C, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3 doi: 10.1128/mBio.00473-12. e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo PC, Wang M, Lau SK, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–85. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Global alert and response: novel coronavirus infection—update. 30 November 2012. http://www.who.int/csr/don/2012_11_30/en/index.html. Accessed 21 December 2012. [Google Scholar]
- 21.ProMED-mail. Novel coronavirus—Saudi Arabia (12): request for information. Archive no. 20121019.1353615. 19 October 2012 Accessed 21 December 2012. [Google Scholar]
- 22.Yuen KY, Chan PK, Peiris M, et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–71. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 23.Beigel JH, Farrar J, Han AM, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–85. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 24.Zheng BJ, Chan KW, Lin YP, et al. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci U S A. 2008;105:8091–6. doi: 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li IW, Chan KH, To KW, et al. Differential susceptibility of different cell lines to swine-origin influenza A H1N1, seasonal human influenza A H1N1, and avian influenza A H5N1 viruses. J Clin Virol. 2009;46:325–30. doi: 10.1016/j.jcv.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 26.To KK, Wong SS, Li IW, et al. Concurrent comparison of epidemiology, clinical presentation and outcome between adult patients suffering from the pandemic influenza A (H1N1) 2009 virus and the seasonal influenza A virus infection. Postgrad Med J. 2010;86:515–21. doi: 10.1136/pgmj.2009.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo PC, Lau SK, Wong BH, et al. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc Natl Acad Sci U S A. 2012;109:5435–40. doi: 10.1073/pnas.1119972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzog P, Drosten C, Müller MA. Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol J. 2008;5:138. doi: 10.1186/1743-422X-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyrc K, Sims AC, Dijkman R, et al. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J Virol. 2010;84:11255–63. doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Chan VS, Zheng B, et al. A novel subset of putative stem/progenitor CD34+Oct-4+ cells is the major target for SARS coronavirus in human lung. J Exp Med. 2007;204:2529–36. doi: 10.1084/jem.20070462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling TY, Kuo MD, Li CL, et al. Identification of pulmonary Oct-4+ stem/progenitor cells and demonstration of their susceptibility to SARS coronavirus (SARS-CoV) infection in vitro. Proc Natl Acad Sci U S A. 2006;103:9530–5. doi: 10.1073/pnas.0510232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshikawa T, Hill TE, Yoshikawa N, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger JT, Voynow JA, Peters KW, Rose MC. Respiratory carcinoma cell lines. MUC genes and glycoconjugates. Am J respire Cell Mol Biol. 1999;20:500–10. doi: 10.1165/ajrcmb.20.3.3383. [DOI] [PubMed] [Google Scholar]
- 34.Woo PC, Lau SK, Tsoi HW, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;192:1898–907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau SK, Woo PC, Yip CC, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–71. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang BS, Chan KH, Cheng VC, et al. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J Virol. 2005;79:6180–93. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeager CL, Ashmun RA, Williams RK, et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–2. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Funk CJ, Wang J, Ito Y, et al. Infection of human alveolar macrophages by human coronavirus strain 229E. J Gen Virol. 2012;93:494–503. doi: 10.1099/vir.0.038414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arbour N, Ekandé S, Côté G, et al. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J Virol. 1999;73:3326–37. doi: 10.1128/jvi.73.4.3326-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arbour N, Côté G, Lachance C, Tardieu M, Cashman NR, Talbot PJ. Acute and persistent infection of human neural cell lines by human coronavirus OC43. J Virol. 1999;73:3338–50. doi: 10.1128/jvi.73.4.3338-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaye M, Druce J, Tran T, et al. SARS–associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng VC, Hung IF, Tang BS, et al. Viral replication in the nasopharynx is associated with diarrhea in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;38:467–75. doi: 10.1086/382681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller MA, Raj VS, Muth D, et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio. 2012;3:e00515–12. doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu KH, Tsang WK, Tang CS, et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler AF, Ladman BS, Dunn PA, et al. Nephropathogenic infectious bronchitis in Pennsylvania chickens 1997–2000. Avian Dis. 2002;45:847–58. doi: 10.1637/0005-2086(2002)046[0847:NIBIPC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 46.Hung IF, Cheng VC, Wu AK, et al. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–7. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau SK, Woo PC, Li KS, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–5. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo PC, Lau SK, Lam CS, et al. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J Virol. 2009;83:908–17. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltavoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau SK, Woo PC, Yip CC, et al. Isolation and characterization of a novel betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol. 2012;86:5481–96. doi: 10.1128/JVI.06927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


