Abstract
Human rhinovirus (HRV) replication triggers exacerbation of asthma and causes most acute respiratory illnesses (ARIs), which may manifest as influenza-like illness. The recent assignment of 60 previously unknown HRV types to a third HRV species, Human rhinovirus C, raised questions about the prevalence of these picornavirus types in the community, the extent of HRV diversity at a single site, and whether the HRVs have an equally diverse clinical impact on their hosts. We quantified HRV diversity, and there was no clinical impact attributable to HRV species and genotypes among a community population of preschool-aged children with ARI who provided respiratory samples during 2003. All HRV species were represented among 138 children with ARI, and 74 distinct HRV types were cocirculating. Fever accompanied 32.8% of HRV-positive ARI cases. HRVs were less likely than DNA viruses to be codetected with another virus, suggesting virus interference at the community level, demonstrated by the inverse correlation between influenza virus detection and HRV detection.
Keywords: Human rhinovirus, Viral epidemiology, Virus:virus interactions, Co-detections, influenza virus
Human rhinovirus (HRV) infections trigger exacerbations of asthma and chronic obstructive pulmonary disease and the majority of acute respiratory illnesses (ARIs), some of which meet criteria for influenza-like illness. These upper and lower respiratory tract illnesses are associated with considerable direct healthcare costs and indirect costs due to time lost from and reduced performance of regular duties [1].
There are 100 known HRV serotypes, with 15–30 circulating simultaneously at a given site [2–7]. Recently, 60 distinct, molecularly defined HRV genotypes were formally assigned to a third species, Human rhinovirus C (HRV-C) [8, 9]. The contribution of HRV species and individual types to the annual burden of circulating HRV is poorly defined. There are 2 apparently distinct phylogenetic clades of HRV-C when typed using the 5′ untranslated region (5′UTR) and adjoining encoding region [10]. It is hypothesized that these clades evolved with or without genetic recombination with Human rhinovirus A (HRV-A) types [10]. Nevertheless, each available majority sequence of an HRV-C type has to date represented a genetically unique, phylogenetically distinct, and globally distributed virus detected in patients with ARIs.
There are specific seasonal and annual variations in respiratory virus circulation and interactions [11–13]. In a retrospective pediatric hospital-based study, HRVs were found to be statistically least likely of 17 examined viruses to be codetected with another virus during 2003 [14]. This aspect of virus-to-virus interaction is described as virus interference and is hypothesized to be the result of the host's response to one virus diminishing the likelihood of infection by another virus [15, 16]. For HRVs, virus-to-virus interaction was once exploited as an in vitro diagnostic tool, whereby successful experimental HRV infection of organ cultures was indicated by blockading the replication of another, superinfecting respiratory virus [17].
We sought to quantify the genetic diversity, epidemiology, and impact of HRV and enterovirus species, conjointly referred to hereafter as picornaviruses, circulating among a community cohort of preschool-aged children who provided respiratory samples over a 1-year period. We also sought to build on our hospital-based virus-to-virus interaction analyses [14] by seeking preliminary observational evidence of virus interference within the community.
METHODS
Community-Based Cohort Study
After receipt of informed consent from the parent or guardian of a potential study subject, we enrolled 234 healthy children <5 years of age into a community-based dynamic cohort study conducted over 12 months in Melbourne, Australia [1]. The study commenced in January 2003, and enrollment was progressive and continued until November 2003, with children observed until January 2004. Parents monitored a set of symptoms in the study child each day, and when the definition of ARI was met we asked parents to collect a combined nose-throat swab specimen [18]. The specimen was couriered to the Victorian Infectious Diseases Reference Laboratory, where it underwent conventional polymerase chain reaction (PCR) testing, with reverse-transcription PCR performed for RNA viruses, including influenza A and B viruses, respiratory syncytial virus, parainfluenza viruses, HRVs, and enteroviruses [19]. PCR was conducted for adenoviruses [19]. Illnesses were classified as ARIs uncomplicated or complicated by fever and/or otitis media.
At the completion of data collection, we transported all available specimens (original nose-throat swab specimens and complementary DNA) on dry ice to the Queensland Paediatric Infectious Diseases (Qpid) Laboratory, where they were tested for metapneumovirus and coronavirus NL63 (hereafter, “coronavirus”), using real-time PCR [19].
Template Preparation and Conventional PCR
HRV templates for PCR amplification at the Qpid Laboratory were complementary DNA, created at the Victorian Infectious Diseases Reference Laboratory during the original studies [19], or fresh RNA extracted by means of the Corbett X-tractor Gene system (Corbett Research, Australia) from the original nose-throat swab specimen. All extracts were amplified using a broadly reactive screening assay that targets the 5′UTR [20], yielding amplicons (length, approximately 380 base pairs) for sequencing by the PRISM BigDye sequencing kit v3.1 (Applied Biosystems). Sequences generated by this study and submitted to GenBank included accession numbers JN861783-92 and JQ406000-184.
HRV Species Designations
Sequence analysis was conducted in Geneious Pro [21]. The picornavirus species was determined by best match, using the following algorithm: when a basic local alignment search tool (BLAST) comparison returned matches with ≥95% sequence identity in the 5′UTR, with characterized members assigned to a given species, our sequence was assigned to that species. For viruses not classified at this stage, we used definitive 5′UTR-derived data from Lee et al [22] as a guide to classify any query sequence that shared >96% nucleotide identity in the 5′UTR with a BLAST match as a variant of that HRV type.
Statistical Analysis
Counts and proportions were recorded for descriptive analyses. Similar to methods we described previously [14], univariate analysis involving the χ2 test or the Fisher exact test, using 2 × 2 contingency tables, was used to evaluate the relationships between picornavirus type, season, and demographic variables, such as age and sex, as well as to study virus codetection with another virus. A P value of <.05 was considered to indicate a statistically significant association.
RESULTS
Community-Based Cohort Study
We observed 56 397 child-days in just over 12 study months. There were 730 ARIs identified; in 563 cases (74%), at least 1 parent-collected combined nose-throat swab specimen was returned [18]. For 269 ARIs (274 specimens; 47.8% of specimens), HRV or enterovirus was identified by conventional PCR at Victorian Infectious Diseases Reference Laboratory [19].
Screening, Sequencing, and Genotype Assignment
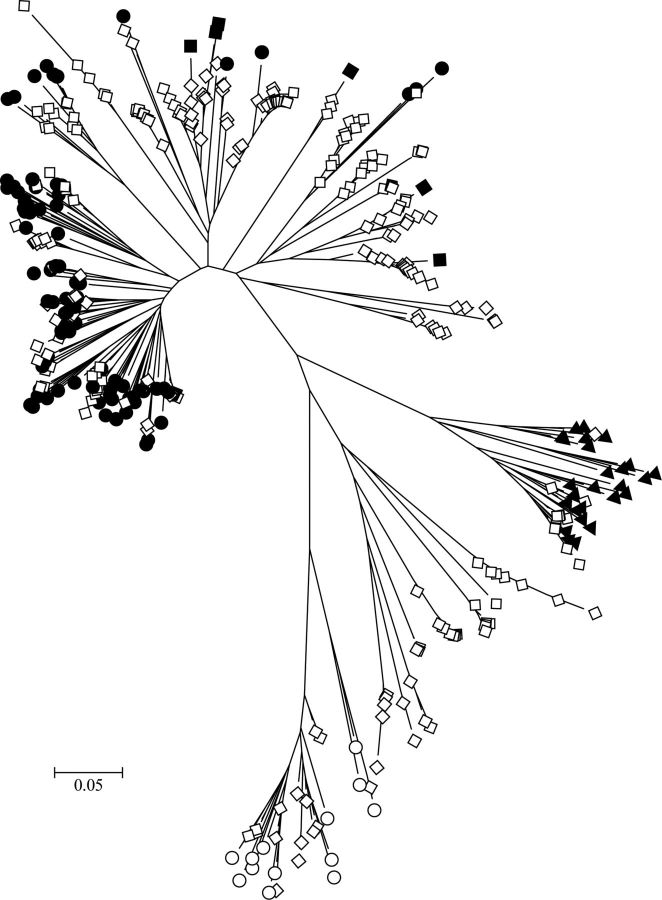
Of 274 specimens previously positive for a picornavirus and shipped to Qpid Laboratory, 238 (86.9%) yielded a genotypable sequence (Figure 1), 14 (5.1%) yielded uninterpretable sequences, and 22 (8.0%) yielded sequences that were not amplifiable [20]. The picornavirus-positive specimens originated from 138 children (mean age, 25.7 months), including 48 from infants (age, 1–12 months), 107 from toddlers (age, 12–24 months), and 119 from older children (age, 2–5 years). A mean of 1.8 picornaviruses (range, 0–6) were detected per child. Sixty picornavirus-positive children (43.5%) had ≥2 picornaviruses detected during study participation, and 32 had ≥3 (Table 1 and Figure 2). No child was positive for the same HRV type during different ARI episodes, although the same species might have been detected. Of the genotyped viruses, 99 (41.5%) were HRV-A, 13 (5.5%) were Human rhinovirus B (HRV-B), 113 (47.5%) were HRV-C, and 13 (5.5%) were enteroviruses (Figure 1).
Figure 1.
Evolutionary relationships among characterized and cohort human rhinovirus (HRV) sequences assigned to the genus Enterovirus. The evolutionary history was inferred using the neighbor-joining method in MEGA5 [37, 38]. The optimal tree is shown drawn to scale, with branch lengths in the same units as those of the evolutionary distances (base substitutions per site; maximum composite likelihood method [39]) The analysis involved 401 nucleotide sequences, including completely sequenced referenced types [40]. Sequences from this study (open diamonds) were included with previously characterized Human rhinovirus A (filled circles), Human rhinovirus B (filled triangles), Human rhinovirus C (filled diamonds), and enterovirus (open circles) types.
Table 1.
Characteristics of Picornavirus (PV)–Positive Specimens
| Characteristic | HRV-A | HRV-B | HRV-C | Enterovirus | Untypable | Overall (n = 563) |
|---|---|---|---|---|---|---|
| Male sex, (%) | 48 (48.5) | 9 (69.2) | 58 (51.4) | 53.8 | 35.7 | 48.0 |
| Viral detections, no. | 99 | 13 | 113 | 13 | … | 395 |
| Age, months, average | 23.5 | 30.8 | 25.4 | 31.7 | 27.1 | 19.9 |
| Peak seasona | Autumn | Autumn | Winter | Winter | Autumn, Spring | Winter |
| Peak monthsb | May, Octc | Apr, May | Jun, Jul, Oct | Mar, Jun, Jul | May, Oct, Nov | Jun, Aug |
| Peak codetection month | Jul, Sep, Dec | Jun | Jun | Jun, Jul, Oct | … | … |
| Wheeze | 0 | 0 | 2 (0.4) | 1 (0.2) | 3 (0.5) | 54 (9.6) |
| ARI, no fever, no OM | 57 (10.1) | 7 (1.2) | 73 (12.9) | 4 (0.7) | 9 (1.6) | 277 (49.2) |
| ARI with fever | 32 (5.7) | 4 (0.7) | 34 (6.0) | 9 (1.6) | 3 (0.5) | 220 (39.1) |
| ARI with OM, no fever | 1 (0.2) | 1 (0.2) | 2 (0.4) | 0 | 2 (0.4) | 46 (8.2) |
| ARI with fever and OM | 9 (1.6) | 1 (0.2) | 4 (0.7) | 0 | 0 | 20 (3.6) |
| Children with 2 PVs | 28 (4.9) | |||||
| Children with 3 PVs | 17 (3.0) | |||||
| Children with 4 PVs | 10 (1.8) | |||||
| Children with 5 PVs | 3 (0.5) | |||||
| Children with 6 PVs | 2 (0.3) |
Data are no. (%), unless otherwise indicated.
Abbreviations: ARI, acute respiratory illness; HRV-A, Human rhinovirus A; HRV-B, Human rhinovirus B; HRV-C, Human rhinovirus C; OM, otitis media; …, Not applicable.
a No significant associations between species and any season were identified (P < .05). Winter months are June, July, and August. Autumn months are March, April, and May. Spring months are September, October, and November.
b Each HRV species exhibited a bimodal distribution.
c Similar totals (±1) were identified in each month.
Figure 2.
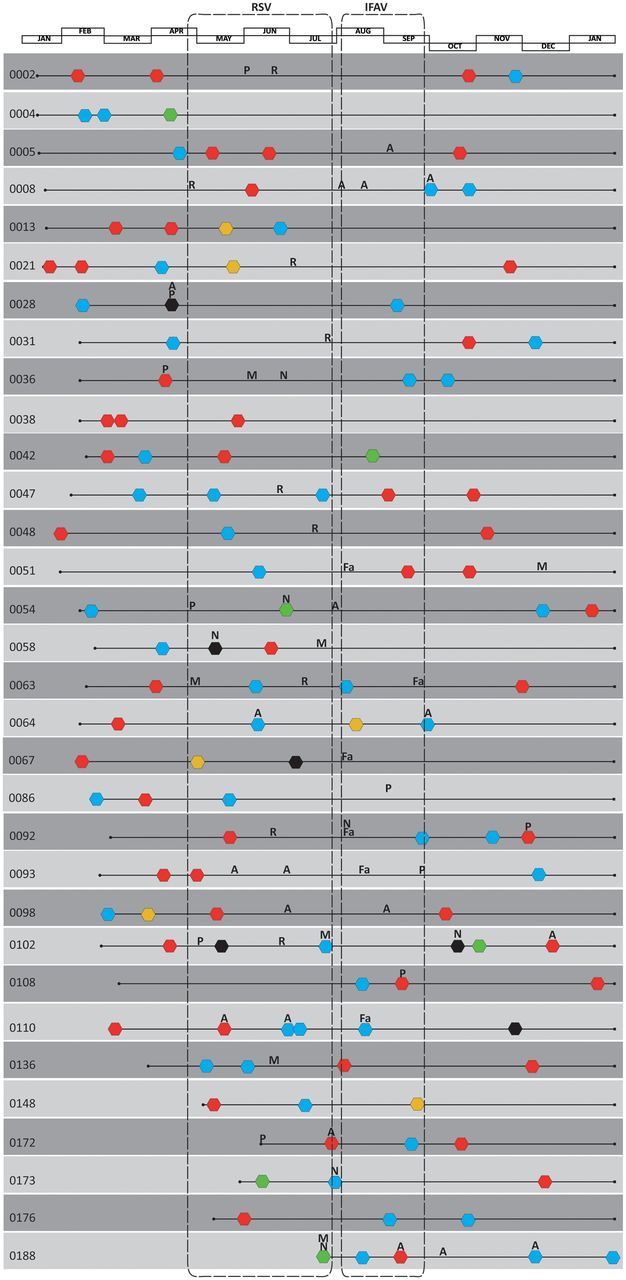
Timeline of all the viruses identified in 60 children (29.6% of all human rhinovirus [HRV]–positive children) who had ≥2 picornaviruses detected during their enrollment. All respiratory syncytial virus (R) and influenza A virus (Fa) detections were contained within periods encapsulated using dashed boxes. Abbreviations: A, adenovirus; black hexagons, untypable picornavirus; blue hexagons, Human rhinovirus C; green hexagons, enterovirus; M, metapneumovirus; N, coronavirus NL63; P, parainfluenzavirus; red hexagons, Human rhinovirus A; yellow hexagons, Human rhinovirus B.
Clinical and Demographic Features and Picornavirus Type
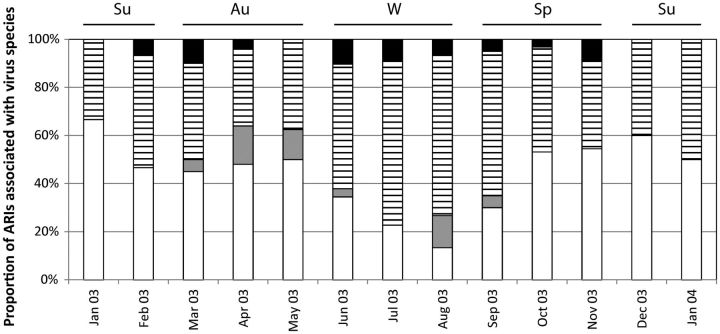
Picornavirus species distributions were not associated with sex (Table 1). There was a decreased likelihood of identifying any of the picornavirus species in infants and an increased risk of identifying HRV-B, HRV-C, and enteroviruses in older children (P < .05 for these associations). The proportion of ARI episodes in which HRV-B and HRV-C were detected was lowest during summer, while the proportion of ARI episodes in which HRV-A was detected was lowest in both summer and winter (Figure 3). Detection of each HRV species declined dramatically when detection of respiratory syncytial virus and metapneumovirus increased and detection of influenza A virus peaked (in August [19]). HRV-A and HRV-C could be detected in subsequent ARI specimens from the same child, but HRV-B positivity did not recur for any child. Peak picornavirus activity occurred in May and October, approximately 2 weeks after school terms commenced. Activity of the species were observed to peak distinctly (species exchange) during the course of the year.
Figure 3.
Proportion of total picornavirus identifications that were Human rhinovirus A (HRV-A; white bars), Human rhinovirus B (HRV-B; grey bars), Human rhinovirus C (stripes), and enterovirus (black bars), by month, during 2003–2004. Abbreviations: ARI, acute respiratory infection; Au, autumn (Mar–May); Su, summer (Dec–Feb); Sp, spring (Sep–Nov); W, winter (Jul–Aug). HRV-C was more common in winter than HRV-A (P = .624).
HRV-positive ARI cases were accompanied by fever in 84 instances (14.9% of all ARIs; 30.7% of HRV detections; Table 2), whereas the majority of cases (9 [69.2%] of 13) in which enterovirus types (including coxsackievirus A1, A2, A4, and B5; echovirus 3, 11, 18, 21, and 27; and poliovirus 1) were identified had fever reported. Only 5 cases of wheeze were reported: 2 were associated with HRV-C, 1 was associated with enterovirus, and 2 were associated with untypable picornaviruses. Otitis media without fever accompanied ARI in 4 picornavirus-positive instances (0.7%), and fever and otitis media co-occurred with ARI in 14 instances (2.5%). HRV-A, HRV-B, and HRV-C were detected in similar proportions of ARI cases with complications (42.0%, 46.2%, and 36.8% respectively) and those without complications (58.0%, 53.8%, and 63.2% respectively).
Table 2.
Measures of Association Between Virus Pairs in Which Codetections Occurred
| Variable | PV | AdV |
PIV |
RSV |
IFAV |
MPV |
CoV |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total detections, no. | 252 | 45 | 33 | 37 | 25 | 33 | 19 | ||||||
| Single detections,a no. | 213 | 20 | 23 | 37 | 21 | 22 | 9 | ||||||
| Codetections,b no. | 39 | 25 | 10 | 4 | 4 | 11 | 10 | ||||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| PV | … | 1.077 (.568–2.039) | .8790 | 0.443 (.195–.983) | .0370 | 0.027 (.001–.184) | <.0001 | 0.150 (.035–.532) | <.0001 | 0.324 (.133–.761) | .0050 | 0.712 (.249–1.979) | .6400 |
| AdV | … | … | 0.731 (.117–3.291) | >.999 | .272 (.14–1.907) | .237 | 0 (0–2.247) | .249 | 1.645 (.466–5.239) | .324 | 0 (0–3.033) | .388 | |
| PIV | … | … | … | 0 (0–1.838) | .159 | 0.659 (.032–4.818) | >.999 | 0.486 (.024–3.497) | .712 | 0 (0–4.281) | .619 | ||
| RSV | … | … | … | … | 0 (0–2.493) | 0.245 | 0.383 (.019–2.724) | .500 | 0.7 (.034–5.176) | >.999 | |||
| IFAV | … | … | … | … | … | 0 (1–6.167) | 1.000 | 1.204 (.058–9.164) | .584 | ||||
| MPV | … | … | … | … | … | … | 1.947 (.296–9.386) | .307 | |||||
Data were determined by 2 × 2 contingency tables with the Fisher exact test. Statistically significant associations (P < .05) between virus pairs are shown in bold.
Abbreviations: AdV, adenovirus; CI, confidence interval; CoV, coronavirus NL63; IFAV, influenza A virus; MPV, metapneumovirus; OR, odds ratio; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
a Sample contained only 1 virus
b Sample contained ≥2 viruses.
Virus Codetection
Of 395 samples with any virus present, 47 (11.9%) included a second virus, whereas 3 (0.8%) were positive for 3 viruses. Among 252 picornavirus-positive specimens, 39 (15.5%) were PCR positive for another virus; 213 (84.5%) were not detected with another virus. Only respiratory syncytial virus was codetected with another virus on fewer occasions (9.8% of respiratory syncytial virus detections). Despite this, among children with ≥1 HRV detection during enrollment (28 children had 2 PVs; 17 had 3 PVs; 10 had 4 PVs, 3 had 5 PVs and 2 had 6 PVs), there was a consistent pattern of a reduced likelihood of codetection with a number of other viruses. For example, an HRV detection was associated with a reduced likelihood of codetecting any of 5 viruses or virus groups (adenovirus, parainfluenza virus, respiratory syncytial virus, influenza A virus, metapneumovirus, or coronavirus), as reflected by an odds ratio of <1 (Table 2). Among picornaviruses, enterovirus was most frequently associated with codetection (31% of enterovirus detections), and HRV-B was least frequently associated with codetection (8% of HRV-B detections). There was an average of 50 days between consecutive ARIs in study children. After each HRV, influenza A virus, respiratory syncytial virus, or adenovirus detection, there was a mean interval of 46, 51, 60, and 45 days, respectively, before the next ARI episode.
Most picornaviruses were detected in toddlers (61 [44.2%] of picornavirus-positive children) and children aged 2–5 years (58 [42.0%]). Picornavirus was most often codetected in infants and toddlers (15 [12.3%] in each age group). Respiratory syncytial virus was involved in the fewest codetections (4 [10.3%] of respiratory syncytial virus–positive children), while adenovirus (25 [55.6%] of adenovirus-positive children) and coronavirus (10 [52.6%] of coronavirus-positive children) were involved in the most. The appearance and disappearance of influenza A virus detection coincided with the disappearance and appearance of picornaviruses, respectively.
DISCUSSION
Our analysis of ARIs among children in the community identified 74 named or uncharacterized rhinovirus types among 138 children followed during this 12-month study. Three-to-five times more distinct HRV types were identified in our single-year cohort than would have been expected from the literature. Recently, van der Zalm et al identified 27 different subtypes among a cohort of 18 children sampled fortnightly during a six-month period [7]. Because a lower sequence threshold was used to identify those viruses, it is likely that the true number of HRV types was underestimated as compared to the higher threshold (96% vs 90%) we applied, which we considered essential when using the highly conserved 5′UTR region. Recent cohort studies that also used the 5′UTR for HRV typing identified 56 [23] distinct HRV types, including 32 new types [23], whereas other cohort studies have not conducted detailed typing analyses [24, 25]. Cohorts with longer follow-up [26] or more-frequent sampling than ours [27] unsurprisingly find greater numbers of HRV types since they span multiple seasons or have a better likelihood of sampling so-called “asymptomatic” infections. Frequently, sampling in longitudinal cohort studies improves our understanding of the role of respiratory viruses in illness because it allows temporal association with signs and symptoms.
Despite the dominance of HRV-C types here and their reported links with asthma elsewhere [28], few new cases of wheezing were reported in our cohort, most likely because of exclusion of children with chronic pulmonary disorders (including diagnosed asthma or frequent use of asthma medication). A frequently sampled, well-controlled longitudinal cohort of children with asthma will be required to answer whether there is a species-specific HRV impact in an asthmatic population. All HRV species were represented among ARI cases with fever, providing further evidence of the confounding capacity of HRV infections during an influenza epidemic or pandemic unless specific diagnostics are included.
Most respiratory viruses were detected in toddlers and children, from whom the majority of specimens were obtained. In our study, the respiratory syncytial virus and metapneumovirus season preceded the influenza A virus season. It was also noteworthy that HRV prevalence declined sharply as respiratory syncytial virus, metapneumovirus, and, particularly, influenza A virus cases peaked. This mirrored the findings from our hospital studies, conducted on specimens from Brisbane, Queensland, Australia, that were also collected during 2003 [14].
We observed that viruses with an RNA genome, in particular the HRVs, were less frequently involved in codetections than those with a DNA genome in this community cohort from Victoria. These numbers might have been different if other virus, such as bocavirus, parainfluenza virus-4, and influenza C virus, had been included, but our previous findings suggest the adjustment would not have changed the significance of the virus-to-virus interaction [14]. Our observation most likely reflects the capacity of RNA viruses to efficiently trigger the early innate interferon response through interactions with a range of pattern-recognition receptors, including Toll-like receptors 3 and 8, MDA5, and RIG-I. Whether DNA viruses are better adapted to interfere with a host interferon response or are better at exploiting the opportunities provided by RNA viruses remains to be defined.
It has been previously suggested that the 5′UTR is unsuitable for HRV genotyping because in silico signs suggest that the region has, at some point, been a site for recombination between some HRV types [29]. However, we have not found, nor has there been published evidence for, false genotyping (a 5′UTR erroneously representing a polyprotein sequence from a different virus) of the ≥100 HRV-A, HRV-B, and HRV-C genomes fully sequenced to date. Even when an HRV-C type falls into the HRV-A clade phylogenetically, to date it exists within a distinct space in that clade. Carefully chosen sequence identity thresholds and experience can successfully produce robust genotyping results, as has been shown here and elsewhere in the literature [23, 26, 27, 30–32]. In this study, we found that each 5′UTR sequence was a unique identifier of HRV type. Others suggest the HRVs are under positive selective pressure to remain conserved [33]. While this may not remain evident as more completed HRV genomes become available, no proof exists to support a theory that the 5′UTR is an unsatisfactory target for genotyping, and it continues to be used successfully elsewhere [34]. Nevertheless, phylogeny based on the 5′UTR region, whether partial or complete, does not accurately discriminate all members of the HRV-A species from those currently considered to be HRV-C but harboring elements of an HRV-A UTR [10]. This is similar to the phylogenetic patterns of enterovirus types derived from use of the 5′UTR, in which the 4 known species are represented by only 2 clades. A longer target encompassing a 5′UTR-VP2 stretch improves phylogenetic tree construction, and these sequences are becoming well represented in the GenBank sequence database [35]. However, until the HRV virome is completely characterized, it is also premature to assume that the 5′UTR-VP2 region's apparent congruity with the rest of an HRV's polyprotein sequence will be maintained by the genomes of future distinct HRV strains. A VP1 target is less sensitive when used directly on clinical specimens [22] and is likely to underrepresent enterovirus diversity [36].
In summary, our screening and 5′UTR-based genotyping study identified extensive HRV genetic diversity as compared to data generated from culture-based diagnostics. There were no associations between sex or clinical outcome and HRV species, but HRVs were less often present in codetections than would have been expected by chance. In particular, the influenza A virus season preceded a precipitous decline in HRV detections. Whether a reduction in HRVs allows an increase in other viruses during the winter months is unknown, and studies to define the immunobiological mechanisms behind these observations are required.
Notes
Acknowledgments. We thank the families and children who participated in this study; Kelly Allen, other VIRGo staff, Maternal and Child Health nurses, and Victorian Infectious Diseases Reference Laboratory and Qpid Laboratory staff who assisted with the original study; and Chloe McIntyre, Peter Simmonds, and Wai-Ming Lee for valuable discussions.
Financial support. This work was supported by the National Health and Medical Research Council (NHMRC; project grant 455905) and the Queensland Children's Medical Research Institute (project grant 10281). The original study was funded by the Victorian Department of Human Services, the Murdoch Children's Research Institute, and the University of Melbourne. Dr Lambert was the recipient of a NHMRC Public Health Postgraduate Scholarship.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed
References
- 1.Lambert SB, Allen KM, Carter RC, Nolan TM. The cost of community-managed viral respiratory illnesses in a cohort of healthy preschool-aged children. Resp Res. 2008;9:1–11. doi: 10.1186/1465-9921-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gwaltney JM, Jr., Hendley JO, Simon G, Jordan WS., Jr Rhinovirus infections in an industrial population. 3. Number and prevalence of serotypes. Am J Epidemiol. 1968;87:158–66. doi: 10.1093/oxfordjournals.aje.a120796. [DOI] [PubMed] [Google Scholar]
- 3.Wisdom A, McWilliam Leitch EC, Gaunt E, Harvala H, Simmonds P. Screening and comprehensive VP4/2-typing of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity HRV species C. J Clin Microbiol. 2009;47:3958–67. doi: 10.1128/JCM.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamre D, Connelly AP, Procknow JJ. Virologic studies of acute respiratory disease in young adults. IV Virus isolations during four years of surveillance. Am J Epidemiol. 1966;83:238–49. doi: 10.1093/oxfordjournals.aje.a120579. [DOI] [PubMed] [Google Scholar]
- 5.Monto AS, Bryan ER, Ohmit S. Rhinovirus infections in Tecumseh, Michigan: frequency of illness and number of serotypes. J Infect Dis. 1987;156:43–9. doi: 10.1093/infdis/156.1.43. [DOI] [PubMed] [Google Scholar]
- 6.Peltola V, Waris M, Österback R, Susi P, Ruuskanen O, Hyypiä T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–9. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 7.van der Zalm MM, Wilbrink B, van Ewijk BE, Overduin P, Wolfs TF, van der Ent CK. Highly frequent infections with human rhinovirus in healthy young children: a longitudinal cohort study. J Clin Virol. 2011;52:317–20. doi: 10.1016/j.jcv.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Lau SKP, Yip CCY, Tsoi H-W, et al. Clinical features and complete genome characterization of a distinct human rhinovirus genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–64. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McErlean P, Shackelton LA, Andrewes E, et al. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, Human rhinovirus C (HRV C) PLoS One. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arden KE, Mackay IM. Newly identified human rhinoviruses: molecular methods heat up the cold viruses. Rev Med Virol. 2010;20:156–76. doi: 10.1002/rmv.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ånestad G. Interference between outbreaks of respiratory syncytial virus and influenza virus infection. Lancet. 1982;1:502. doi: 10.1016/s0140-6736(82)91466-0. [DOI] [PubMed] [Google Scholar]
- 12.Ånestad G, Vainio K, Hungnes O. Interference between outbreaks of epidemic viruses. Scand J Inf Dis. 2007;39:653–4. doi: 10.1080/00365540701253860. [DOI] [PubMed] [Google Scholar]
- 13.Glezen WP, Paredes A, Taber LH. Influenza in children relationship to other respiratory agents. JAMA. 1980;243:1345–9. doi: 10.1001/jama.243.13.1345. [DOI] [PubMed] [Google Scholar]
- 14.Greer RM, McErlean P, Arden KE, et al. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol. 2009;45:10–5. doi: 10.1016/j.jcv.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennette EH. Interference between animal viruses. Annu Rev Microbiol. 1951;5:277–94. doi: 10.1146/annurev.mi.05.100151.001425. [DOI] [PubMed] [Google Scholar]
- 16.DaPalma T, Doonan BP, Trager NM, Kasman LM. A systematic approach to virus-virus interactions. Vir Res. 2010;149:1–9. doi: 10.1016/j.virusres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrewes CH. Rhinoviruses and common colds. Annual Review of Medicine. 1966;17:361–70. doi: 10.1146/annurev.me.17.020166.002045. [DOI] [PubMed] [Google Scholar]
- 18.Lambert SB, Allen KM, Nolan TM. Parent-collected respiratory specimens—a novel method for respiratory virus and vaccine efficacy research. Vaccine. 2008;26:1826–31. doi: 10.1016/j.vaccine.2008.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert SB, Allen KM, Druce JD, et al. Community epidemiology of human metapneumovirus, human coronavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent-collected specimens. Pediatrics. 2007;120:e929–37. doi: 10.1542/peds.2006-3703. [DOI] [PubMed] [Google Scholar]
- 20.Gama RE, Horsnell PR, Hughes PJ, et al. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. J Med Virol. 1989;28:73–7. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- 21.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2011). Geneious v5.4 [computer program] doi: 10.1093/bioinformatics/bts199. http://www.geneious.com . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee W-M, Kiesner C, Pappas T, et al. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illness in infants. PLoS One. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bizzintino J, Lee WM, Laing IA, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–42. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson KG, Kent J, Hammersley V, Cancio E. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. BMJ. 1996;313:1119–23. doi: 10.1136/bmj.313.7065.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vesa S, Kleemola M, Blomqvist S, Takala A, Kilpi T, Hovi T. Epidemiology of documented viral respiratory infections and acute otitis media in a cohort of children followed from two to twenty-four months of age. Pediatr Infect Dis J. 2001;20:574–81. doi: 10.1097/00006454-200106000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Annamalay AA, Khoo SK, Jacoby P, et al. Prevalence of and risk factors for human rhinovirus infection in health aboriginal and non-aboriginal Western Australian children. Pediatr Infect Dis J. 2012;31(7):673–79. doi: 10.1097/INF.0b013e318256ffc6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jartti T, Lee W-M, Pappas T, Evans M, Lemanske RF, Gern JE. Serial viral infections in infants with recurrent respiratory illnesses. Eur Respir J. 2008;32:314–20. doi: 10.1183/09031936.00161907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller EK, Edwards KM, Weinberg GA, et al. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol. 2009;123:98–104. doi: 10.1016/j.jaci.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang T, Wang W, Bessaud M, et al. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiang D, Kalra I, Yagi S, et al. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–45. doi: 10.1128/JCM.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloepfer KM, Olenec JP, Lee WM, et al. Increased H1N1 infection rate in children with asthma. Am J Respir Crit Care Med. 2012;185(12):1275–79. doi: 10.1164/rccm.201109-1635OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochkov YA, Palmenberg AC, Lee W-M, et al. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17(5):627–32. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kistler A, Webster DR, Rouskin S, et al. Genome-wide diversity and selective pressure in the human rhinovirus. Virol J. 2007;4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denlinger LC, Sorkness RL, Lee WM, et al. Lower airway rhinovirus burden and the seasonal risk of asthma exacerbation. Am J Respir Crit Care Med. 2011;184:1007–14. doi: 10.1164/rccm.201103-0585OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmonds P, McIntyre CL, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. Proposals for the classification of human rhinovirus species C into genotypically-assigned types. J Gen Virol. 2010;91:2409–19. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- 36.Tapparel C, Junier T, Germann D, et al. New respiratory enterovirus and recombinant rhinoviruses among circulating strains. Emerg Infect Dis. 2009;15:719–26. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–5. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmenberg AC, Spiro D, Kuzmickas R, et al. Sequencing and analyses of all known human rhinovirus genomes reveals structure and evolution. Science. 2009;324:55–9. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]