Abstract
Background. Analysis of clinical samples from patients with new viral infections is critical to confirm the diagnosis, to specify the viral load, and to sequence data necessary for characterizing the viral kinetics, transmission, and evolution. We analyzed samples from 112 patients infected with the recently discovered Middle East respiratory syndrome coronavirus (MERS-CoV).
Methods. Respiratory tract samples from cases of MERS-CoV infection confirmed by polymerase chain reaction (PCR) were investigated to determine the MERS-CoV load and fraction of the MERS-CoV genome. These values were analyzed to determine associations with clinical sample type.
Results. Samples from 112 individuals in which MERS-CoV was detected by PCR were analyzed, of which 13 were sputum samples, 64 were nasopharyngeal swab specimens, 30 were tracheal aspirates, and 3 were bronchoalveolar lavage specimens; 2 samples were of unknown origin. Tracheal aspirates yielded significantly higher MERS-CoV loads, compared with nasopharyngeal swab specimens (P = .005) and sputum specimens (P = .0001). Tracheal aspirates had viral loads similar to those in bronchoalveolar lavage samples (P = .3079). Bronchoalveolar lavage samples and tracheal aspirates had significantly higher genome fraction than nasopharyngeal swab specimens (P = .0095 and P = .0002, respectively) and sputum samples (P = .0009 and P = .0001, respectively). The genome yield from tracheal aspirates and bronchoalveolar lavage samples were similar (P = .1174).
Conclusions. Lower respiratory tract samples yield significantly higher MERS-CoV loads and genome fractions than upper respiratory tract samples.
Keywords: Middle East, MERS-CoV, RT-PCR, molecular, diagnosis, coronavirus, clinical, screening, viral load, Ct value, genome fraction
A range of clinical specimens from patients with respiratory tract infections (RTIs) [1–3] are sent to the laboratory by clinicians for making a diagnosis and monitoring disease activity. Sputum and nasopharyngeal swab specimens are commonly used for patients who are seen as outpatients or at points of care, and deeper respiratory tract samples, such as tracheal aspirates and bronchoalveolar lavage samples, are frequently obtained from patients who are seriously ill and require admission to the intensive care unit [1–3]. Analysis of clinical samples of patients with new viral infections is critical to confirm the diagnosis, undertake genome sequence analysis, and study the transmission dynamics and evolution of the virus [4–9].
The proportion of the virus genome sequence obtained is dependent on collection of good-quality clinical specimens from relevant disease sites that can yield higher levels of the virus. Measuring the concentration of viral genome in the patients' clinical samples (ie, the viral load) during the course of the illness is also important for estimating the period of infectiousness and for defining guidelines on the duration of isolation precautions. Viral load measurements can also reflect active replication and are used in severe viral RTIs for monitoring disease activity, clinical progress, response to therapy, cure, and relapse. Studies of diverse viral RTIs have found that maximal viral shedding occurs in the first few days after onset of symptoms and then declines with time [3, 4, 7, 9]. Thus, depending on the site of pathology and viral replication, the most appropriate clinical sample for obtaining the maximal viral genome yield can be ascertained.
Several community- and hospital-based studies of the recently discovered novel Middle East respiratory syndrome coronavirus (MERS-CoV), a novel species of the genus Betacoronavirus with positive-sense, single-stranded RNA [10, 11], have shown that infection with this virus is associated with respiratory tract disease ranging in severity from mild to severe, rapidly fulminant disease in patients with comorbid medical conditions [11–19]. Although a real-time reverse-transcription polymerase chain reaction (RT-PCR) technique for detecting MERS-CoV was developed [20, 21] and approved by the World Health Organization soon after the first case of MERS-CoV infection was reported from the Kingdom of Saudi Arabia (KSA), in September 2012, there are scant data on the yield of MERS-CoV genome sequences from various respiratory tract specimens. Molecular studies of viral infections are crucially dependent on obtaining good-quality clinical specimens yielding adequate quantities of intact viral nucleic acid for sequence analysis.
We conducted a study of the relationship between respiratory tract sample type, MERS-CoV genome load, the proportion of the virus genome sequence obtained using a range of respiratory tract samples obtained from laboratory-confirmed cases of MERS-CoV infection reported from the KSA.
METHODS
Collection of Clinical Specimens
The following respiratory tract samples were analyzed for MERS-CoV load (by determining threshold cycle [Ct] values) and the proportion of MERS-CoV genome obtained: sputum samples, nasopharyngeal swab samples, tracheal aspirates, and bronchoalveolar lavage specimens. Sputum specimens were collected directly into a sterile, leak-proof, screw cap container; nasopharyngeal swabs specimens were collected using sterile, synthetic (Dacron)–tipped flocked swabs. Swabs were inserted through the nostril, parallel to the palate, into the nasopharynx, and left in place for a few seconds to absorb secretions. All swabs were placed immediately into sterile tubes containing 2–3 mL of viral transport medium. For lower respiratory tract samples, 2–3 mL of tracheal aspirates or bronchoalveolar lavage fluid were obtained and placed into a dry sterile, leak-proof, screw cap container.
Storage and Transport of Specimens
Transport of specimens was performed as previously described [19]. In brief, for short periods (≤48 hours) of transport, specimens were kept at 2°C–8°C. If the transport duration was >48 hours, specimens were shipped frozen on dry ice as soon as possible after collection. Each specimen container was labeled with the patient identifier, specimen type, and the sample collection date. Packaging was performed to prevent breakage and spillage, containers were sealed with parafilm and placed in ziplock bags with sufficient absorbent material to absorb the entire contents if spillage occurred, and the primary container was placed inside a secondary container [19].
RNA Extraction
RNA extraction was performed as described previously [19], using the Roche Magna Pure LC (RNA Viral isolation Kit). Sputum samples were pretreated with 2× lysis buffer for 30 minutes in a shaking incubator. Swabs were placed in lysis buffer. A total of 200 µL of each sample was added to a MagNA pure LC plate, which contains 96 wells. Reaction reagents were then loaded and checked before running the samples according to the manufacturer's instructions for nucleic acid extraction in a specimen preparation area.
MERS-CoV PCR Testing
Clinical samples were screened by a real-time RT-PCR amplification test as previously described [20, 21], with amplification targeting the upstream E region (upE) and the ORF1a for confirmation. Results were considered positive only if both assays were positive. When the first and second assays were discordant or if the real-time RT-PCR result was ambiguous, an additional clinical sample was requested and analyzed.
Genome Extraction and Sequence Generation
MERS-CoV deep sequencing was conducted as previously described [22, 23]. The MERS-CoV genome sequences that have been analyzed have all been published and described previously [13, 22–24].
Statistical Analyses
MERS-CoV load and genome fraction were recorded for each set of sputum specimens, nasopharyngeal swab specimens, tracheal aspirates, or bronchoalveolar lavage samples. Standard box and whisker plots with a median value for each set were calculated for MERS-CoV load and fraction of MERS-CoV genome obtained. The fraction of MERS-CoV genome obtained was plotted by sample viral Ct value against sample type. Mann–Whitney U tests were performed for all comparisons; a P value of <.05 was considered to be statistically significant.
RESULTS
Respiratory tract samples obtained from 112 individuals with positive results of real-time RT-PCR for MERS-CoV were analyzed. This set includes 13 sputum samples, 64 nasopharyngeal swabs, 30 tracheal aspirates, and 3 bronchoalveolar lavage samples. Two samples received by the laboratory had no sample type indication and were not included in the analysis.
Viral Load Ct Values
A comparison of the MERS-CoV real-time RT-PCR Ct values as a function of sample type was performed (Figure 1, upper panel). Table 1 shows the P values for the comparison of sample type to Ct value. Tracheal aspirates yielded significantly lower MERS-CoV Ct values (ie, a higher viral load) than nasopharyngeal swab specimens (P = .0005) and sputum specimens (P = .0001). There was no significant difference in viral load Ct values when tracheal aspirates were compared to bronchoalveolar lavage specimens (P = .3079).
Figure 1.
Clinical sample type and Middle East respiratory syndrome coronavirus (MERS-CoV) threshold cycle (Ct) values. The Ct of real-time reverse-transcription polymerase chain reaction, a measurement of MES-CoV viral load (upper panel) or fraction of MERS-CoV genome obtained by deep sequencing (lower panel) were plotted by clinical sample type (tracheal aspirates [TAs], nasopharyngeal swab specimens [NPs], sputum specimens, and bronchoalveolar lavage specimens [BALs]). Box and whisker plots were prepared using the Python/Matplotlib box plot module (http://matplotlib.org/examples/pylab_examples/boxplot_demo.html). Data are for 110 specimens collected through 14 November 2014. Gray boxes indicate the lower to upper quartile values of each subset, blue lines indicate median values, and whiskers indicate ranges, with outlier points falling above or below 1.5 times the interquartile range indicated individually.
Table 1.
Statistical Analyses of Clinical Sample Type Comparisons for Yield of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Load and of the MERS-CoV Genome Fraction
| Comparison | P, Viral Loada | P, Genome Fractionb |
|---|---|---|
| TA vs NP | .0005 | .0002 |
| TA vs Sputum | .0001 | .0001 |
| TA vs BAL | .3079 | .1174 |
| NP vs Sputum | .0113 | .0092 |
| NP vs BAL | .0298 | .0095 |
| Sputum vs BAL | .0074 | .0009 |
Abbreviations: BAL, bronchoalveolar lavage fluid; Ct, threshold cycle; NP, nasopharyngeal swab specimen; TA, tracheal aspirate.
a Values denote results of the comparison of Ct real-time reverse-transcription polymerase chain reaction findings, using the Mann–Whitney U test.
b Values denote results of the comparison of deep sequencing findings, using the Mann–Whitney U test.
Genome Fraction Values
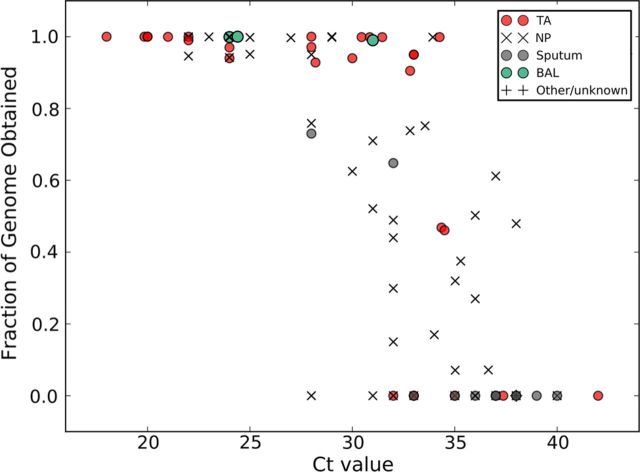
Figure 1 (lower panel) shows the MERS-CoV genome fraction obtained from each sample type. Figure 2 shows the correlation of the fraction of the MERS-CoV genome obtained with the sample Ct value relative to the sample type. Higher MERS-CoV genome fractions were obtained from bronchoalveolar lavage samples and tracheal aspirates than from nasopharyngeal swab specimens (P = .0095 and P = .0002, respectively) and sputum samples (P = .0009 and P = .0001, respectively; Table 1). There was no significant difference in genome yield between tracheal aspirates and bronchoalveolar lavage samples (P = .1174; Table 1).
Figure 2.
Fraction of the sequenced Middle East respiratory syndrome coronavirus (MERS-CoV) genome obtained as a function of MERS-CoV load. Samples were stratified by clinical sample type and the fraction of MERS-CoV genome obtained by deep sequencing was plotted as a function of the MERS-CoV load (presented in terms of the threshold cycle [Ct] value). Data are for 110 specimens collected through 14 November 2014. Tracheal aspirates (TAs) are indicated by red circles, nasopharyngeal swab samples (NPs) are indicated with black Xs, sputum samples are indicated with gray circles, bronchoalveolar lavage samples (BAL) are indicated with green circles, and samples with an unknown type (n = 2) are indicated with black crosses.
DISCUSSION
This study presents the largest data set available to date on molecular analyses of several types of respiratory tract samples and describes the distribution of the MERS-CoV genome load and fraction of the virus genome sequence obtained from these samples. Varying amounts of MERS-CoV load and fractions of MERS-CoV genomes were obtained from all clinical sample types received from 110 Saudi Arabian patients with MERS-CoV infection. When stratified by site of sample origin, samples from deeper in the respiratory tract (ie, tracheal aspirates and bronchoalveolar lavage specimens) yielded significantly higher MERS-CoV genome loads and genome sequenced fractions than samples from other respiratory tract (ie, sputum and nasopharyngeal swab specimens), although samples from all anatomical sites appear to be suitable for viral load determination and virus genome sequencing studies.
Conventionally, tests to detect viral infections in the respiratory tract are performed on sputum or nasopharyngeal swab specimens from patients not requiring admission to the intensive care unit [1, 2, 19] and on tracheal aspirates and bronchoalveolar lavage samples from patients in the intensive care unit [3, 4, 7, 13], and the samples are therefore accessible. A limitation of this study is the lack of multiple samples from multiple compartments of a single patient. However, given this limitation, it appears that viral load is good predictor of MERS-CoV sequencing success.
The viral load in a clinical sample at any given time reflects the dynamic interaction between MERS-CoV replication and the ability of the host's immune system to eliminate the virus. Thus, MERS-CoV load measurements can be clinically useful for monitoring disease activity, clinical progress, response to therapy, and cure, and they can also be used as a marker of prognosis [7–11]. The ideal approach to determining the most appropriate clinical sample for making a diagnosis, ascertaining the viral load, and obtaining the optimal genome fraction requires understanding of the natural history of the viral infection [9–11]. Data from the severe acute respiratory syndrome (SARS) epidemic showed low SARS-CoV loads in the upper respiratory tract and high viral loads in the lower respiratory tract [9–11]. The natural history of SARS-CoV infection was unique in that test results for nasopharyngeal specimens were often negative during the first week of infection, and the highest positivity rates occurred during the second week of illness, peaking at approximately day 10. This allowed definition of which and when during the course of infection clinical specimens will test positive [5, 11]. The low virus detection rate in nasopharyngeal specimens early in the course of SARS-CoV infection illustrates the importance of optimal timing of specimen collection and the optimal specimen type for diagnosis.
Definition of the natural history of the virus may indicate possible sites in the respiratory tract and other parts of the body where the virus causes inflammation and damage. In other common respiratory viral infections, such as influenza, the viral load peaks soon after the onset of symptoms [4]. There were several features of SARS that distinguished it from other viral causes of RTIs [7, 11]. The pathogenic potential, natural history, and transmission dynamics of MERS-CoV require definition before the optimal sample type can be ascertained. Dipeptidyl peptidase 4 was identified as the receptor for MERS-CoV [25], and these receptors are expressed on primary human bronchiolar lung tissue; thus, the virus is able to infect lower respiratory tract tissues. Gastrointestinal symptoms are also present in patients infected with MERS-CoV [13].
Diagnostic tests for respiratory viral infections or screening of close contacts have traditionally been performed on upper respiratory tract samples, particularly nasopharyngeal swab specimens, and it is no different for MERS-CoV. For detecting MERS-CoV, the choice of the most appropriate respiratory tract specimen for diagnostic purposes remains to be determined and requires further study of several respiratory tract sample types obtained at the same time from the same patient. Further studies are required to (1) define the natural history of MERS-CoV infection in humans and the viral load kinetics over time in various respiratory tract samples; 2) determine MERS-CoV shedding in various nonrespiratory clinical sample types, such as urine, stool, blood, or effusions from the time of infection to recovery or death; and 3) correlate viral load with intensive care unit admission and death as a composite end point. These data are required to shed further light on MERS-CoV pathogenesis, ascertain the optimal clinical samples for diagnosis, and guide optimal infection control measures. Viral load measurements could also serve as biomarkers for monitoring response to therapy, disease activity, and predict prognosis.
Notes
Acknowledgments. We are grateful to all staff of the Ministry of Health, Kingdom of Saudi Arabia (KSA); and to Mr Adam Zumla, University College London School of Pharmacy, for kindly providing technical and administrative support.
This study was initiated, designed, and conducted as a major priority issue under the auspices of The Global Center for Mass Gatherings Medicine (GCMGM), Ministry of Health, KSA. Board members of the GCMGM, Abdullah Al-Rabeeah (chair), Ziad A Memish (vice chair), Alimuddin Zumla, Jaffar A. Al-Tawfiq, Abdullah Assiri, Ali Albarrak, and Rafaat Al-Hakeen, initiated a range of MERS-CoV studies.
H. Q. M., A. Z., J. Al-T., and M. C. collected data on viral load and genome fraction and finalized the database. All KSA authors were involved in sample collection and patient management. M. C., P. K., and S. J. W. were responsible for generating the MERS-CoV genome sequences analyzed, and M. C. developed the figures and tables. Z. A. M., A. Z., H. Q. M., J. Al-T., and M. C. wrote the first and final drafts of the manuscript. All authors contributed to the finalization of the manuscript.
Financial support. This work was supported by the Saudi Ministry of Health. The following authors acknowledge the University College London Hospitals NHS Foundation Trust (to A. Z.); the National Institute of Health Research, Biomedical Research Centre, UCL Hospitals (to A. Z.); the EDCTP (to A. Z.); the EC-FW7 (RiD-RTI to A. Z.); the Wellcome Trust (to P. K., M. C., and S. W.); the European Community Seventh Framework Programme (FP7/2007-2013), under the projects EMPERIE (grant 223 498 to P. K., M. C., and S. W. and contract 223 498 to C. D.) and ANTIGONE (contract 278 976 to C. D.); the German Centre for Infection Research (to C. D.); the German Ministry for Research and Education (to C. D.); and the German Research Council (grants 01KIO701 and DR 772/3-1 to C. D.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lieberman D, Shimoni A, Keren-Naus R, et al. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling. J Clin Microbiol. 2009;47:3439–43. doi: 10.1128/JCM.00886-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loens K, Van Heirstraeten L, Malhotra-Kumar S, et al. Optimal sampling sites and methods for detection of pathogens possibly causing community-acquired lower respiratory tract infections. J Clin Microbiol. 2009;47:21–31. doi: 10.1128/JCM.02037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rello JA, Rodríguez P, Ibañez L, et al. Intensive care adult patients with severe respiratory failure caused by influenza A (H1N1)v in Spain. Crit Care. 2009;13:R148. doi: 10.1186/cc8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To KK, Chan KH, Li IW, et al. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol. 2010;8:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung IFN, Lau SKP, Woo PCY, et al. Viral loads in clinical specimens and SARS manifestations. Hong Kong Med J. 2009;15:S20–2. [PubMed] [Google Scholar]
- 6.Liu CL, Lu YT, Peng MJ, et al. Clinical and laboratory features of severe acute respiratory syndrome vis-a-vis onset of fever. Chest. 2004;126:509–17. doi: 10.1378/chest.126.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan CK, Yieh KM, Peng MY, et al. Clinical and laboratory features in the early stage of severe acute respiratory syndrome. J Microbiol Immunol Infect. 2006;39:45–53. [PubMed] [Google Scholar]
- 8.Chan PKS, To W-K, Ng K-C, et al. Laboratory diagnosis of SARS. Emerg Infect Dis. 2004;10:825–31. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui DS, Chan PK. Severe acute respiratory syndrome and coronavirus. Infect Dis Clin North Am. 2010;3:619–38. doi: 10.1016/j.idc.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rainer TH, Lee N, Ip M, et al. Features discriminating SARS from other severe viral respiratory tract infections. Eur J Clin Microbiol Infect Dis. 2007;26:121–9. doi: 10.1007/s10096-006-0246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 12.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;9:752–61. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assiri A, McGeer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–16. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bermingham A, Chand MA, Brown CS, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 15.Mailles A, Blanckaert K, Chaud P, et al. First cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Euro Surveill. 2013;18:20502. [PubMed] [Google Scholar]
- 16.Guery B, Poissy J, El Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–74. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchholz U, Müller MA, Nitsche A, et al. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October–November 2012. Euro Surveill. 2013;18:20406. [PubMed] [Google Scholar]
- 18.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–51. doi: 10.1016/S1473-3099(13)70154-3. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memish ZA, Al-Tawfiq JA, Makhdoom HQ, et al. Screening for Middle East respiratory syndrome coronavirus infection in hospital patients and their healthcare worker and family contacts: a prospective descriptive study. Clin Microbiol Infect. 2014 doi: 10.1111/1469-0691.12562. . [Epub ahead of print]. PMID: 24460984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman VM, Eckerle I, Bleicker T, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 21.Corman VM, Muller MA, Costabel U, et al. Assay for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 22.Cotten M, Watson SJ, Kellam P, et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotten M, Watson SJ, Zumla AI, et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5:e01062-13. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Memish ZA, Cotten M, Meyer B, et al. Human infection with MERS Cornavirus after exposure to infected camels, Saudi Arabia. Emerg Infect Dis. 2014;20:1012–5. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]