Using ferret models of influenza virus and human respiratory syncytial virus (hRSV), we demonstrate that infection with influenza virus can prevent or limit infection with hRSV, whereas infection with hRSV reduces morbidity attributed to influenza virus infection.
Keywords: Influenza, RSV, viral interference, ferret
Abstract
Epidemiological studies have observed that the seasonal peak incidence of influenza virus infection is sometimes separate from the peak incidence of human respiratory syncytial virus (hRSV) infection, with the peak incidence of hRSV infection delayed. This is proposed to be due to viral interference, whereby infection with one virus prevents or delays infection with a different virus. We investigated viral interference between hRSV and 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) in the ferret model. Infection with A(H1N1)pdm09 prevented subsequent infection with hRSV. Infection with hRSV reduced morbidity attributed to infection with A(H1N1)pdm09 but not infection, even when an increased inoculum dose of hRSV was used. Notably, infection with A(H1N1)pdm09 induced higher levels of proinflammatory cytokines, chemokines, and immune mediators in the ferret than hRSV. Minimal cross-reactive serological responses or interferon γ–expressing cells were induced by either virus ≥14 days after infection. These data indicate that antigen-independent mechanisms may drive viral interference between unrelated respiratory viruses that can limit subsequent infection or disease.
Viral interference is a phenomenon whereby infection with one virus limits or delays infection with a second virus. It has been described in human epidemiological studies observing viral epidemic peaks [1–4], vaccine efficacy studies [5], studies assessing virus infections in clinical samples [6–8], animal studies [9–13] and in vitro infectivity studies [14]. Viral interference has been observed between a range of viruses, including between arboviruses, such as yellow fever and dengue virus [15]; between different respiratory viruses [9, 13, 16]; and between influenza viruses of different types [10] and subtypes/lineages [10, 11].
At a population level, respiratory virus infections may display distinct epidemic peaks. Observational studies from the Netherlands, France, and Hong Kong showed that emergence of 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) delayed infections with human respiratory syncytial virus (hRSV) [1, 3, 4]. Influenza A virus infections also interrupted peak incidences of hRSV infections in Japan during 2000–2002 [2] and in the Netherlands during 2003–2012 [3]. Negative associations between respiratory viruses have been reported when analyzing the proportion of coinfections with different respiratory viruses, using swab specimens from patients [6, 7, 17]. A(H1N1)pdm09 was least likely to be detected with any of the other respiratory viruses tested, including hRSV, in samples from all age groups [8, 18]. Taken together, these data suggest that interference may occur between A(H1N1)pdm09 and hRSV.
The ferret provides an ideal model of human influenza because animals can be directly infected with virus without adaptation and display similar disease symptoms to those in humans [19, 20]. Historically, the ferret has also been used to study hRSV infection [21–23], with recent studies assessing the pathogenesis, immunity, and transmission of hRSV [24, 25]. Clinical symptoms are mild in ferrets infected with hRSV strains described to date [24, 25]. Previously, we used the ferret model to demonstrate that viral interference can occur following infection with human influenza A and B viruses and will prevent, delay, or limit subsequent infection with an influenza virus of a different type, subtype, or lineage [10, 11]. Notably, this effect depends on the virus combinations and the order and timing of sequential infections [10, 11, 26]. We have established complementary influenza viral dynamics models that explain these observations via the innate immune response [27] and cross-reactive adaptive immune responses [28].
Ecological data suggest that infection with A(H1N1)pdm09 can prevent or delay infection with hRSV. Using our ferret models of influenza and hRSV, we have systematically investigated this hypothesis.
MATERIALS AND METHODS
Ferrets
Adult ferrets were housed at the Peter Doherty Institute for Infection and Immunity Bioresources Facility. Experiments were conducted with approval from the University of Melbourne Microbiology and Immunology Animal Ethics Committee, in accordance with the Australian National Health and Medical Research Council code of practice for the care and use of animals for scientific purposes. All ferrets were seronegative for antibodies to currently circulating influenza viruses and hRSV (Long and A2 strains) before use in experiments.
Viruses
A/Tasmania/2004/2009 (A[H1N1]pdm09) virus was passaged allantoically in embryonated hen’s eggs and stored at −80°C. The infectious influenza virus titer was measured by a 50% tissue culture infectious dose (TCID50) assay [29], read by hemagglutination with turkey red blood cells. hRSV Long and A2 strains were passaged [24]. Infectious hRSV titers were determined by plaque assay [24].
Virus Infection, Sampling, and Monitoring of Ferrets
Ferrets were infected intranasally with 103.5 TCID50 A(H1N1)pdm09 in 500 µL and 105 plaque-forming units (PFU) of Long hRSV or 106 PFU of Long or A2 hRSV in 500 µL and monitored [24, 26]. Ferrets were housed in pairs, by infection group. Nasal wash specimens were collected and stored [24]. On the day of collection, viral RNA was extracted from 140-µL nasal wash specimens for quantitative polymerase chain reaction (qPCR) analysis. Blood samples were obtained from ferrets before primary virus infection and immediately before and 14 days after challenge, and serum was isolated. The proportional change in weight was calculated as the percentage difference from the weight on the day of challenge.
Reverse Transcription (RT)–qPCR Quantification of Viral Load in Ferret Nasal Wash Specimens
Four microliters of viral RNA [24] was assayed by RT-qPCR with A(H1N1)pdm09 hemagglutinin–specific primers/probes from the CDC Influenza Virus RT-qPCR Influenza A (H1/H3/H1pdm09) Subtyping Panel, obtained from the Influenza Reagent Resource (available at: http://www.influenzareagentresource.org/) and hRSV N–specific primers/probes [24]. Copy numbers for A(H1N1)pdm09 viral RNA were calculated relative to plasmid pHW2000-A/Tasmania/2004/2009 hemagglutinin; copy numbers for RSV RNA were calculated relative to a hRSV RNA standard [24].
qPCR Analysis of Ferret Cytokine and Chemokine Messenger RNA (mRNA)
mRNA was isolated from nasal wash samples [30]. mRNA expression of cytokines, chemokines, and housekeeping genes was quantified by qPCR [30, 31].
ViroSpot (VS) Assay
Infectious hRSV in nasal wash samples was measured using the VS assay [24].
Interferon γ (IFN-γ) Enzyme-Linked Immunospot (ELISpot) Assay
IFN-γ–producing cells were detected by a ferret IFN-γ ELISpotPlus assay (Mabtech). Single cell suspensions were prepared from ferret retropharyngeal lymph nodes [31]. A total of 5 × 104 lymph node cells were cultured with or without live influenza virus, hRSV, or 5 µg/mL concanavalin A (Sigma) for 48 hours at 37oC in 5% CO2 [11].
Hemagglutination Inhibition (HI) Assay
Titers of antibodies to A/Tasmania/2004/2009 were measured using HI assays [31, 32]. Titers were expressed as the reciprocal of the highest dilution of serum for which hemagglutination was prevented. Geometric mean titers (GMTs) were calculated, with undetectable titers expressed as having a value of “5.” Seroconversion was defined as a titer of ≥40 at the end of the experiment and at least a 4-fold rise from baseline.
VS Microneutralization (VS MN) Assay
Titers of antibodies that neutralize hRSV Long and A2 were measured using VS MN assays [24]. Seroconversion was defined as a titer ≥160 at the end of the experiment and an increase of at least 4-fold from the baseline titer.
Enzyme-Linked Immunosorbent Assay (ELISA)
Antibodies that bind to the F glycoprotein of hRSV were detected by an ELISA [24].
Definitions of Infection Measurements and Statistics
Viral kinetics were assessed in viral RNA from nasal wash specimens. For A(H1N1)pdm09, >106 copies of hemagglutinin/100 µL of nasal wash were positively correlated with replicating virus, based on the TCID50 assay [10] and the level of infectious virus as measured by transmission in ferrets [33]. For hRSV, 103.8 copies of N/100 µL of nasal wash corresponded to a 50% chance of a sample being positive by the ViroSpot assay, as determined using a probit regression model (Supplementary Figure 1). Accordingly, samples were considered to be infectious for hRSV when the amount of viral RNA exceeded 103.8 copies/100 μL nasal wash and infectious for A(H1N1)pdm09 when viral RNA exceeded 106 copies/100 µL of nasal wash for at least 1 measurement. Clinical signs (ie, weight loss and fever) were assessed daily, and seroconversion was measured 14 days after challenge.
Statistical Analysis
Statistical analysis was conducted using Prism, version 6.0g, unless otherwise indicated and is described in the figure legends.
RESULTS
A(H1N1)pdm09 Infection Can Prevent or Alter the Kinetics of hRSV Infection
Ferrets were first infected with A(H1N1)pdm09 virus then challenged with hRSV 3, 7, or 11 days later, or vice versa (Figure 1A). The intervals between inoculations spanned the times of peak titer and clearance of both virus infections [24, 30] and induction of humoral immunity (Figure 2A).
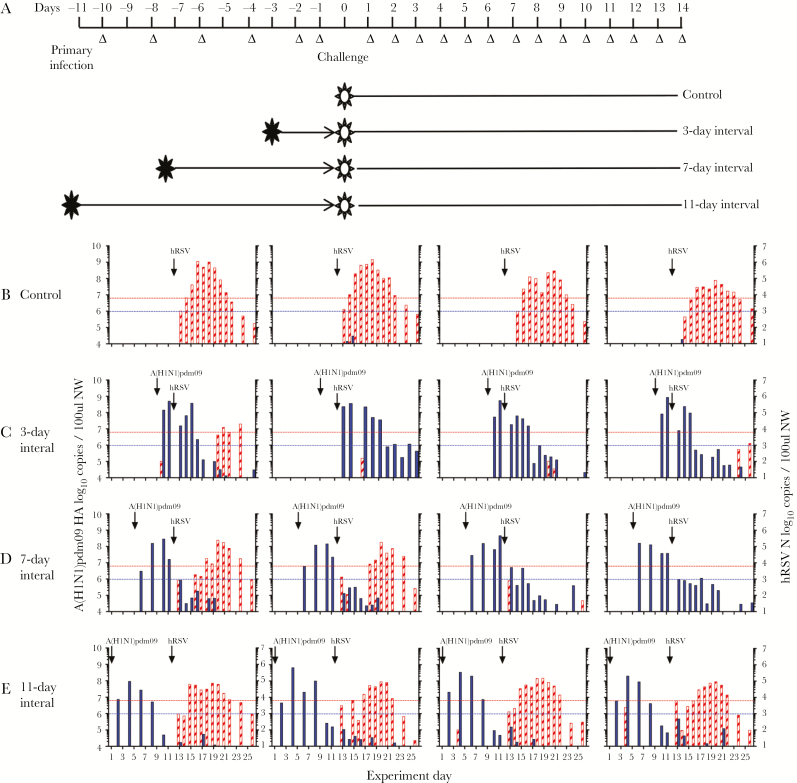
Figure 1.
Virus shedding among ferrets infected with 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09), followed at intervals of 3, 7, or 11 days by human respiratory syncytial virus (hRSV). A, Experimental plan and outcomes. Ferrets were infected via the intranasal route with A(H1N1)pdm09 and then challenged at various intervals (3, 7, or 11 days later) with hRSV, or vice versa. Control ferrets were not infected with the primary infecting virus. Virus shedding in nasal wash specimens was assessed every second day after primary infection and daily after challenge. B–E, Ferrets underwent primary infection with 103.5 50% tissue culture infectious doses of A(H1N1)pdm09, 3.100 followed by challenge with 105 plaque-forming units of hRSV strain Long 3 (C), 7 (D), or 11 (E) days later. Control animals were infected with hRSV alone (B). Quantitative reverse-transcription polymerase chain reaction analysis was used to detect the A(H1N1)pdm09 hemagglutinin gene (filled) and the hRSV N gene (striped) in viral RNA recovered from nasal wash samples. The lower dotted lines indicate the limit of detection of infectious A(H1N1)pdm09, and the upper dotted lines indicate the limit of detection of infectious hRSV, as defined in Materials and Methods.
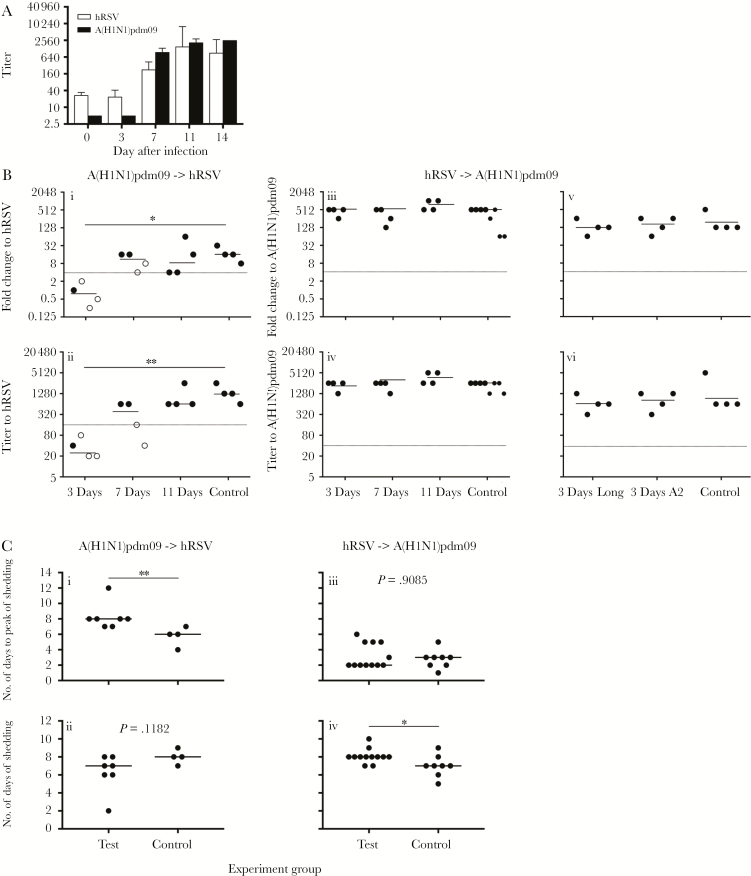
Figure 2.
Neutralizing antibody responses following challenge virus infection, and kinetics of virus shedding following challenge infection. A, Ferrets were infected via the intranasal route with either 103.5 50% tissue culture infectious doses (TCID50) of 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) or 105 plaque-forming units (PFU) of human respiratory syncytial virus (hRSV), and sera, collected on the days indicated, was assayed for neutralizing antibodies to the infecting virus, using hemagglutination inhibition (HI) or ViroSpot (VS) microneutralization (MN) assays. Data are geometric mean titers (GMTs) and 95% confidence intervals from 4 ferrets. B, After undergoing primary infection with either A(H1N1)pdm09 or hRSV, ferrets were challenged 3, 7, or 11 days later with the alternate virus. Control animals in each experiment received only the challenge virus. Sera were collected 14 days after challenge, and neutralizing antibodies to the challenge virus were measured by the HI assay, for influenza virus, and by the VS MN assay, for hRSV. Primary and challenge infections are as follows: 103.5 TCID50 of A(H1N1)pdm09 (primary infection) and 105 PFU of hRSV Long (challenge infection; Bi and Bii), 105 PFU of hRSV Long (primary infection) and 103.5 TCID50 of A(H1N1)pdm09 (challenge infection; Biii and Biv), and 106 PFU of hRSV Long/A2 (primary infection) and 103.5 TCID50 of A(H1N1)pdm09 (challenge infection; Bv and Bvi). Fold changes (Bi, Biii, and Bv) were calculated by dividing the titer of the serum sample collected 14 days after challenge by the titer of the serum sample collected prior to primary infection. Horizontal lines indicate the median of each group, samples above the dotted line are positive for seroconversion. Titers (Bii, Biv, and Bvi) were measured in serum samples collected 14 days after challenge, with horizontal lines indicating GMT, and samples above the dotted line considered seropositive. Closed circles and open circles indicate animals that did or did not, respectively, shed detectable challenge virus, as determined by quantitative reverse-transcription polymerase chain reaction analysis of viral RNA from nasal wash (NW) samples. For statistical analysis, titers or fold changes were compared between test and control groups, using 1-way Kruskal-Wallis analysis of variance with the Dunn multiple comparison test. *P < .05 and **P < .01. C, The kinetics of shedding was analyzed for all ferrets that shed challenge virus in Figures 1 and 3. Data from ferrets obtained at the 3-day, 7-day, and 11-day intervals were pooled into the test group. The number of days from challenge inoculation to the peak level of challenge virus shedding (Ci and Ciii) and the number of days the challenge virus was shed (Cii and Civ) was determined for each ferret in the indicated groups. Horizontal lines indicate median values. The number of days of virus shedding were compared between test and control groups, using the Mann-Whitney test. *P < .05 and **P < .01.
Primary infection with A(H1N1)pdm09 prevented subsequent infection with hRSV in 3 of 4 ferrets when primary infection and challenge were separated by 3 days. Shedding of hRSV was minimal in the single ferret infected, compared with control animals (Figure 1B and 1C). No ferrets in this group seroconverted to hRSV (Figure 2Bi and 2Bii). Primary infection with A(H1N1)pdm09 prevented infection with hRSV in 2 of 4 ferrets when infections were separated by 7 days (Figure 1D). Ferrets that did not shed virus did not seroconvert (Figure 2Bi and 2Bii), while ferrets that shed virus seroconverted to hRSV (Figure 2Bi and 2Bii). Prior infection with A(H1N1)pdm09 did not prevent infection with hRSV 11 days later (Figure 1E), with all ferrets showing a similar pattern of virus shedding (Figure 1B) and antibody titers to control animals that received hRSV alone (Figure 2Bi and 2Bii).
The kinetics of hRSV shedding was examined in animals not protected from hRSV challenge. The peak of hRSV shedding was delayed in ferrets infected with A(H1N1)pdm09 followed by hRSV as compared to control animals infected with hRSV alone (median, 8 vs 6 days; P = .0091 by the Mann-Whitney test; Figure 2Ci). There was no change in the duration of virus shedding (Figure 2Cii).
Clinical signs following hRSV challenge were minimal (Supplementary Figure 2), consistent with our previous study [21]. All ferrets, except 1 control ferret infected with hRSV, maintained or gained weight (Supplementary Figure 2A–D).
hRSV Infection Can Reduce Morbidity Attributed to A(H1N1)pdm09 Infection
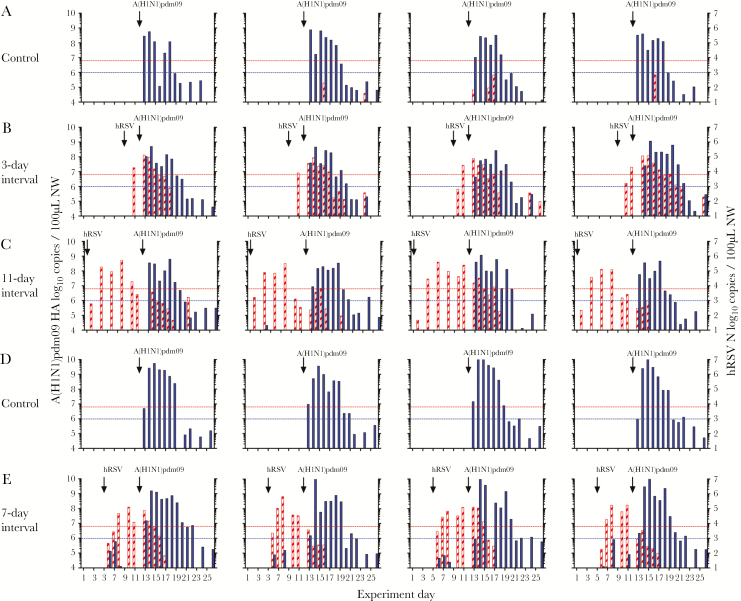
Primary infection with hRSV did not prevent infection with A(H1N1)pdm09 at any interval; rather, animals shed both hRSV and A(H1N1)pdm09, indicative of coinfection (Figure 3). All animals seroconverted to A(H1N1)pdm09 (Figure 2Biii) with similar GMTs as control animals (2153 [95% confidence interval {CI}, 1526–2779] at 3 days, 2153 [95% CI, 1526–2779] at 7 days, and 3620 [95% CI, 2172–5069] at 11 days, compared with 2152 [95% CI, 1742–2563] in the control group; Figure 2Biv). The median duration of A(H1N1)pdm09 shedding was increased in ferrets infected with hRSV followed by A(H1N1)pdm09 as compared to control animals infected with A(H1N1)pdm09 alone (8 vs 7 days; P = .0196 by the Mann-Whitney test; Figure 2Civ). There was no change in the peak day of shedding (Figure 2Ciii).
Figure 3.
Virus shedding among ferrets infected with human respiratory syncytial virus (hRSV), followed at intervals of 3, 7, or 11 days by 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09). A and D, Control ferrets infected with A(H1N1)pdm09 alone. B, C, and E, Ferrets underwent primary infection with 105 plaque-forming units (PFU) of hRSV strain Long following by challenge with 103.5 50% tissue culture infectious doses of A(H1N1)pdm09 3 (B), 7 (E), or 11 (C) days later. Note that control animals in panel A were in the same experiment as the test ferrets in the 3-day interval (B) and 11-day interval (C) groups, whereas control animals in panel D were in the same experiment as the test ferrets in the 7-day interval group (E). Quantitative reverse-transcription polymerase chain reaction analysis was used to detect the A(H1N1)pdm09 hemagglutinin gene (filled) and the hRSV N gene (striped) in viral RNA from nasal wash (NW) samples. The lower dotted lined indicate the limit of detection of infectious A(H1N1)pdm09, and the upper dotted lines indicate the limit of detection of infectious hRSV.
Prior infection with hRSV did reduce disease following infection with A(H1N1)pdm09. The mean maximum weight loss (±SD) among 8 control ferrets infected with A(H1N1)pdm09 was 10.6% ± 3.7% (Supplementary Figure 3A and 3D). The mean maximum weight loss (±SD) for ferrets in all test groups (n = 12) was 4.1% ± 2.3% (Supplementary Figure 3B, 3C, and 3E). Thus, prior infection with hRSV significantly reduced morbidity, as measured by weight loss, after challenge with A(H1N1)pdm09 (P = .0002 by the Mann-Whitney test). No fever was detected following A(H1N1)pdm09 infection (Supplementary Figure 3F–J).
An Increased Dose of Infectious hRSV or a Different Strain of hRSV Does Not Prevent or Limit Subsequent A(H1N1)pdm09 Infection
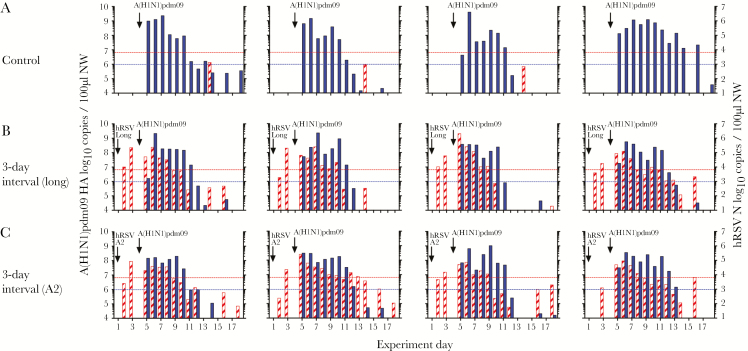
Ferrets were infected (1) with an increased viral dose of the same hRSV strain, Long, or (2) with an alternate hRSV strain, A2 (also at an increased viral dose), then challenged with A(H1N1)pdm09 3 days later. A2 is a laboratory-adapted strain that is shed at similar levels to Long in ferrets and transmits between cohoused animals [24]. Infection of ferrets with a 10-fold higher inoculum (ie, 106 PFU) of hRSV Long led to a small increase in virus shedding on days 2–6 after infection, compared with animals infected with 105 PFU of hRSV Long, although these differences were not significant (Supplementary Figure 4).
Primary infection with 106 PFU hRSV Long or A2 did not prevent infection with A(H1N1)pdm09 when infections were separated by 3 days (Figure 4). All animals seroconverted to A(H1N1)pdm09 at similar levels (Figure 2Bv and 2Bvi). Most ferrets lost weight after A(H1N1)pdm09 infection. The mean maximum weight loss (±SD) among 4 ferrets infected with A(H1N1)pdm09 alone was 7.1% ± 3.0%, whereas the mean maximum weight loss (±SD) for ferrets (n = 8) that received primary infection with hRSV prior to A(H1N1)pdm09 challenge was 4.9% ± 3.2% (P = .2828 by the Mann-Whitney test; Supplementary Figure 5A–C). There was no difference in fever (Supplementary Figure 5D–F) and no change to the kinetics of infection between animals that received a prior hRSV infection, compared with those that did not (data not shown).
Figure 4.
Virus shedding among ferrets infected with human respiratory syncytial virus (hRSV), followed at an interval of 3 days by 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09). A, Control ferrets infected with A(H1N1)pdm09 alone. B and C, Ferrets underwent primary infection with 106 plaque-forming units (PFU) of hRSV strain Long (B) or strain A2 (C), followed by challenge with 103.5 50% tissue culture infectious doses of A(H1N1)pdm09 3 days later. Quantitative reverse-transcription polymerase chain reaction analysis was used to detect the A(H1N1)pdm09 hemagglutinin gene (filled) and the hRSV N gene (striped) in viral RNA obtained from nasal wash (NW) samples. The lower dotted lines indicate the limit of detection of infectious A(H1N1)pdm09, and the upper dotted lines indicate the limit of detection of infectious hRSV.
A(H1N1)pdm09 Infection Induces Increased Levels of Proinflammatory Cytokines, Compared With hRSV Infection
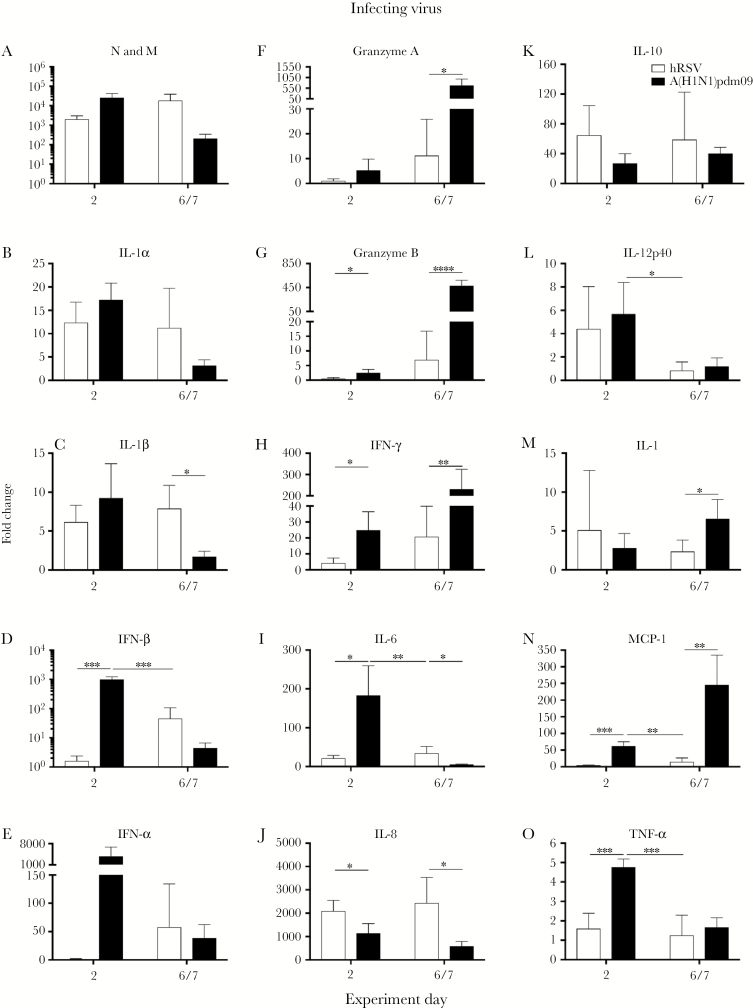
Inflammation induced by viral infection may contribute to viral interference [10, 11]. We investigated the localized immune response following infection with hRSV or A(H1N1)pdm09. Nasal wash specimens were collected early (day 2) and later (day 6/7) after infection, because the pattern of inflammatory mediators changes throughout H(H1N1)pdm09 [30] and hRSV [24] infections. Expression of influenza virus matrix (M) mRNA was highest on day 2 after infection, whereas expression of hRSV nucleoprotein (N) mRNA was highest on day 6 (Figure 5A). Two days after infection, animals infected with A(H1N1)pdm09 had significantly higher levels of interferon β (IFN-β), granzyme B, IFN-γ, interleukin 6 (IL-6), monocyte chemoattractant protein 1 (MCP-1), and tumor necrosis factor α (TNF-α) mRNA as compared to animals infected with hRSV (Figure 5D, 5G–I, 5N, and 5O). Expression of IFN-α and granzyme A mRNA was increased but not significantly (IFN-α, P = .097; granzyme A, P = .18; Figure 5E and 5F). On day 6/7 after infection, levels of granzyme A, granzyme B, IFN-γ, interleukin 17, and MCP-1 mRNA were significantly higher in animals infected with A(H1N1)pdm09 as compared to those infected with hRSV (Figure 5F–H, 5M, and 5N). There was significant increase in expression of interleukin 8 (IL-8) mRNA 2 days after infection and of interleukin 1β, IL-6, and IL-8 mRNAs 6/7 days after infection in ferrets infected with hRSV as compared to A(H1N1)pdm09 (Figure 5C, 5I, and 5J). This suggests a localized inflammatory response was induced after hRSV infection, which coincided with the increase in hRSV virus replication (Figure 5A).
Figure 5.
Expression of inflammatory mediator genes in messenger RNA from nasal wash samples from ferrets after infection with 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) or human respiratory syncytial virus (hRSV). Ferrets were infected with 105 plaque-forming units (PFU) of hRSV strain Long or 103.5 50% tissue culture infectious doses of A(H1N1)pdm09 (n = 4 ferrets/virus). Nasal wash specimens were collected after challenge from ferrets on days 2 and 6 after infection, and mRNA was assayed for the indicated genes, using quantitative polymerase chain reaction (qPCR) assays. For each graph, qPCR data are expressed as fold changes relative to values for nasal wash specimens from uninfected animals and normalized to ATF4 and GAPDH housekeeping genes. In panel A, expression of N is shown for hRSV-infected ferrets, and expression of M is shown for influenza virus–infected ferrets. For statistical analyses, inflammatory mediators were compared between (1) hRSV-infected and A(H1N1)pdm09-infected animals sampled on day 2 after infection, (2) hRSV-infected and A(H1N1)pdm09-infected animals sampled on day 6/7 after infection, and (3) A(H1N1)pdm09-infected animals sampled on day 2 and hRSV-infected animals sampled on day 6 after infection. Fold changes were compared between viruses, using the Mann-Whitney U test. IFN, interferon; IL-1, interleukin 1; IL-6, interleukin 6; IL-8, interleukin 8; IL-10, interleukin 10; IL-12p40, interleukin 12p40; IL-17, interleukin 17; MCP-1, monocyte chemoattractant protein 1; TNF-α, tumor necrosis factor α. *P < .05, **P < .01, ***P < .001, and ****P < .0001.
To directly compare the magnitude of expression of cytokines and chemokines induced by both virus infections, we assessed mRNA expression on the day after infection at which the level of virus shedding was highest (ie, day 2 for A(H1N1)pdm09 and day 6 for hRSV). When assessed at these times, an equivalent fold change in mRNA expression was observed for RSV N and influenza virus M (Figure 5A). Infection with A(H1N1)pdm09 induced significantly higher levels of IFN-β (P = .00021; Figure 5D), IL-6 (P = .0088; Figure 5I), interleukin 12p40 (P = .0137; Figure 5L), MCP-1 (P = .0018; Figure 5N), and TNF-α (P = .00078; Figure 5O) mRNA expression in ferrets, compared with hRSV. These data suggests there is increased inflammation in nasal tissues of animals infected with A(H1N1)pdm09 as compared to hRSV. There was no difference in expression of any cytokines or chemokines between ferrets infected with 105 or 106 PFU of hRSV Long (data not shown).
There Is Minimal Cross-reactive Immunity Between A(H1N1)pdm09 and hRSV
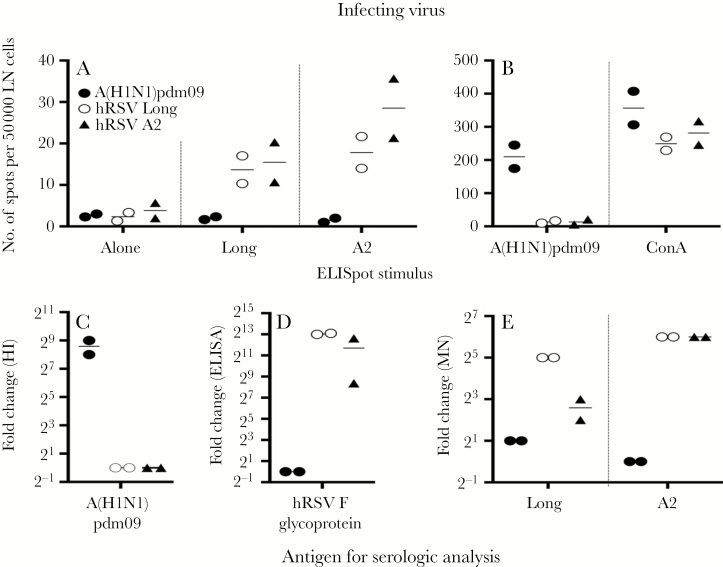
We have demonstrated that cross-reactive IFN-γ cellular responses can be detected between influenza B virus lineages and may contribute to viral interference [11]. Thus, we assessed whether cellular immunity induced by infection with A(H1N1)pdm09 showed any cross-reactivity to hRSV. Whereas retropharyngeal lymph node cells from A(H1N1)pdm09-infected ferrets were restimulated with A(H1N1)pdm09 (Figure 6B), few cells produced IFN-γ when stimulated with hRSV (Figure 6A). Lymph node cells from hRSV-infected ferrets were restimulated with hRSV in vitro, although at much lower levels (Figure 6A), and were not restimulated by A(H1N1)pdm09 (Figures 6B). Responses to concanavalin A were similar for all ferrets regardless of infection (Figure 6B). Moreover, there was limited serological cross-reactivity. Animals infected with A(H1N1)pdm09 had high levels of influenza virus–specific neutralizing antibodies (Figure 6C), yet minimal total serum or neutralizing antibodies to hRSV (Figure 6D and 6E). Similarly, infection with hRSV induced total serum and neutralizing antibodies to hRSV but few antibodies that were reactive with A(H1N1)pdm09 (Figure 6C–E).
Figure 6.
Assessment of cross-reactive immunological responses to with 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) and human respiratory syncytial virus (hRSV). Ferrets were infected with A(H1N1)pdm09 (black circles), hRSV strain Long (white circles), or hRSV strain A2 (black triangles), and retropharyngeal lymph nodes (LNs) and sera were collected 17 and 14 days after infection, respectively. A and B, Single-cell suspensions of LNs were restimulated in vitro with live hRSV (A), A(H1N1)pdm09, or concanavalin A (ConA; B). The number of interferon γ (IFN-γ)–producing cells was determined by an enzyme-linked immunospot (ELISpot) assay. C–E, Sera were tested for antibodies to A(H1N1)pdm09, by a hemagglutination inhibition (HI) assay (C); for total serum antibody binding to hRSV F protein, by an enzyme-linked immunosorbent assay (ELISA; D); and for neutralizing serum antibody to hRSV Long or A2, by a ViroSpot microneutralization (MN) assay (E). Data were obtained from 2 ferrets per group.
DISCUSSION
We have demonstrated that infection with A(H1N1)pdm09 can prevent infection and replication of hRSV in a ferret model of human disease for up to 7 days. Infection with hRSV did not prevent subsequent infection with A(H1N1)pdm09; rather, animals were coinfected, albeit with reduced morbidity. Infection with A(H1N1)pdm09 leads to increased levels of proinflammatory cytokines in the respiratory tract as compared to infection with hRSV. Overall, these data support the ecological observation that viral interference induced by A(H1N1)pdm09 infection delayed infection with hRSV in the winter of 2009–2010.
Infection with A(H1N1)pdm09 induced higher expression of MCP-1, IL-6, type I IFNs, TNF-α, IFN-γ, and granzyme A/B mRNAs as compared to hRSV infection. MCP-1 and TNF-α regulate the migration of macrophages/monocytes and natural killer (NK) cells into the respiratory tract. Macrophages produce MCP-1, TNF-α, and IL-6; thus, upregulation of these genes suggests an influx of macrophages and NK cells into the respiratory tissues [34]. NK cells produce IFN-γ, which activates macrophages and neutrophils and promotes T-cell proliferation and killing of virus-infected cells [34]. Because cytotoxic T lymphocytes and NK cells also produce granzymes A/B, increased expression of IFN-γ and granzyme A/B mRNAs on day 6 after infection suggests recruitment/activation of these cells to the site of infection. IL-6 and type I IFNs are produced by respiratory epithelial cells, monocytes/macrophages, and dendritic cells [34, 35]. IL-6 is a proinflammatory cytokine, whereas type I IFNs induce an antiviral state that may also limit replication and spread of hRSV [34, 35] It would be useful to explore the cellular infiltrate following A(H1N1)pdm09 and hRSV infections to gain further insight into potential differences in the level and cellular composition present in local inflammation. Infection with A(H1N1)pdm09 induced a 7-fold higher cellular IFN-γ recall response as compared to infection with hRSV in our study. Because there was no significant difference in IFN-γ responses to concanavalin A between the groups, this observation was not due to a difference in overall T-cell numbers but, instead, was due to an increase in the reactivation of A(H1N1)pdm09-specific cells. Taken together, these data suggest that infection with A(H1N1)pdm09 induces a robust cytokine and chemokine response that strongly stimulates the adaptive and memory immune responses. Conversely, infection with hRSV elicited a weaker and more limited cytokine and chemokine response that led to a reduced antigen-specific cellular response. However, it is possible that hRSV may not infect the ferret respiratory tract as efficiently as A(H1N1)pdm09 does, and this could result in reduced inflammatory responses. Yet, infected animals seroconverted at titers consistent for sterilizing immunity, indicating a productive infection (data not shown) [24]. Furthermore, increasing the inoculum of hRSV did not significantly affect the pattern or amount of virus shedding nor the expression of inflammatory mediators, suggesting that the hRSV level was already maximal in this ferret model. Notably, increased expression of inflammatory mediators following infection with influenza virus as compared to hRSV has been observed in studies assessing human clinical samples and in vitro airway epithelial cell cultures [36–39].
What is the mechanism of viral interference induced by A(H1N1)pdm09? The increased antiviral state and inflammation observed after A(H1N1)pdm09 infection has the potential to prevent subsequent infection or delay shedding of hRSV, as was observed here. Both viruses predominantly infect ciliated airway epithelial cells, and we have shown that A(H1N1)pdm09 and hRSV Long replicated in the upper and lower respiratory tracts of ferrets [24, 30]. Infection with A(H1N1)pdm09 can also prevent infection with an influenza B/Yamagata virus [10]. There are minimal shared epitopes between influenza A and B viruses [40], and we showed that minimal cross-reactive IFN-γ–producing cells were induced between hRSV and influenza virus. These data suggest that short-lived mechanisms drive this effect, as no effect was detected in ferrets after one week or, as shown by others, in mice, when infections were separated by 35 days [41]. The timing of interference indicate that interactions between different viruses may also be important. It is possible that different mechanisms act on different virus combinations. Gene expression analysis of early markers of the immune response of respiratory epithelium infected with the virus strains used in these studies may provide further insight.
It is notable that infection with hRSV reduced morbidity induced by A(H1N1)pdm09 infection. Although virus loads were not decreased in nasal wash specimens, virus shedding may be reduced in the lower respiratory tract, limiting clinical disease. IL-8 mRNA expression was elevated in nasal wash samples of ferrets infected with hRSV as compared to A(H1N1)pdm09. Increased IL-8 expression has been associated with milder disease in ferrets infected with pathogenic influenza virus strains, potentially mediated by rapid recruitment of neutrophils, which assist in clearing virus [42]. Analysis of the lung influenza virus loads in animals that have been infected with hRSV prior to challenge with A(H1N1)pdm09 would be of interest.
Epidemiological data reported in France described a 3–4-week delay in the peak incidence of hRSV infections following the emergence of the A(H1N1)pdm09, compared with previous years [1]. Similarly, a delay of 2-4 weeks of the expected peak of hRSV was reported following an early influenza A season in the Netherlands. [3]. These population-level observations of viral interference arise from the interplay between (1) immunodynamics (ie, host-level viral interference), (2) heterogeneity between hosts (ie, differences in immunity to virus strains between individuals), and (3) transmission dynamics (ie, within or between different age groups) [43]. For influenza, these processes have been investigated in some detail. Others have demonstrated that a short period (ie, days rather than weeks) of viral interference at the host level may result in substantial separation between epidemic waves at the population level [43]. Our results provide the first host-level immunodynamic evidence in support of these processes driving the epidemiological interactions observed previously in Europe [1, 2].
Our study has limitations. We used a circulating strain of A(H1N1)pdm09 from early 2009 and laboratory strains of hRSV, Long and A2. The Long and A2 strains induce consistent infections and disease in ferrets, with characterized cytokine profiles [24]. Use of a circulating clinical isolate of hRSV may provide more-realistic data but was not available for these experiments. Interference between influenza virus and hRSV has been reported in epidemiological studies in various years [2, 3], suggesting that influenza viruses other than A(H1N1)pdm09 may also prevent/limit infection and replication of hRSV.
There are currently no licensed vaccines for hRSV. Identification of host- and/or virus-encoded factors that contribute to viral interference provides a platform to facilitate development of novel prophylactic or therapeutic strategies to prevent or ameliorate respiratory infections, such as hRSV infection, that have a significant burden in the community, especially among young children.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Rubaiyea Farrukee and Celeste Tai for technical assistance.
Financial support. This work was supported by the Australian Government Department of Health (to the Melbourne WHO Collaborating Centre for Reference and Research on Influenza) and the Royal Melbourne Hospital Home Lottery.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Casalegno JS, Ottmann M, Bouscambert-Duchamp M, Valette M, Morfin F, B L. Impact of the 2009 influenza A(H1N1) pandemic wave on the pattern of hibernal respiratory virus epidemics, France, 2009. Eurosurveillance 2010;15:19485. [PubMed] [Google Scholar]
- 2. Nishimura N, Nishio H, Lee MJ, Uemura K. The clinical features of respiratory syncytial virus: lower respiratory tract infection after upper respiratory tract infection due to influenza virus. Pediatr Int 2005; 47:412–6. [DOI] [PubMed] [Google Scholar]
- 3. van Asten L, Bijkerk P, Fanoy E et al. Early occurrence of influenza A epidemics coincided with changes in occurrence of other respiratory virus infections. Influenza Other Respir Viruses 2016; 10:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang L, Chan KH, Suen LKP et al. Impact of the 2009 H1N1 pandemic on age-specific epidemic curves of other respiratory viruses: a comparison of pre-pandemic, pandemic and post-pandemic periods in a subtropical city. PLoS One 2015; 10:e0125447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cowling BJ, Fang VJ, Nishiura H et al. Increased risk of noninfluenza respiratory virus infections associated with receipt of inactivated influenza vaccine. Clin Infect Dis 2012; 54:1778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karppinen S, Toivonen L, Schuez-Havupalo L, M W, Peltola V. Interference between respiratory syncytial virus and rhinovirus in respiratory tract infections in children. Clin Microbiol Infect 2015:1.e1–1.e6. [DOI] [PubMed] [Google Scholar]
- 7. Martin ET, Pairchok MP, Stednicj ZJ, Kuypers J, Englund JA. Epidemiology of multiple respiratory viruses in childcare attendees. JID 2013; 207:9182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanner H, Boxall E, Osman H. Respiratory viral infections during the 2009–2010 winter season in Central England, UK: incidence and patterns of multiple virus co-infections. Eur J Clin Microbiol Infect Dis. 2012; 31:3001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costa-Hurtado M, Afonso CL, Miller PJ et al. Virus interference between H7N2 low pathogenic avian influenza virus and lentogenic Newcastle disease virus in experimental co-infections in chickens and turkeys. Vet Res 2014; 45:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laurie KL, Guarnaccia TA, Carolan LA et al. Interval between infections and viral hierarchy are determinants of viral interference following influenza virus infection in a ferret model. J Infect Dis 2015; 212:1701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laurie KL, Horman W, Carolan LA et al. Evidence for viral interference and cross-reactive protective immunity between influenza B virus lineages. 2017. Submitted. [DOI] [PMC free article] [PubMed]
- 12. Selin LK VS, Wong IC, Welsh RM. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J Exp Med 1998; 188:1705–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen HD, Fraire AE, Joris I, Welsh RM, Selin LK. Specific history of heterologous virus infections determines anti-viral immunity and immunopathology in the lung. Am J Pathol 2003; 163:1341–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wanitchang A, Narkpuk J, Jaru-ampornpan P, Jengarn J, Jongkaewwattana A. Inhibition of influenza A virus replication by influenza B virus nucleoprotein: an insight into interference between influenza A and B viruses. Virology 2012; 432:194–203. [DOI] [PubMed] [Google Scholar]
- 15. Salas-Benito JS, De Nova-Ocampo M. Viral Interference and persistence in mosquito-borne flaviviruses. J Immunol Res 2015; 2015:873404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Reeth K, Nauwynck H, Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol 1996; 48:325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Casalegno JS, Ottmann M, Bouscambert Duchamp M et al. Rhinoviruses delayed the circulation of the pandemic influenza A(H1N1) 2009 virus in France. Clin Microbiol Infect 2010; 16:326–9. [DOI] [PubMed] [Google Scholar]
- 18. Manicassamy B, Medina RA, Hai R et al. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog 2010; 6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Belser JA, Katz JM, Tumpey TM. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 2011; 4:575–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Enkirch T, vM V. Ferret models of viral pathogenesis. Virology 2015; 479–480:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coates HV, Chanock RM. Experimental infection with respiratory syncytial virus in several species of animals. Am J Hyg 1962; 76:302–12. [DOI] [PubMed] [Google Scholar]
- 22. Hsu KH, Lubeck MD, Bhat BM et al. Efficacy of adenovirus-vectored respiratory syncytial virus vaccines in a new ferret model. Vaccine 1994; 12:607–12. [DOI] [PubMed] [Google Scholar]
- 23. Prince GA, Porter DD. The pathogenesis of respiratory syncytial virus infection in infant ferrets. Am J Pathol 1976; 82:339–52. [PMC free article] [PubMed] [Google Scholar]
- 24. Chan KF, Carolan LA, Druce J et al. Pathogenesis, humoral immune responses and transmission between co-housed animals in a ferret model of human RSV infection. J Virol 2017; 92:e01322–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stittelaar KJ, de Waal L, van Amerongen G et al. Ferrets as a novel animal model for studying human respiratory syncytial virus infections in immunocompetent and immunocompromised hosts. Viruses 2016; 8:168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J Infect Dis 2010; 202:1011–20. [DOI] [PubMed] [Google Scholar]
- 27. Cao P, Yan AWC, Heffernan JM et al. Innate immunity and the inter-exposure interval determine the dynamics of secondary influenza virus infection and explain observed viral hierarchies. PLoS Comput Biol 2015; 11:e1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan AW, Cao P, Heffernan JM et al. Modelling cross-reactivity and memory in the cellular adaptive immune response to influenza infection in the host. J Theor Biol 2017; 413:34–49. [DOI] [PubMed] [Google Scholar]
- 29. Oh DY, Barr IG, Mosse JA, Laurie KL. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol 2008; 46:2189–94. doi:JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carolan LA, Rockman S, Borg K et al. Characterisation of the localised immune response in the respiratory tract of ferrets following infection with influenza A and B viruses. J Virol 2015; 90:2838–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carolan LA, Butler J, Rockman S et al. TaqMan real time RT-PCR assays for detecting ferret innate and adaptive immune responses. J Virol Methods 2014; 205:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. World Health Organization. Global Influenza Surveillance NetworkManual for the laboratory diagnosis and virological surveillance of influenza, 1982. http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf. Accessed 17 April 2018. [Google Scholar]
- 33. Guarnaccia T, Carolan LA, Maurer-Stroh S et al. Antigenic drift of the pandemic 2009 A(H1N1) influenza virus in a ferret model. PLoS Pathog 2013; 9:e1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. La Gruta NL, Kedzierska K, Stambas J, Doherty PC. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol 2007; 85:85–92. [DOI] [PubMed] [Google Scholar]
- 35. Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 2009; 29:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Domachowske JB, Rosenberg HF. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin Microbiol Rev 1999; 12:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hall CB, Douglas RG Jr, Simons RL, Geiman JM. Interferon production in children with respiratory syncytial, influenza, and parainfluenza virus infections. J Pediatr 1978; 93:28–32. [DOI] [PubMed] [Google Scholar]
- 38. Ioannidis I, McNally B, Willette M et al. Plasticity and virus specifcity of airway epithelial cell immune response during respiratory virus infection. Clin Microbiol Rev. 1999;12:298–309.10194461 [Google Scholar]
- 39. McIntosh K. Interferon in nasal secretions from infants with viral respiratory tract infections. J Pediatr 1978; 93:33–6. [DOI] [PubMed] [Google Scholar]
- 40. Terajima M, Babon JA, Co MD, Ennis FA. Cross-reactive human B cell and T cell epitopes between influenza A and B viruses. Virol J 2013; 10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walzl G, Tafuro S, Moss P, Openshaw PJ, Hussell T. Influenza virus lung infection protects from respiratory syncytial virus-induced immunopathology. J Exp Med 2000; 192:1317–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Svitek N, Rudd PA, Obojes K, Pillet S, von Messling V. Severe seasonal influenza in ferrets correlates with reduced interferon and increased IL-6 induction. Virology 2008; 376:53–9. [DOI] [PubMed] [Google Scholar]
- 43. Camacho A, Cazelles B. Does homologous reinfection drive multiple-wave influenza outbreaks? Accounting for immunodynamics in epidemiological models. Epidemics 2013; 5:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.