We report a second case of pigeon paramyxovirus type 1 infection, which has pigeons and doves as reservoir, causing fatal respiratory disease in a human patient under immunosuppressive therapy. We recommend testing for this virus in respiratory disease cases where common respiratory pathogens cannot be identified.
Keywords: communicable diseases, emerging, immunocompromised host, Paramyxoviridae, respiratory distress syndrome, adult, zoonoses
Abstract
The characteristics and risk factors of pigeon paramyxovirus type 1 (PPMV-1) infection in humans are poorly known. We performed virological, pathological, and epidemiological analyses of a Dutch case, and compared the results with those of a US case. Both infections occurred in transplant patients under immunosuppressive therapy and caused fatal respiratory failure. Both virus isolates clustered with PPMV-1, which has pigeons and doves as reservoir. Experimentally inoculated pigeons became infected and transmitted the virus to naive pigeons. Both patients were likely infected by contact with infected pigeons or doves. Given the large populations of feral pigeons with PPMV-1 infection in cities, increasing urbanization, and a higher proportion of immunocompromised individuals, the risk of severe human PPMV-1 infections may increase. We recommend testing for avian paramyxovirus type 1, including PPMV-1, in respiratory disease cases where common respiratory pathogens cannot be identified.
Changes in socioeconomic, environmental, and ecological factors in recent decades are probably responsible for the significant increase of emerging infectious diseases, the majority of which are zoonotic [1]. Of particular concern are emerging respiratory viruses of the families Orthomyxoviridae [2, 3], Coronaviridae [4, 5], and Paramyxoviridae [6, 7]. The use of novel molecular techniques has made it possible to identify previously unsuspected or unknown viruses [8]. However, it is important to assess the clinical and public health relevance of these viruses by determining their origin and establishing their impact in the human population.
The impetus for the current study was the identification of a virus related to avian paramyxovirus type 1 (APMV-1) from a fatal human case of unknown cause in the Netherlands by viral metagenomics analysis [8]. APMV-1 is classified in the genus Avulavirus, family Paramyxoviridae [9]. Infection with APMV-1 is mainly restricted to birds, but occasionally causes mild disease—usually transient conjunctivitis—in humans [10]. So far, there is only one case report, from New York State, of a person who died with APMV-1 infection [11].
In this study, we fully characterized the Dutch clinical isolate of APMV-1–like virus, determined its phylogenetic relationship to other APMV-1 strains, and correlated presence of this virus with lesions in tissues obtained from the patient at autopsy. We also examined virulence and excretion pattern of this virus in chickens and domestic pigeons.
METHODS
Case Description
A 54-year-old woman with a history of multiple myeloma received an allogenic bone marrow transplant in September 2002, following preparation with a nonmyeloablative regimen (cyclosporin) and while on antifungal therapy (intravenous itraconazole). She initially did well, but was hospitalized 8 weeks after transplantation with symptoms of malaise, anorexia, fever, and abdominal complaints (including abdominal pain, diarrhea, and food intolerance). She lived in the country near to a city, but was not employed in agriculture, and there was no evidence of contact with animals in her clinical history. Gastroscopic examination showed no abnormalities. Infiltrative lesions in the thorax were detected by radiological and computed tomographic examinations, and there was progressive swelling of cervical lymph nodes. No pathogens were detected in bronchoalveolar lavage (BAL) specimens (virus culture on HEL, LLC-MK2, HEp-2, and human rhabdomyosarcoma cells; bacterial culture on blood agar, chocolate agar, Columbia blood agar containing colistin and nalidixic acid, and Sabouraud agar; cytological examination for bacteria, fungi, and Pneumocystis carinii), and a biopsy of the peripheral right lower lung lobe was histologically normal. Epstein-Barr virus (EBV) antigen expression and lymphoproliferation were detected in a biopsy of the right jugular lymph node, and a blood sample was positive for EBV DNA by polymerase chain reaction (PCR). Reactivation of EBV and associated lymphoproliferation were treated successfully with rituximab (anti-CD20) and discontinuation of cyclosporin therapy, but other clinical signs remained. Enteroscopic examination of the small intestine showed no abnormalities, and no pathogens were detected in fecal samples by bacterial culture.
Five weeks after hospitalization, the patient developed high fever and progressive worsening of respiratory signs. Treatment with cotrimoxazole was initiated for suspected P. carinii infection, and with prednisone for suspected bronchiolitis obliterans organizing pneumonia or graft-vs-host disease of the lung. Two weeks later, the patient developed respiratory failure. There was crepitation over both lung fields and bilateral, progressively expanding, blotchy infiltrates in the thorax by radiological examination. Gram-negative, rod-shaped bacteria (Pseudomonas species) were detected by microscopic examination of BAL fluid, and treatment with ceftazidime and tobramycin was initiated. Despite intubation and artificial respiration at increasing pressure, respiratory function deteriorated further and circulatory failure developed. Artificial respiration was stopped after 3 days, after which the patient died.
Autopsy was performed 1 day after death. There were extensive areas of bronchopneumonia associated with Pseudomonas species infection in both lungs by bacterial culture. In addition, there was marked diffuse alveolar damage, characterized by abundant hyaline membranes lining the alveolar septa. The mucosa of the colon ascendens was lined by a hemorrhagic exudate, but autolysis was too advanced to confirm or rule out graft-vs-host disease, and no satisfactory explanation was found for the abdominal complaints. In conclusion, the progressive respiratory failure was explained by severe diffuse alveolar damage of undetermined cause, and extensive bilateral bronchopneumonia caused by terminal Pseudomonas species infection.
Metagenomic Analysis of Suspect Cell Culture Specimens
As part of an in-depth investigation, archived cell culture samples with an unexplained cytopathic effect were subjected to metagenomic analysis as described previously [8]. These samples included human rhabdomyosarcoma cells inoculated with a BAL specimen collected in 2002 from the above patient. Metagenomic analysis revealed the presence of short nucleotide fragments with homology to APMV-1. To complete the genome of the clinical isolate, fragments obtained through whole transcriptome amplification were assembled against the APMV-1 genome with the highest similarity (Supplementary Methods). In brief, the assembly was performed using Sequencher software (version 4.1; Gene Codes). Because the whole-transcriptome amplification fragments did not overlap into a complete genome, the assembly was used to design primers for genome walking, PCR assays were performed with these primers, and products were sequenced. These efforts resulted in the genome completion of the Dutch clinical virus isolate.
Phylogenetic Analysis of the Dutch Clinical Virus Isolate
Nucleotide sequences were aligned with the Clustal W program running within the BioEdit software package, version 5.0.9 [12]. With the nucleotide sequence alignment, the best-fit model of nucleotide substitution was determined by jModelTest [13]. Maximum-likelihood (ML) phylogenetic trees were generated using the GTR+Γ4+I model of nucleotide substitution and the PhyML package version 3.0 [14]. The reliability of all phylogenetic groupings was determined through a bootstrap resampling analysis of a 1000 replicates with PhyML. Trees were visualized through the FigTree program version 1.4. (http://tree.bio.ed.ac.uk/software/figtree/) and rooted on the APMV-1 with the highest BLAST similarity in GenBank (APMV-1/NL/152608/1993).
Virological and Pathological Analyses of Biopsies and Autopsy Specimens
The following tissue biopsies and autopsy tissue specimens had been fixed in 10% neutral-buffered formalin and embedded in paraffin blocks: heart, lung, jugular lymph node, esophagus, thyroid gland, stomach, pancreas, liver, kidney, skin, diaphragm, and muscle. These specimens were used for detection of APMV-1 RNA by real-time reverse-transcription PCR (RT-PCR), of APMV-1 antigen by immunoperoxidase method, and of histological lesions by hematoxylin and eosin stain (Supplementary Methods).
Characterization of Virulence of the Dutch Clinical Virus Isolate for Chickens and Pigeons
Chickens were inoculated intracerebrally with the Dutch clinical virus isolate to determine the intracerebral pathogenicity index (Supplementary Methods). Domestic pigeons were inoculated intratracheally with the Dutch clinical virus isolate to determine infectivity and transmissibility, clinical signs, and pathological changes (Supplementary Methods).
RESULTS
Phylogenetic Analysis
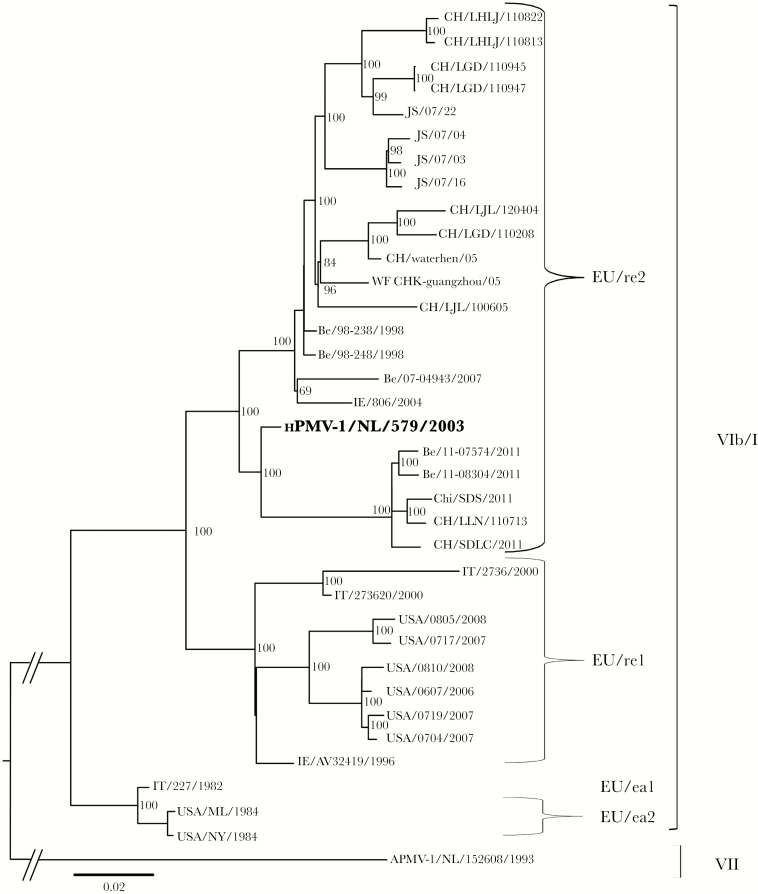
The sequence of the full-length genome of the Dutch clinical virus isolate displayed 97% nucleotide sequence homology with an APMV-1 isolated from pigeons in Belgium (APMV-1/Belgium/98–238/1998; JX901109). Phylogenetic analysis of full-length genomic sequences of closely related APMV-1 isolates demonstrated that the Dutch clinical isolate clustered among Belgian and Chinese avian strains in genotype VIb/1 (Figure 1). Viruses in this genotype are also called pigeon paramyxovirus type 1 (PPMV-1), because pigeons and doves are the animal reservoir. Therefore, the Dutch clinical virus isolate was named human PPMV-1 (hPPMV-1/NL/579/2003; accession number KJ544861).
Figure 1.
Maximum likelihood phylogenetic tree depicting the relationship of the Dutch clinical isolate (hPPMV-1/NL/579/2003) with related avian paramyxovirus type 1 (APMV-1) strains based on analysis of full-length genomes. Assignment of APMV-1 genotypes (VIb/1 or VII) and sublineages (EUre1, EUre2, EUea1, or EUea2) was based on the studies of Ujvári et al [16] and Czegledi et al [35] and the sequence comparison shown in Table 1. GenBank accession numbers are given in parentheses: hPPMV-1/NL/579/2003 (KJ544861), Be/98–238/1998 (JX901109), Be/98–248/1998 (JX901110), WF CHK-guangzhou/2005 (HM063425), IE/806/2004 (JN986839), CH/waterhen/05 (HM063423), CH/SDLC/2011 (JQ979176), CH/LLN/110713 (JX486552), Be/07-04943/2007 (JX901121), Chi/SDS/2011 (JQ993431), Be/11-08304/2011 (JX901123), JS/07/03 (FJ766531), JS/07/22 (FJ766526), JS/07/16 (FJ766527), Be/11-07574/2011 (JX901122), JS/07/04 (FJ766530), CH/LGD/110947 (JX486556), CH/LGD/110945 (JX486555), IE/AV32419/1996 (GQ429292), CH/LJL/100605 (JX486551), CH/LHLJ/110813 (JX486553), CH/LGD/110208 (JX486550), CH/LHLJ/110822 (JX486554), CH/LJL/120404 (JX486557), IT/273620/2000 (GQ429293), USA/0607/2006 (KC013033), IT/227/1982 (AJ880277), USA/0704/2007 (KC013034), USA/0810/2008 (KC013038), USA/0719/2007 (KC013036), USA/0717/2007 (KC013035), USA/0805/2008 (KC013037), USA/NY/1984 (FJ410145), USA/ML/1984 (FJ410147), IT/2736/2000 (AY562989), APMV-1/NL/152608/1993 (JN986837).
Comparison of predicted amino acid sequences revealed that all of the hPPMV-1/NL/579/2003 encoded proteins displayed >95% identity with those of other PPMV-1. In general, the fusion protein (F) cleavage site of medium/high virulent APMV-1s contain the sequence 113RQ(K/R)R*F117, while those of low virulent APMV-1s usually have the sequence 113(K/R)Q(G/E)R*L117. The cleavage site of hPPMV-1/NL/579/2003 (113RQKR*F117) was consistent with a virulent pathotype, similar to that observed in all PPMV-1 (113R(Q/K) KR*F117) [15–17].
Based on phylogenetic analysis of sequences of partial F protein sequences, Ujvári et al identified 4 sublineages of PPMV-1 (ie, APMV-1 genotype VIb/1) strains. These subgroups, designated Iraqi (IQ), early European (EU/ea), North American (NA), and recent European (EU/re), corresponded partly to the times of isolation and/or geographical origin [16]. Alignment of the F protein amino acid sequences from hPPMV-1/NL/579/2003 with those of Ujvári and others revealed that it had identical amino acids substitutions as isolates belonging to sublineage EUR/re2 (Table 1). No sequence information was available for this partial F protein sequence of the only other APMV-1 clinical isolate, from a fatal human case in New York state [11]. The available partial sequence for this New York clinical virus isolate demonstrated the closest relationship with a PPMV-1 isolated from pigeons in New York in 2006 (KC013033.1), and clustering in sublineage VIb/1 NA1 (data not shown).
Table 1.
Amino Acid Sequence (Position on Top) Comparison Around the Proteolytic Cleavage Site of the Fusion Protein Between the Dutch Clinical Isolate (hPPMV-1/NL/579/2003) and Representative Avian Paramyxovirus Type 1 Strains Used for Differentiation of Sublineages
| Strain | Accession No.a | Geno-type | Sub- lineage | 8 | 11 | 13 | 14 | 19 | 20 | 22 | 27 | 28 | 49 | 50 | 60 | 63 | 90 | 111 | 112 | 113 | 114 | 115 | 117 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken/NL/152608/93 | JN986837 | VII | R | V | L | M | I | M | I | C | L | A | V | S | V | T | G | R | R | Q | K | F | |
| Iraq’78 | AY150092 | VIb/1 | IQ | … | … | … | … | … | T | … | … | P | … | … | … | … | … | … | … | … | … | … | … |
| IT-227/1982 | AY150093* | VIb/1 | EU/ea1 | … | … | … | T | T | … | … | … | P | … | … | … | … | … | … | G | … | … | … | … |
| SE-2/83 | AY150100 | VIb/1 | EU/ea1 | … | … | … | T | T | … | … | … | P | … | … | … | … | … | … | G | … | … | … | … |
| DE-209/83 | AY150095 | VIb/1 | EU/ea1 | … | … | … | T | T | … | … | … | P | … | … | … | … | … | … | G | … | … | … | … |
| HU-491/87 | AY150114 | VIb/1 | EU/ea2 | M | … | … | T | T | … | … | … | P | … | … | … | … | … | … | G | … | … | … | … |
| HU-655/89 | AY150120 | VIb/1 | EU/ea2 | M | … | … | T | T | … | L | … | P | … | … | … | … | … | … | G | … | … | … | … |
| USA/NY/1984 | FJ410145* | VIb/1 | M | … | … | T | T | … | … | … | P | … | … | … | … | … | … | … | … | … | … | … | |
| USA/ML/1984 | FJ410147* | VIb/1 | M | … | … | T | T | … | … | … | P | … | … | … | … | … | … | … | … | … | … | … | |
| US-17/87 | AY150116 | VIb/1 | NA 1 | … | … | P | … | … | … | … | … | Q | … | I | … | … | … | E | … | … | … | … | … |
| CA-24/88 | AY150117 | VIb/1 | NA 1 | … | … | P | … | … | … | … | … | Q | … | I | … | … | … | E | … | … | … | … | … |
| MX-11/98 | AY150152 | VIb/1 | NA 1 | … | … | P | … | … | V | … | … | Q | … | I | … | … | … | E | … | … | … | … | … |
| HU-61/83 | AY150099 | VIb/1 | NA 2 | … | … | P | … | … | … | … | … | P | … | I | … | … | … | E | … | … | … | … | … |
| DE-47/87 | AY150110 | VIb/1 | NA 2 | … | … | P | … | … | … | … | … | P | … | I | … | … | … | E | … | … | … | … | … |
| DK-12/93 | AY150138 | VIb/1 | NA 2 | … | … | P | … | … | … | … | … | P | … | I | … | … | … | E | … | … | … | … | … |
| ES-3/92 | AY150132 | VIb/1 | EU/re1 | K | A | … | … | … | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … |
| DE-48/92 | AY150129 | VIb/1 | EU/re1 | … | A | … | … | … | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … |
| DE-51/93 | AY150133 | VIb/1 | EU/re1 | … | A | … | … | V | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … |
| DE-57/95 | AY150143 | VIb/1 | EU/re1 | … | A | … | … | … | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … |
| DE-2653/98 | AY150149 | VIb/1 | EU/re1 | … | A | … | … | … | T | … | … | … | V | I | … | … | … | … | … | … | K | … | … |
| IE/AV324/1996 | GQ429292* | VIb/1 | … | A | … | … | … | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … | |
| USA/0607/2006 | KC013033* | VIb/1 | … | A | … | … | … | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … | |
| USA/0704/2007 | KC013034* | VIb/1 | … | A | … | … | … | T | … | … | … | V | I | … | … | … | V | … | … | K | … | … | |
| IT/2736/2000 | GQ429293* | VIb/1 | … | A | … | … | … | T | T | … | … | V | I | … | … | … | V | … | … | K | … | … | |
| HU-1G/00 | AY150155 | VIb/1 | EU/re2 | … | A | … | … | … | T | … | … | P | … | I | … | … | … | … | … | … | … | … | … |
| YU(Vo)-595/01 | AY150164 | VIb/1 | EU/re2 | … | A | … | … | … | T | … | … | P | … | I | … | … | … | … | … | … | … | … | … |
| IT-125/87 | AY150115 | VIb/1 | EU/re2 | … | A | … | … | … | T | … | … | P | … | I | … | … | … | … | … | … | … | … | … |
| DE-95/92 | AY150130 | VIb/1 | EU/re2 | … | T | … | … | … | T | … | … | P | … | I | … | … | … | … | … | … | … | … | … |
| hPPMV-1/NL/579/2003 | KJ544861* | VIb/1 | … | A | P | … | … | T | V | … | P | … | I | … | … | … | … | … | … | … | … | … | |
| CH/SDLC/2011 | JQ979176* | VIb/1 | … | A | P | T | … | T | V | … | S | … | I | … | … | … | … | … | … | … | … | … | |
| Be/11-08304/2011 | JX901123* | VIb/1 | … | A | … | T | … | I | V | … | S | … | I | … | … | … | … | … | … | … | … | … | |
| DE-690/00 | AY150154 | VIb/1 | EU/re2 | M | A | … | … | … | T | V | … | … | … | I | … | … | … | … | … | … | … | … | … |
| Be/98–248/1998 | JX901110* | VIb/1 | … | A | … | … | … | T | V | … | … | … | I | … | … | … | … | … | … | … | … | … | |
| Be/07-04943/2007 | JX901121* | VIb/1 | … | A | … | … | … | T | V | … | … | … | I | … | … | … | … | … | … | … | … | … | |
| CH/LGD/110947 | JX486556* | VIb/1 | G | A | P | … | … | T | V | R | … | … | I | … | … | … | … | … | … | … | … | … | |
| HR-65/95 e | AY150144 | Vib/2 | … | A | … | I | … | T | … | R | … | … | … | L | I | A | … | K | … | … | … | … | |
| HR-111/01 | AY150162 | Vib/2 | … | A | … | … | … | T | … | R | … | … | … | L | I | A | … | K | … | … | … | … | |
| HR-3/02 | AY150165 | Vib/2 | … | A | … | … | … | T | … | R | … | … | … | L | I | A | … | K | … | … | … | … |
Virological and Pathological Analyses of Biopsies and Autopsy Specimens
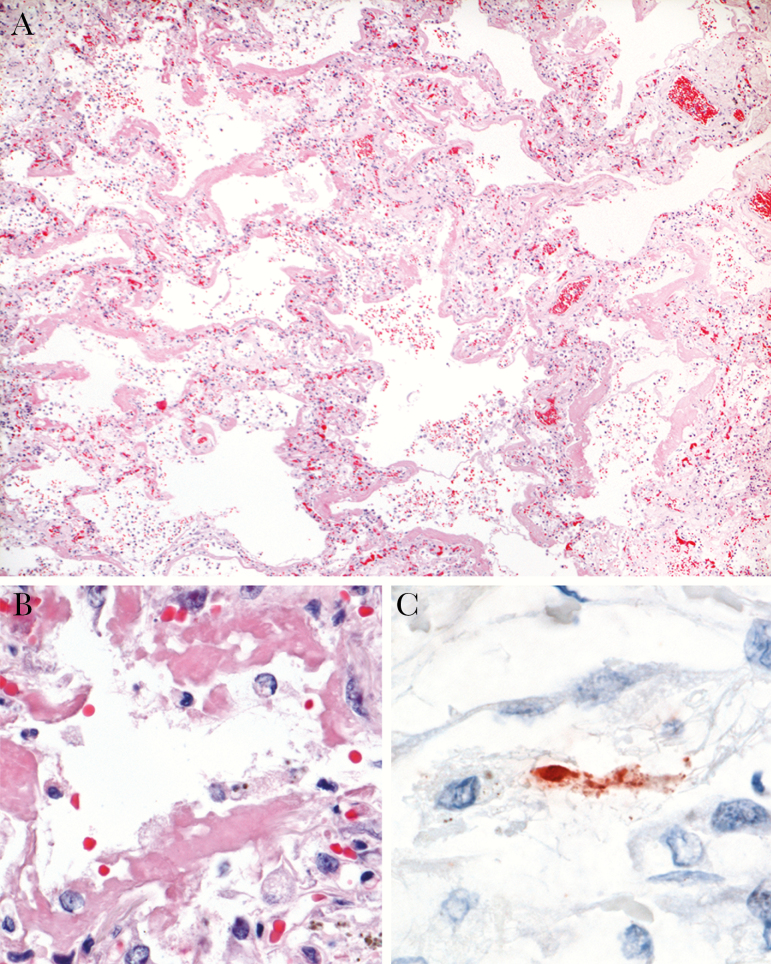
Real-time RT-PCR analysis revealed the presence of high levels of PPMV-1 RNA in the left and right lung (Table 2). Lower levels were detected in liver, kidney, and bone marrow, while the remaining tissues tested negative. Immunohistochemistry analysis revealed the expression of PPMV-1 antigen, visible as red-brown granular staining, in the same tissues that tested positive with real-time RT-PCR (Table 2). The exact localization of these granules was not clear, although some aggregates of granules appeared to be located in the cytoplasm of degenerate cells (Figure 2). PPMV-1 antigen expression was present in the positive control tissue and absent in the isotype control and negative control tissues.
Table 2.
Detection of Pigeon Paramyxovirus Type 1 by Real-time Reverse-Transcription Polymerase Chain Reaction and Immunohistochemistry in Tissues Collected From the Patient
| Tissue (No.) | PPMV-1 RNA (Ct) | PPMV-1 Antigen |
|---|---|---|
| Left lung (n = 2) | 31 | + |
| 21 | ++ | |
| Right lung (n = 3) | 25 | ++ |
| 23 | + | |
| 27 | ++ | |
| Liver (n = 1) | 31 | + |
| Left kidney (n = 1) | 33 | + |
| Bone marrow (n = 1) | 37 | + |
Heart, esophagus, stomach, pancreas, right kidney, skin, diaphragm, and muscle tested negative for PPMV-1 RNA by real-time reverse-transcription polymerase chain reaction and for PPMV-1 antigen by immunohistochemistry; lung biopsy and lymph node biopsy tested negative for PPMV-1 antigen by immunohistochemistry.
Abbreviations: +, rare positive granules; ++, occasional positive granules; Ct, cycle threshold; PPMV-1, pigeon paramyxovirus type 1.
Figure 2.
Pulmonary immunohistochemistry and histopathology of the patient with hPPMV-1/NL/579/2003 infection. A, Diffuse alveolar damage, characterized by thickening of alveolar septa, flooding of alveolar lumina, and presence of hyaline membranes (hematoxylin and eosin [H&E] staining; original magnification ×4.) B, Higher magnification of A, showing single pulmonary alveole, where epithelial lining is replaced by hyaline membranes (H&E; original magnification ×40.) C, Avian paramyxovirus type 1 antigen visible as granular staining in cytoplasm of degenerate cell, suggestive of alveolar epithelial cell (immunoperoxidase stain for pigeon paramyxovirus type 1 [PPMV-1]; original magnification ×100.).
Histopathological examination demonstrated that PPMV-1 antigen–expressing sections of lung had diffuse alveolar damage, characterized by loss of alveolar epithelium, rare hypertrophic type II pneumocytes, widened alveolar septa, and fibrin thrombi in alveolar capillaries (Figure 2). No histological lesions were apparent in other tissues that expressed PPMV-1 antigen, or in PPMV-1-negative tissues.
Characterization of hPPMV-1/NL/579/2003 in Chickens and Pigeons
Based on intracerebral inoculation into 1-day-old chickens, the intracerebral pathogenicity index of hPPMV-1/NL/579/2003 was 0.164 (Supplementary Table 2). This corresponds with a lentogenic pathotype. In pigeons, all 5 intratracheally inoculated animals developed a productive infection, with shedding from the pharynx between 2 and 10 dpi, and from the cloaca between 7 and 17 dpi (Supplementary Figure 1). Virus was transmitted to 1 of the 2 naive pigeons. None of the pigeons displayed clinical signs at any time point during the 3-week observation period. No gross lesions were observed in any pigeons at autopsy at 21 dpi. However, histopathological analysis showed that all 5 intratracheally inoculated pigeons plus the 1 infected naive pigeon had diffuse interstitial nonpurulent nephritis, which was absent in 2 negative control pigeons from the same flock (Supplementary Figure 2). Also, 1 inoculated pigeon had multifocal nonpurulent pancreatitis. Real-time RT-PCR analysis revealed that hPPMV-1/NL/579/2003 was present in the kidney sample of 3 inoculated pigeons with nephritis, and in the pancreas sample of 2 inoculated pigeons, including the 1 with pancreatitis (Supplementary Figure 3).
DISCUSSION
We diagnosed PPMV-1 infection as the cause of diffuse alveolar damage in the Dutch female patient, based on colocalization of PPMV-1 antigen, high loads of PPMV-1 RNA, and characteristic histological lesions in autopsy specimens (Figure 2 and Table 2). The PPMV-1–associated diffuse alveolar damage was exacerbated by bronchopneumonia due to Pseudomonas species infection, probably from intravenous catheterization. The combination of these 2 pulmonary diseases likely explains the severe respiratory failure leading to the death of this immunosuppressed patient.
It is rare to diagnose PPMV-1 infection as the cause of severe respiratory disease in humans. To our knowledge, the only other known case was reported by the New York State Department of Health in 2007 [11]. It is not clear whether this paucity of reports is because the disease is rare or because APMV-1, including PPMV-1, is not suspected in such cases. We suggest that APMV-1, including PPMV-1, should be included in the differential diagnosis of cases of respiratory disease that test negative for more common respiratory pathogens. Samples could be tested by various molecular methods, such as a family-wide RT-PCR assay for paramyxoviruses or metagenomics approaches [18, 19].
When they became infected with PPMV-1, the Dutch and New York patients were receiving immunosuppressive therapy to improve the success of peripheral blood stem cell or bone marrow transplantation. There is an increase in the proportion of immunocompromised people in the population [20]. As these immunocompromised people are at high risk of severe disease from other opportunistic infections [20], they also may be at risk of severe disease from infection with APMV-1, including PPMV-1.
PPMV-1 appears to target the lower respiratory tract epithelium in humans. This is based on the demonstration of PPMV-1 antigen expression in the alveolar epithelium of autopsy lung samples from both human cases (Figure 2) [11]. The tissue tropism of PPMV-1 in these 2 human cases resembles that of other emerging zoonotic respiratory viruses—H5N1 influenza virus and the severe acute respiratory syndrome and Middle East respiratory syndrome coronaviruses—that also target the lower respiratory tract [20, 22–24].
There was evidence of extrarespiratory spread of PPMV-1 in the Dutch case. This was based on evidence of PPMV-1 infection in liver, kidney, and bone marrow (Table 2). This is consistent with the New York case, where evidence of PPMV-1 infection in feces and urine also suggested extrarespiratory spread [11]. The immunosuppressive state of the Dutch and New York patients may have allowed spread of PPMV-1 beyond the respiratory tract.
Pigeons and doves are the animal reservoir of PPMV-1 (Figure 1 and Table 1). In Europe, these viruses are circulating predominantly in domestic pigeons (Columba livia domestica) or their wild relatives (rock pigeons, C. livia), and other members of the family Columbidae, especially Eurasian collared doves (Streptopelia decaocto) and European turtle doves (Streptopelia turtur) [15]. The tropism of hPPMV-1/NL/579/2003 for the kidney and pancreas of experimentally infected pigeons (Supplementary Figures 2 and 3) matches the tissue tropism of PPMV-1 in naturally infected pigeons [25]. The low pathogenicity of this isolate in chickens fits with the phenotype of other PPMV-1 strains [26].
The number of feral pigeons—free-living birds that have descended from domestic pigeons—in European and North American cities increased substantially from the 1940s to the 1970s as a result of changes in agricultural practices and the rapid human population increase after World War II. Feral pigeon numbers then stabilized, likely due to reaching the carrying capacity of the urban environment. However, population growth of feral pigeons is still likely to occur in recently colonized cities and at the newly built outskirts of cities [27].
First reported in Italy in 1981 [28], PPMV-1 subsequently spread across Europe and has become endemic in feral pigeons, with regular spread to wild pigeons and doves [15]. In North America, PPMV-1 was introduced in the 1980s and now is maintained endemically in pigeons and doves [29], including the Eurasian collared dove [29, 30]. This invasive species, first reported in Florida in the 1980s, has rapidly spread across most of North America (http://www.audubon.org/field-guide/bird/eurasian-collared-dove), and could facilitate virus dispersal throughout North America [29, 30].
The clinical histories of the Dutch and New York cases do not readily explain how they became infected. The Dutch patient lived in a rural area near a city, but was not employed in agriculture, and there was no evidence of contact with animals (this study). The New York patient was an urban dweller, but it is unknown whether he had pets or was exposed to birds in other settings [11]. Based on phylogenetic analysis of the viruses, the most probable route of transmission was contact with infected pigeons or doves. PPMV-1 is environmentally stable in bird feces and can be spread by direct contact or by windborne dust [31].
The route of transmission of other zoonotic pathogens from pigeons to humans may be instructive. Five pathogen species (Chlamydia psittaci, Histoplasma capsulatum, Aspergillus species, Candida parapsilosis, and Cryptococcus neoformans) have been reported to be routinely transmitted from feral pigeons to people [32], mostly by inhalation of airborne excreta, including dried feces, ocular discharges, and crop milk. Contact was sometimes brief, and patients did not always recall any encounters with birds. It is relevant for these PPMV-1 cases that the risk of 2 pigeon-associated diseases—chlamydiosis and cryptococcosis—was largely a function of the immune status of patients, rather than contact with infected birds [32, 33].
Close contact between feral pigeons and humans commonly occurs in squares, public gardens, parks, markets, and railway stations in urban areas [33]. Therefore, people in urban areas are likely to have a higher rate of contact with feral pigeons than in rural areas. The proportion of the global human population living in urban areas is increasing. In 1950, 30% of the world’s population lived in urban areas; this increased to 54% in 2014, and by 2050, 66% of the world’s population is projected to be urban [34]. This suggests that the number of people at risk of contracting zoonotic infections, including PPMV-1, from feral pigeons will increase in coming decades. The combination of the above factors—large populations of feral pigeons with endemic PPMV-1 infection, increasing urbanization, and a higher proportion of immunocompromised individuals—may increase the risk of severe human cases of PPMV-1 infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the Department of Pathology, Leiden University Medical Center, The Netherlands, for providing biopsies and autopsy specimens from the Dutch patient.
Financial support. This work was supported by European FP7 programme ANTIGONE (ANTIcipating the Global Onset of Novel Epidemics, project number 278976) and by the European Commission H2020 Programme under contract number 643476 (www.compare_europe.eu).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Jones KE, Patel NG, Levy MA, et al. . Global trends in emerging infectious diseases. Nature 2008; 451:990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reperant LA, Kuiken T, Osterhaus AD. Adaptive pathways of zoonotic influenza viruses: from exposure to establishment in humans. Vaccine 2012; 30:4419–34. [DOI] [PubMed] [Google Scholar]
- 3. Richard M, Schrauwen EJ, de Graaf M, et al. . Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 2013; 501:560–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. N Engl J Med 2003; 349:2431–41. [DOI] [PubMed] [Google Scholar]
- 5. Reusken CB, Raj VS, Koopmans MP, Haagmans BL. Cross host transmission in the emergence of MERS coronavirus. Curr Opin Virol 2016; 16:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clayton BA. Nipah virus: transmission of a zoonotic paramyxovirus. Curr Opin Virol 2017; 22:97–104. [DOI] [PubMed] [Google Scholar]
- 7. Field HE. Hendra virus ecology and transmission. Curr Opin Virol 2016; 16:120–5. [DOI] [PubMed] [Google Scholar]
- 8. Svraka S, Rosario K, Duizer E, van der Avoort H, Breitbart M, Koopmans M. Metagenomic sequencing for virus identification in a public-health setting. J Gen Virol 2010; 91:2846–56. [DOI] [PubMed] [Google Scholar]
- 9. Lamb RA, Parks GD. Paramyxoviridae. In: Knipe DM, Howley PM, eds. Fields Virology. Philadelphia: Wolters Kluwer, 2013:957–95. [Google Scholar]
- 10. Swayne DE, King DJ. Avian influenza and Newcastle disease. J Am Vet Med Assoc 2003; 222:1534–40. [DOI] [PubMed] [Google Scholar]
- 11. Goebel SJ, Taylor J, Barr BC, et al. . Isolation of avian paramyxovirus 1 from a patient with a lethal case of pneumonia. J Virol 2007; 81:12709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41:95–8. [Google Scholar]
- 13. Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol 2008; 25:1253–6. [DOI] [PubMed] [Google Scholar]
- 14. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696–704. [DOI] [PubMed] [Google Scholar]
- 15. Alexander DJ. Newcastle disease in the European Union 2000 to 2009. Avian Pathol 2011; 40:547–58. [DOI] [PubMed] [Google Scholar]
- 16. Ujvári D, Wehmann E, Kaleta EF, et al. . Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res 2003; 96:63–73. [DOI] [PubMed] [Google Scholar]
- 17. Dortmans JC, Koch G, Rottier PJ, Peeters BP. Virulence of pigeon paramyxovirus type 1 does not always correlate with the cleavability of its fusion protein. J Gen Virol 2009; 90:2746–50. [DOI] [PubMed] [Google Scholar]
- 18. van Boheemen S, Bestebroer TM, Verhagen JH, et al. . A family-wide RT-PCR assay for detection of paramyxoviruses and application to a large-scale surveillance study. PLoS One 2012; 7:e34961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Briese T, Kapoor A, Mishra N, et al. . Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio 2015; 6:e01491–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van den Brand JM, Smits SL, Haagmans BL. Pathogenesis of Middle East respiratory syndrome coronavirus. J Pathol 2015; 235:175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 2010; 36:1–53. [DOI] [PubMed] [Google Scholar]
- 22. Kuiken T, Fouchier RA, Schutten M, et al. . Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 2003; 28S:S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine 2008; 26:D59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Short KR, Kroeze EJBV, Fouchier RAM, Kuiken T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis 2014; 14:57–69. [DOI] [PubMed] [Google Scholar]
- 25. Barton JT, Bickford AA, Cooper GL, Charlton BR, Cardona CJ. Avian paramyxovirus type 1 infections in racing pigeons in California. I. Clinical signs, pathology, and serology. Avian Dis 1992; 36:463–8. [PubMed] [Google Scholar]
- 26. Fuller CM, Collins MS, Easton AJ, Alexander DJ. Partial characterisation of five cloned viruses differing in pathogenicity, obtained from a single isolate of pigeon paramyxovirus type 1 (PPMV-1) following passage in fowls’ eggs. Arch Virol 2007; 152:1575–82. [DOI] [PubMed] [Google Scholar]
- 27. Giunchi D, Albores-Barajas YV, Baldaccini NE, Vanni L, Soldatini C. Feral pigeons: problems, dynamics and control methods. Integrated pest management and pest control: current and future tactics. Rijeka, Croatia: InTech, 2012:215–40. [Google Scholar]
- 28. Biancifiori F, Fioroni A. An occurrence of Newcastle disease in pigeons: virological and serological studies on the isolates. Comp Immunol Microbiol Infect Dis 1983; 6:247–52. [DOI] [PubMed] [Google Scholar]
- 29. Chong YL, Lam TT, Kim O, Lu H, Dunn P, Poss M. Successful establishment and global dispersal of genotype VI avian paramyxovirus serotype 1 after cross species transmission. Infect Genet Evol 2013; 17:260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schuler KL, Green DE, Justice-Allen AE, et al. . Expansion of an exotic species and concomitant disease outbreaks: pigeon paramyxovirus in free-ranging Eurasian collared doves. Ecohealth 2012; 9:163–70. [DOI] [PubMed] [Google Scholar]
- 31. Alexander DJ. Newcastle disease: methods of spread. In: Alexander DJ, ed. Newcastle disease. Boston, MA: Springer US, 1988:256–72. [Google Scholar]
- 32. Haag-Wackernagel D, Moch H. Health hazards posed by feral pigeons. J Infect 2004; 48:307–13. [DOI] [PubMed] [Google Scholar]
- 33. Magnino S, Haag-Wackernagel D, Geigenfeind I, et al. . Chlamydial infections in feral pigeons in Europe: review of data and focus on public health implications. Vet Microbiol 2009; 135:54–67. [DOI] [PubMed] [Google Scholar]
- 34. Population Division, Department of Economic and Social Affairs, United Nations. World urbanization prospects: 2014 revision, highlights (ST/ESA/SER. A/352). New York: United Nations, 2014:27. [Google Scholar]
- 35. Czegledi A, Herczeg J, Hadjiev G, Doumanova L, Wehmann E, Lomniczi B. The occurrence of five major Newcastle disease virus genotypes (II, IV, V, VI and VIIb) in Bulgaria between 1959 and 1996. Epidemiol Infect 2002; 129:679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.