Summary
IFITM3 and TLR3 SNPs are associated with fatal clinical outcome of Chinese patients with avian (H7N9) or pandemic (H1N1pdm09) influenza virus infections, and the risks are cumulative. Our findings pose important public health and clinical implications in the at-risk populations.
Keywords: IFITM3, influenza, outcomes, SNP, TLR3.
Abstract
Background.
We examined associations between single-nucleotide polymorphisms (SNPs) of IFITM3, TLR3, and CD55 genes and influenza clinical outcomes in Chinese.
Methods.
A multicenter study was conducted on 275 adult cases of avian (H7N9) and pandemic (H1N1pdm09) influenza. Host DNA was extracted from diagnostic respiratory samples; IFITM3 rs12252, TLR3 rs5743313, CD55 rs2564978, and TLR4 rs4986790/4986791 were targeted for genotyping (Sanger sequencing). The primary outcome analyzed was death.
Results.
IFITM3 and TLR3 SNPs were in Hardy–Weinberg equilibrium; their allele frequencies (IFITM3/C-allele 0.56, TLR3/C-allele 0.88) were comparable to 1000 Genomes Han Chinese data. We found over-representation of homozygous IFITM3 CC (54.5% vs 33.2%; P = .02) and TLR3 CC (93.3% vs 76.9%; P = .04) genotypes among fatal cases. Recessive genetic models showed their significant independent associations with higher death risks (adjusted hazard ratio [aHR] 2.78, 95% confidence interval [CI] 1.29–6.02, and aHR 4.85, 95% CI 1.11−21.06, respectively). Cumulative effects were found (aHR 3.53, 95% CI 1.64−7.59 per risk genotype; aHR 9.99, 95% CI 1.27−78.59 with both). Results were consistent for each influenza subtype and other severity indicators. The CD55 TT genotype was linked to severity. TLR4 was nonpolymorphic.
Conclusions.
Host genetic factors may influence clinical outcomes of avian and pandemic influenza infections. Such findings have important implications on disease burden and patient care in at-risk populations.
Avian and pandemic influenza are major public health threats. The H7N9 virus, emerged in 2013, has caused more than 880 human infections in China, with a case fatality of 20%–30% in hospitalized cases [1, 2]. The H1N1pdm09 virus, which caused a pandemic in 2009, has continued to circulate globally, resulting in seasonal outbreaks; fatality among the hospitalized has been reported to exceed 6%–10% [3, 4]. Senility, chronic medical and immunocompromising conditions, obesity, and pregnancy are known risk factors for severe influenza; however, in a substantial proportion of avian/pandemic virus infections, no apparent predisposition could be identified [3, 5, 6]. The reasons why ineffective viral clearance, progression to fulminant pneumonitis, and excessive inflammatory response and tissue damage occur in some but not other individuals have remained incompletely understood [2, 3, 6, 7].
Recently, evidence has emerged to suggest host genetic factors may play important roles in determining the clinical course and/or susceptibility to influenza [8–10]. Of these, single-nucleotide polymorphisms (SNPs) of several innate immunity-related genes, including interferon-induced transmembrane protein 3 (IFITM3), the toll-like receptors (TLRs), and complement decay-accelerating factor (CD55), have been described experimentally to affect protein function, and there is clinical observational data, albeit limited, to support associations with severity. IFITM3 restricts a wide range of RNA viruses, including influenza A; and it is suggested that the SNP rs12252/C-allele may lead to truncation of the protein, impairing its viral replication restriction function [11–17]. TLRs are pattern-recognition receptors, which signal type I interferons against viral replication and dissemination, and mediate the proinflammatory cytokine responses [7, 18, 19]. Genetic variants of TLR3, including SNP rs5743313 (intronic, but located near exon 4, the region encoding transmembrane signal induction), have been linked to impaired signaling function and weakened host responses [8, 20, 21]. Likewise, SNPs of the TLR4 gene have been linked to impaired innate immunity against respiratory viruses [22, 23]. CD55 inhibits the formation of C3 and C5 convertases responsible for complement activation and inflammation, mechanisms implicated in fulminant influenza pneumonitis. The SNP of CD55 rs2564978 has been associated (indirectly) with decreased promotor activity and lowered protection level [8, 24, 25]. Results of these SNPs’ associations with clinical severity, however, appear to be inconsistent, and likely confounded by patients’ ethnicity, clinical characteristics, antiviral treatment, and small sample sizes; further, the impacts on survival had not been examined [17, 20, 21, 25–27]. Notably, some of these genetic variants are much more prevalent among the East Asian populations, thus potentially exerting stronger impacts than in populations where their frequencies are low (eg, those with European ancestries) [15, 17, 24]. Also, it is unknown if their effects are cumulative, which will have important implications on genetic risk evaluation.
In this study, we have adopted a targeted approach to simultaneously investigate the associations of the innate-immunity-related IFITM3, TLR, and CD55 gene SNPs (which had preliminary human data support) with influenza clinical outcomes in a large Chinese cohort. Avian (H7N9) and pandemic (H1N1pdm09) influenza virus infections were studied. Impacts on survival were examined under different genetic models; effects of important clinical confounders were adjusted in multivariate analyses. Joint effects of the genetic variants were investigated with the use of a genetic risk score. Our data may improve the understanding of the genetic risk for severe influenza, and provide an important basis for future research.
METHODS
We studied SNPs of the targeted host genes in H7N9 and H1N1pdm09 influenza cases that were consecutively diagnosed in 4 participating institutes in mainland China (Guangdong, Shanghai, Beijing) and Hong Kong during 3 respective seasonal outbreaks (H7N9, 2013–2015; H1N1pdm09, 2011−2014; Table 1 footnotes). Virological features, and procedures related to laboratory diagnosis and clinical management of influenza in these medical units (all being urban hospital settings) has been described in detail [2, 5, 28–31]. Briefly, patients presented with symptoms of acute respiratory tract infections during the seasonal outbreaks were tested for influenza virus infections (ie, prospectively diagnosed) with molecular assays, regardless of perceived severity. The inclusion criteria were: polymerase chain reaction (PCR)–confirmed H7N9 or H1N1pdm09 virus infection, age ≥18 years, and Chinese ethnicity (Chinese populations residing in Hong Kong, Guangdong, Shanghai, and Beijing are mostly of Han ancestry; >95%–99%); there was no exclusion criterion. Patients’ original respiratory tract samples collected at presentation for diagnostic purpose, which contained virus-infected epithelial cells, were retrieved for host gene SNP and viral RNA load studies; identical methodologies were used across institutes (see below). Clinical data, including patient characteristics, disease severity, and outcomes, were collected using a standardized research database (to ensure identical definitions of variables), as previously described [5]. All patient identifying information was removed from the dataset, and only anonymous data were used for analysis. Ethics approvals for this study were obtained from the Institution Review Boards (IRB) of all participating institutes.
Table 1.
Descriptive Data on Clinical Characteristics, Severity, and Outcomes of 275 Chinese Patients with Confirmed H7N9 or H1N1pdm09 Influenza Infections
| Variable | H7N9 (n = 51) |
H1N1pdm09 (n = 224) |
|---|---|---|
| Age, mean ± SD, years | 56.7 ± 22.7 | 50.8 ± 19.4 |
| Sex, male (%) | 36 (70.6) | 109 (48.7) |
| Major comorbidity (%)a | 16 (34.0) | 86 (38.6) |
| Onset-to-presentation, mean ± SD, days | 7.2 ± 3.3 | 3.0 ± 3.3 |
| Hospitalization (%) | 47 (92.2) | 169 (75.4) |
| Pneumonia, radiographic (%) | 42 (82.4) | 113 (50.7) |
| Supplemental oxygen use (%) | 37 (77.1) | 120 (53.8) |
| Acute respiratory failure (%)b | 26 (55.3) | 57 (25.4) |
| Death (%) | 10 (22.2) | 23 (10.3) |
| NAI treatment within 2 days from onsetc |
5 (10.6) | 135 (60.8) |
All H7N9 cases were enrolled in mainland China institutes (2013, 2014, 2015 outbreaks); H1N1pdm09 cases were from both Hong Kong and mainland China institutes (2011, 2013, 2014 seasonal peaks of H1N1pdm09; low-level circulation in 2012). Vaccines against human H7N9 infection were unavailable; a low (18%) vaccination rate among H1N1pdm09 patients was noted in the available records, consistent with our previous reports [4, 5].
Abbreviations: H1N1pdm09, pandemic influenza virus; H7N9, avian influenza virus; NAI, neuraminidase inhibitor.
aDefined as congestive heart failure; cerebrovascular, neoplastic, or chronic liver or renal diseases; chronic lung diseases; and diabetes mellitus, chronic cardiovascular, autoimmune or neurological diseases; or immunosuppression (not hypertension or hypercholesterolemia alone). Obesity is uncommon in our Chinese cohort [2, 4, 5].
bDefined as acute hypoxemic respiratory failure, requiring mechanical ventilation or noninvasive positive-pressure ventilation, and/or oxygen therapy for vital life support [4, 5].
cAltogether, 258/275 (93.8%) patients had received oseltamivir treatment; 140/275 (50.9%) were treated within 2 days from onset.
SNP Genotyping and Quality Control
Viral ribonucleic acid (RNA) and host DNA were coextracted directly from the samples using the PureLink Viral RNA/DNA Mini kit (Thermo Fisher) as per manufacturer’s instructions. No DNase/RNase digestion step was performed [32]. Genotyping of 5 SNPs, including IFITM3 rs12252, CD55 rs2564978, TLR3 rs5743313, and TLR4 rs4986790 and rs4986791, was performed by Sanger sequencing of PCR amplicons. In brief, host DNA was subjected to gene-specific PCR amplification using Phusion High-Fidelity DNA polymerase (New England Biolabs). The PCR primers used are provided in Supplementary material 1 [15, 20, 23, 24]. PCR amplicons (300–600 base-pairs in size) were column-purified with QIAquick PCR Purification Kit (Qiagen), followed by Sanger sequencing in both directions using PCR primers. DNA chromatograms were inspected and genotypes were called manually. With this method, genotyping results were successfully obtained in 90.5%, 88.4%, 84.4%, 87.6%, and 87.6% of cases for IFITM3 rs12252, TLR3 rs5743313, CD55 rs2564978, and TLR4 rs4986790 and rs4986791, respectively. The success rates were not significantly different among the target genes and participating institutes. At the time of writing, a study on IFITM3 SNP was published, reporting a similar genotyping success rate of 87% using archived respiratory tract samples collected from patients with influenza-like illnesses [27]. Genotyping accuracy with respiratory samples was confirmed by the results obtained using paired peripheral blood mononuclear cell samples (n = 10), which showed 100% concordance.
The SNP results were tested for Hardy–Weinberg equilibrium by the exact test using PLINK (a Bonferroni-corrected P value of >.01 indicated no significant deviation) [33]. To examine for representativeness, the observed allele frequencies in this cohort were compared to the 1000 Genomes general population data on Han Chinese (and East Asians) (www.1000genomes.org/home; last accessed on 18 September 2016) (Supplementary material 2).
Influenza viral RNA Quantification and Standardization
A 1-step, probe-based real-time quantitative reverse transcription–polymerase chain reaction assay targeting the matrix (M)–gene of the virus genome was used to measure the viral RNA load in the same respiratory samples, using methods that were standardized across the participating institutes (Supplementary material 1) [34, 35].
Outcome Measures and Data Analysis
The primary outcome of this study was all-cause death. The secondary outcome was acute respiratory failure, defined as acute hypoxemic respiratory failure requiring mechanical ventilation, noninvasive positive-pressure ventilation, and/or oxygen therapy for vital life support. We also analyzed hospitalization requirement as an indicator for severity among the H1N1pdm09 influenza patients; this was not performed for the H7N9 patients, as nearly all were admitted for clinical care and isolation.
The Student t test, χ2 test with continuity correction, and the Fisher exact test were used for univariate comparisons based on data distribution. Cox proportional hazards regression analyses were used to examine independent associations between the genetic variants and death (censored at 30 days from time of presentation), which were tested under the additive, dominant, and recessive genetic models. Multivariate models were constructed to adjust for the effects of confounders, including age, gender, comorbidity, use of neuraminidase inhibitor (NAI) treatment, and influenza subtype (see Table 1 footnotes for definitions) [34, 35]. The adjusted hazard ratio (aHR) and the 95% confidence interval (95% CI) were reported for each explanatory variable; an aHR >1 indicated a higher probability of death. The largest test statistic (MAX statistic) among the 3 genetic models was chosen as the best-fitting model; the experiment-wise significance of the MAX statistic was estimated from its empirical distribution under the null hypothesis after performing 10000 permutations of genotypes of the SNPs (Pperm), to correct for multiple comparisons [36, 37]. Pairwise gene–gene interactions were evaluated by including the main effects, the interaction term (product) of the SNPs, and the covariates in the Cox regression models. In addition, we tested for the joint effect of all significant loci (under corresponding genetic models) on the risk of death by computing a genetic risk score (GRS) for each individual using the simple count method. Assuming a similar and independent effect between loci, the GRS was calculated by summing the score of the risk genotype for each SNP based on the most significant genetic model. The significance of the trend was tested by the Cox regression model, using the GRS as an independent variable with adjustment for clinical confounders [36, 37]. The adjusted survival curve stratified according to the GRS was constructed for graphical presentation. Statistical analyses were performed using SPSS for Windows v.22 (SPSS, Chicago, IL).
RESULTS
Descriptions of Patients and Genotype Distributions
Altogether, 275 Chinese patients confirmed with H7N9 (n = 51) or H1N1pdm09 (n = 224) influenza infections were studied (Table 1). Their mean ± SD age was 56.7 ± 22.7 and 50.8 ± 19.4 years, respectively; 92.2% and 75.4%, respectively, were hospitalized; and 55.3% and 25.4%, respectively, developed acute respiratory failure. There were 10 (22.2%) and 23 (10.3%) deaths among the H7N9 and H1N1pdm09 patients, respectively.
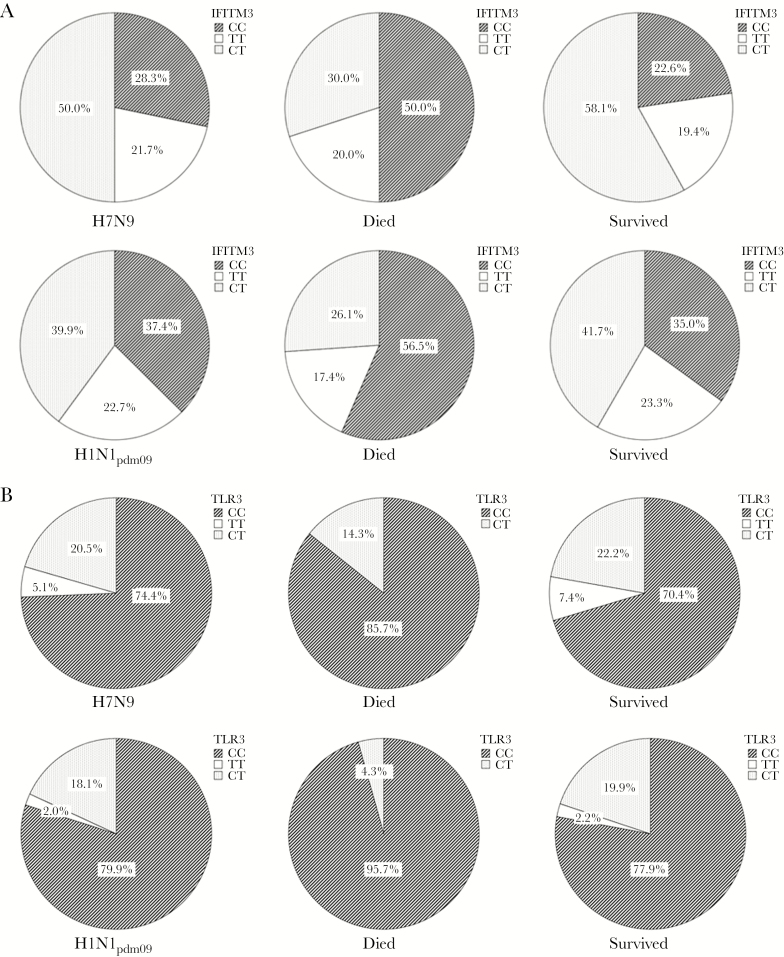
Overall, the genotype distributions were: IFITM3 rs12252—CC (35.7%), CT (41.8%), TT (22.5%); TLR3 rs5743313—CC (79.0%), CT (18.5%), TT (2.5%); CD55 rs2564978—TT (51.7%), CT (33.2%), CC (15.1%); TLR4 rs4986790—AA (100%); and TLR4 rs4986791—CC (100%) (Figure 1). IFITM3 and TLR3 SNPs were in Hardy–Weinberg equilibrium (both P > .01), and their allele frequencies (IFITM3/C-allele 0.56, TLR3/C-allele 0.88) were comparable to those reported for the general population of Han Chinese (1000 Genome data; Table 2 and Supplementary material 2). The allele frequency of CD55 was similar to that previously reported in Hong Kong. The TLR4 loci were nonpolymorphic, thus precluding their analysis for clinical associations. There was no significant difference in genotype distribution between H7N9 and H1N1pdm09 influenza patients (Figure 1 and Supplementary material 2).
Figure 1.
IFITM3 rs12252 and TLR3 rs5743313 genotype distribution in Chinese patients with confirmed H7N9 (n = 51) and H1N1pdm09 (n = 224) influenza infections. A, IFITM3 rs12252 genotype distribution (whole cohort: CC [35.7%], CT [41.8%], TT [22.5%)]. B, TLR3 rs5743313 genotype distribution (whole cohort: CC [79.0%], CT [18.5%], TT [2.5%]). Comparisons of genotype distribution between H7N9 and H1N1pdm09 patients did not show significant difference: IFITM3 (P = .40), TLR3 (P = .46), CD55 (P = .90, figure not shown); fatal infections occurred in 22.2% and 10.3%, respectively. IFITM3 and TLR3 genotype distributions were significantly different between fatal and nonfatal infections (IFITM3 CC vs CT/TT, 54.5% vs 33.2%, P = .02; TLR3 CC vs CT/TT, 93.3% vs 76.9%, P = .04; whereas for CD55 TT vs CT/CC, 48.4% vs 52.5%, P = .67). Consistent results were found within each virus subtype: H1N1pdm09 (IFITM3, P = .02; TLR3, P = .03); H7N9 (IFITM3, P = .03). These risk genotypes are less frequent in populations of European ancestry (IFITM3 CC 0.1%−0.3%, CT 2.3%−8.1%; TLR3 CC, 51.0%−63.5%, CT 31.4%−41.2%) [14, 15, 20, 21, 38, 43]. Abbreviations: H1N1pdm09, pandemic influenza virus; H7N9, avian influenza virus.
Table 2.
Associations of IFITM3, TLR3, and CD55 SNPs With the Primary Outcome of Death in Additive, Dominant, and Recessive Genetic Models
| RAF | Additive | Dominant | Recessive | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Genes (SNP) | Risk/ Nonrisk Allele |
Death | Survival | Model | HR (95% CI) |
P Value | HR (95% CI) |
P Value | HR (95% CI) |
P Value |
P
perm
(MaxStat) |
| 1 |
CD55
(rs2564978) |
T/C | 0.692 | 0.683 | I | 1.04 (0.61−1.76) | .88 | 1.40 (0.42−4.66) |
.58 | 0.93 (0.43−2.01) |
.86 | --- |
| II | 0.94 (0.54 −1.64) |
.83 | 1.24 (0.36−4.27) |
.73 | 0.80 (0.36−1.79) |
.58 | --- | |||||
| 4 |
TLR3
(rs5743313) |
C/T | 0.962 | 0.873 | I | 3.18 (0.80−12.69) |
.10 | --- | --- | 3.32 (0.79−14.06) |
.10 | .19 |
| II | 4.55 (1.09 −19.03) |
.04* | --- | --- | 4.85 (1.11−21.06) |
.04* | .04* | |||||
| 11 |
IFITM3
(rs12252) |
C/T | 0.679 | 0.556 | I | 1.57 (0.92−2.67) |
.10 | 1.05 (0.43−2.59) |
.92 | 2.54 (1.20−5.38) |
.02* | .05* |
| II | 1.62 (0.93−2.82) |
.09 | 1.00 (0.40−2.48) |
.99 | 2.78 (1.29−6.02) |
.01* | .04* | |||||
The HRs and 95% CI were reported for the “risk allele,” as calculated using Cox regression models. Analyses were based on data from 275 patients, because no significant difference in allele distribution was found between the 2 influenza subtypes.
P perm: experiment-wise significance of the MAX statistic (of the best-fitting “recessive model”) was calculated using the permutation method to correct for multiple comparisons (see text) [37].
*P < .05.
The allele frequencies reported in this study are comparable to those reported in the 1000 Genomes for Han Chinese and East Asians general populations: IFITM3 C-allele, 0.47−0.64; TLR3 C-allele, 0.78−0.97; CD55 T-allele, 0.55−0.64 (previous study in Hong Kong Chinese 0.64) [15, 24]. In populations of European ancestry, the reported allele frequencies are 0.03−0.08, 0.56−0.80, and 0.25−0.29, respectively [14, 20, 21, 43].
Abbreviations: Chr, chromosome; CI, confidence interval; HRs, hazards ratios; Model I, no adjustment for confounders; Model II, adjusted for age, sex, comorbidity, influenza subtype and NAI treatment (initiated within 2 days from onset, yes vs no); NAI, neuraminidase inhibitor; RAF, risk allele frequency; SNP, single-nucleotide polymorphism.
Associations With the Primary Clinical Outcome
We observed significantly higher proportions of IFITM3 CC (54.5% vs 33.2%; P = .02), and the TLR3 CC (93.3% vs 76.9%; P = .04) genotypes in fatal influenza infections (Figure 1A and 1B). Their allele frequencies were analyzed according to the primary outcome measure of death/survival, and tested under different genetic models (Table 2). Our results showed that the IFITM3 homozygous CC genotype was significantly associated with increased risk of death in the unadjusted recessive model, which remained significant after adjusting for major clinical confounders (aHR 2.78, 95% CI 1.29−6.02; P = .01). Although the TLR3 homozygous CC genotype was insignificant in the unadjusted models, it was found to be significantly associated with increased death risk in both recessive (aHR 4.85, 95% CI 1.11−21.06; P = .04) and additive (aHR 4.55, 95% CI 1.09−19.03; P = .04) genetic models in multivariate analyses. The MAX statistic indicated that the recessive models were the “best-fitting” genetic model for both SNPs (Pperm < 0.05). We did not find significant association between the CD55 alleles and primary outcome. Findings were consistent between the 2 influenza subtypes (Supplementary material 3).
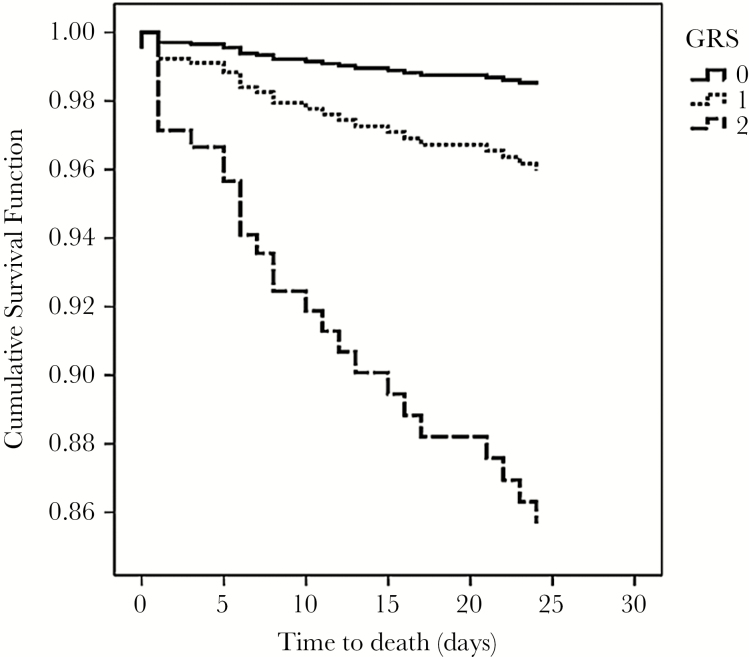
No pairwise interaction effect between loci on the primary outcome of death was detected (data not shown). To examine the joint effects of the 2 significant SNPs under the “best-fitting” genetic model (ie, recessive models of IFITM3 and TLR3 SNPs), a simple GRS was computed for each individual. Our results showed that an increasing GRS was independently associated with cumulative risk for death (aHR 3.53, 95% CI 1.64−7.59 per risk genotype; P = .001 for trend) (Figure 2). The presence of both IFITM3 CC and TLR3 CC genotypes conferred almost a 10-fold increase in death risk (aHR 9.99, 95% CI 1.27−78.59; P = .03) (Table 3a). Descriptively, we found that 22.7% of patients harboring 2 (H7N9 40.0%, H1N1pdm09 21.3%), 10.8% harboring 1 (H7N9 21.7%, H1N1pdm09 8.4%), and 3.1% (H7N9 0.0%, H1N1pdm09 3.7%) with none of these risk genotypes had died.
Figure 2.
Cumulative survival function of influenza patients stratified according to the GRS.
Two significant SNPs (IFITM3 rs12252 and TLR3 rs5743313, both in recessive model) were selected to calculate the GRS for survival. The survival functions were adjusted for age, sex, comorbidity, influenza subtype, and NAI use (see Table 3a). Comparison groups: GRS 0 (14%), GRS 1 (57%), GRS 2 (29%), representing the presence of neither, either, and both recessive risk genotypes, respectively.
Abbreviations: GRS, genetic risk score; NAI, neuraminidase inhibitor; SNPs, single-nucleotide polymorphisms.
Table 3.
Independent Variables Associated With Death and Acute Respiratory Failure, As Shown in the Final Multivariate Models
| Variable | Adjusted Hazards Ratio (95% CI) | P Value |
|---|---|---|
| Deatha | ||
| Age, per 20 years | 2.55 (1.50−4.31) | <.001 |
| NAI treatment | 0.33 (0.13−0.83) | .02 |
| Risk genotype, overall | … | .004 |
| No. of risk genotype (GRS) |
||
| 0 | … | … |
| 1 | 2.65 (0.34−20.82) | .36 |
| 2 | 9.99 (1.27−78.59) | .03 |
| Acute respiratory failureb | ||
| Age, per 20 year | 1.61 (1.14−2.28) | .01 |
| Comorbidity | 1.86 (0.93−3.69) | .08 |
| H7N9 subtype | 2.71 (1.11−6.63) | .03 |
| IFITM3 CC genotype | 2.10 (1.10−4.01) | .03 |
| NAI treatment | 0.38 (0.20−0.72) | .003 |
Abbreviations: CI, confidence interval; GRS, genetic risk score; H1N1pdm09, pandemic influenza virus; H7N9, avian influenza virus; NAI, neuraminidase inhibitor; SNPs, single-nucleotide polymorphisms.
aThe 2 significant SNPs (IFITM3 rs12252 and TLR3 rs5743313, both in the recessive model; ie, IFITM3 CC and TLR3 CC) were selected to calculate the GRS for death.
bIFITIM3 CC vs CT/TT genotypes (as dichotomous variable). Age, gender, comorbidity, influenza virus subtype (H7N9 vs H1N1pdm09), and NAI treatment (initiated within 2 days from onset, yes vs no) were included in these regression models as covariates (backward, stepwise); the explanatory variables are being shown in the tables.
Associations With Clinical Severity and Viral Load
Consistent with findings on the primary outcome, the IFITM3 CC genotype was shown to be independently associated with the development of acute respiratory failure in hospitalized patients (overall, 44.3% vs 25.5%; P = .01; adjusted odds ratio [OR] 2.10, 95% CI 1.10−4.01; P = .03) (Table 3b and Table 4). For the TLR3 CC genotype, an insignificant trend was observed among the H7N9 influenza patients (80.0% vs 61.5%). There was an association between the CD55 TT genotype and requirement for hospitalization (55.6% vs 38.9%; P = .04; adjusted OR 2.77, 95% CI 1.21−6.36; P = .02) (Table 4). There was a tendency of higher viral load in patients harboring the risk genotypes, even when presenting late in the course of illness; however, the analysis results did not reach statistical significance (P > .1) (Supplementary material 4).
Table 4.
Univariate Associations of IFITM3, TLR3, and CD55 Genotypes With Clinical Severity
| IFITM3 Genotypes | TLR3 Genotypes | CD55 Genotypes | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CC (%) | CT/TT (%) | P Value | CC (%) | CT/TT (%) | P value | TT (%) | CT/CC (%) | P Value | |
| H7N9 influenza: | |||||||||
| Acute respiratory failure | |||||||||
| Yes | 34.8 | 65.2 | .26 | 80.0 | 20.0 | .43 | 53.3 | 46.7 | .87 |
| No | 13.3 | 86.7 | 61.5 | 38.5 | 57.1 | 42.9 | |||
| H1N1pdm09 influenza: | |||||||||
| Hospitalization | |||||||||
| Yes | 35.1 | 64.9 | .24 | 77.5 | 22.5 | .15 | 55.6 | 44.4 | .04* |
| No | 44.2 | 55.8 | 86.8 | 13.2 | 38.9 | 61.1 | |||
| Acute respiratory failure | |||||||||
| Yes | 48.2 | 51.8 | .01* | 74.1 | 25.9 | .45 | 58.2 | 41.8 | .63 |
| No | 27.4 | 72.6 | 79.4 | 20.6 | 54.2 | 45.8 | |||
*P value < .05.
Abbreviations: H1N1pdm09, pandemic influenza virus; H7N9, avian influenza virus.
DISCUSSION
We found significant associations between SNPs of IFITM3 rs12252 and TLR3 rs5743313 genes and outcomes of avian (H7N9) and pandemic (H1N1pdm09) influenza in our Chinese cohort. There was increased death risk with the respective homozygous CC genotype of IFITM3 and TLR3, and the effects were cumulative. Our results provide evidence for genetic risks for severe influenza disease, which may have important public health and clinical implications.
Everitt and colleagues [14] had first described over-representation of IFITM3 CC (5.7%), an uncommon genotype in populations of European ancestry (0.3%), among patients hospitalized for H1N1pdm09 influenza in the United Kingdom (n = 53). Several subsequent studies from Europe reported insignificant associations with severe H1N1pdm09 infection, but the number of cases detected with the genotype were very small (0.7%−2.4%) [17, 25–27]. In Chinese and East Asian populations, however, the IFITM3 CC genotype is known to be much more prevalent (25%−44%); and a study from China had reported its over-representation (69%) among hospitalized patients with H1N1pdm09 pneumonia (n = 32) [15]. Our study added that this genotype might actually predict a fatal outcome of influenza patients. Its frequency was about 21% higher among the fatal cases (54.5%, vs 33.2% in survivors); the death risk was found to be significantly increased by nearly 3-fold, even after adjustment for clinical confounders. The recessive model for the C-allele was confirmed to be the “best-fitting” genetic model [15, 26]. Consistent result was also shown for clinical severity, as indicated by development of acute respiratory failure. The mechanism of how this SNP would affect the disease course is incompletely understood and likely complex. It has been suggested that the C-allele homozygosity can impair IFITM3 function, leading to reduced viral clearance, which, in turn, aggravates host inflammatory responses [14, 15]. Viral load coupled with increased proinflammatory cytokines has been described in patients harboring the CC genotype with progressive H7N9 infections, but the study was small (n = 18) [16].
In this study, we first report possible association between the TLR3 gene’s SNP with influenza clinical outcomes. There was over-representation of the TLR3 homozygous CC genotype in fatal infections (93.3%, vs 76.9% among survivors; in European populations its prevalence was lower at 50%−65%); and a significant increase in death risk was found in recessive and additive models for the major C-allele after adjustment for confounders. To the best of our knowledge, there was only 1 small-scale study (n = 51) describing a univariate association of TLR3 rs5743313 CT genotype with the risk of developing influenza pneumonia in 18 European children [20]. The apparent discrepant result may be attributable to ethnicity difference, as observed in other diseases linked to this SNP, and/or the patients’ age and disease stage [38]. Intriguingly, TLR3 function had been shown to be detrimental to the survival of mice with influenza virus–induced pneumonia despite a lowered viral load, owing to the induction of exuberant inflammatory responses and excessive lung damage [39]. A mechanistic study on how the SNP could have affected TLR3 signaling function, and its impact on different phases of infection is indicated [8, 8, 19, 40]. Consistent with a recent report, we also noted a relationship between CD55 T-allele homozygosity (more common among Asians) and clinical severity (hospitalization), though there was no significant association with survival [24, 25]. The TLR4 loci were shown to be nonpolymorphic in this cohort; seemingly, emerging data have suggested a lack of association between TLR4 SNP and influenza severity, unlike in pediatric respiratory syncytial virus disease [20, 23].
Although the risk associated with individual genetic variant may appear moderate, our data showed that the effects can be cumulative. Fatality of patients with TLR3 CC plus the IFITM3 CC genotype was 23%, whereas patients with either or none of these risk genotypes had fatality rates of 11% and 3%, respectively. The death risk was shown to be incremental in GRS analysis (aHR 3.5, per risk genotype); the presence of both genotypes conferred an almost 10-fold higher risk than those with neither. These results strongly suggest that a combination of host genetic factors (“genetic background”) may influence the clinical course and outcomes of influenza, along with known clinical and virologic factors [7, 8, 9, 41, 42]. Our list of candidate genes/SNPs is not exhaustive; as experimental and genome-wide association studies continue to uncover these determinants, more variables/combinations, including those related to adaptive immunity are expected to be considered in genetic risk evaluation in severe influenza [7, 8, 9, 10, 21, 25, 43, 44].
Our data have important implications. Owing to their high frequencies in Chinese and East Asian populations, the proportion of disease burden attributable to these genetic variants could be substantial. An earlier study on 83 Chinese influenza patients had put the estimate of the population-attributable risk percentage (PAR%) of IFITM3 CC for severe nonfatal/fatal infections at 54% (in contrast to 5% in European populations) [15]. We had calculated the PAR% of IFITM3 CC for influenza death with this mostly hospitalized Chinese cohort, which was around 36% (95% CI 10–70%); the combined risk with TLR3 CC was higher (Supplementary material 5). Precise PAR estimation would require a much larger sample size; and within/cross-population epidemiological studies are necessary to verify the plausibility. Interestingly, emerging data seem to suggest significant regional heterogeneity in influenza mortality across continents (Europe, Americas, Asia), even accounting for country income status and comorbidity [45]. The knowledge on genetic risk can better inform public health-care planning, pandemic preparedness, and the development of preventive strategies against avian/pandemic influenza in our region [15, 16, 19]. At the individual patient level, we indicate the potential of assessing genetic risk factors, together with virologic parameters in the same respiratory sample, to assist prognostication and treatment decisions (eg, regimen intensification), which warrant exploration [15, 16, 19, 35]. As genetic/ethnic factors can confound outcomes, future clinical trials on influenza therapeutics should consider these variables; our study has provided useful information for their design/planning.
The strengths of our study include a larger sample size (n = 275; >200 hospitalized and >150 influenza pneumonia cases), unbiased sampling (original diagnostic respiratory specimens from consecutive patients with fatal/nonfatal disease courses), data on pandemic and avian influenza, analyses of well-defined outcome measures with different genetic models, and adjustment for potential confounders. Notably, early NAI treatment was significantly associated with improved outcomes independent of genotype, age, and comorbidity (Table 3) [3, 5, 6, 35]. Interaction and cumulative effects were examined. Unlike genome-wide association studies, however, our approach had limited the number of candidate genes/SNPs for study, and a more detailed analyses on haplotypes and population substructure are infeasible with this design. The SNPs’ effects on protein function, and how this might impact on viral clearance and/or signaling of proinflammatory responses at different disease stages, would require further elucidation (eg, serial viral load and cytokine/chemokine measurements) [14, 15, 16, 18, 19, 20, 28]. D222G, H275Y, and R292K mutations should have a minimal impact on results because of their reported rarity [25, 28, 29, 31, 35]. The effects on host susceptibility to initial infection (compared with uninfected patients) [14, 15, 20, 26], vaccine response, and the reasons why these immune-related genetic variants (thus implicated in infective/noninfective inflammatory diseases) are selected and gained prevalence among the Asian populations should warrant investigation [15, 19, 37, 38, 42, 43, 45]. Studies on seasonal influenza where preexisting immunity may exist, and other potential genetic determinants for influenza severity are in progress.
In conclusion, our results suggest that host genetic factors may influence clinical outcomes of pandemic and avian influenza virus infections, and the effects are cumulative. The impacts on disease burden in populations where such risk genotypes are common should deserve evaluation. Our findings may pose important implications on public health-care planning, patient care, and future designs of clinical trials in the at-risk populations.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. Concept and design: Lee Nelson, Chan C. W. Martin, Chan K. S. Paul, and Hui S. C. David. Data acquisition: Chan C. W. Martin, Lee Nelson, Cao Bin, Ke Changwen, Lu Hongzhou, Hu Yunwen, Wong Y. K. Rity, and Yung M. H. Irene. Laboratory analysis: Chan C. W. Martin, Chan K. S. Paul, Li Hui, Ke Changwen, Guan Dawei, Hu Yunwen, Zhu Zhaoqin, and Lin Mulei. Data analysis and interpretation: Lee Nelson, Chan C. W. Martin, Tam H. T. Claudia, and Ma C. W. Ronald. Drafting of article: Lee Nelson. Critical revision for important intellectual content and final approval of article: Lee Nelson, Ma C. W. Ronald, Horby Peter, Hui S. C. David, and Chan K. S. Paul.
Financial support. This work was supported by the Health and Medical Research Fund (RRG-09), Government of the Hong Kong SAR, PRC.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. WHO. Influenza at the human-animal interface Summary and assessment, 22 November to 19 December 2016. http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_12_19_2016.pdf?ua=1 Accessed 10 January 2017.
- 2. Gao HN, Lu HZ, Cao B et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 2013; 368:2277–85. [DOI] [PubMed] [Google Scholar]
- 3. Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM et al. ; Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med 2010; 362:1708–19. [DOI] [PubMed] [Google Scholar]
- 4. Lee N, Chan PK, Lui GC et al. Complications and outcomes of pandemic 2009 influenza A (H1N1) virus infection in hospitalized adults: how do they differ from those in seasonal influenza? J Infect Dis 2011; 203:1739–47. [DOI] [PubMed] [Google Scholar]
- 5. Lee N, Leo YS, Cao B et al. Neuraminidase inhibitors, superinfection and corticosteroids affect survival of influenza patients. Eur Respir J 2015; 45:1642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan PK, Lee N, Zaman M et al. Determinants of antiviral effectiveness in influenza virus A subtype H5N1. J Infect Dis 2012; 206:1359–66. [DOI] [PubMed] [Google Scholar]
- 7. Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol 2014; 14:315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin TY, Brass AL. Host genetic determinants of influenza pathogenicity. Curr Opin Virol 2013; 3:531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horby P, Nguyen NY, Dunstan SJ, Baillie JK. An updated systematic review of the role of host genetics in susceptibility to influenza. Influenza Other Respir Viruses 2013; 7(Suppl 2):37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ciancanelli MJ, Abel L, Zhang SY, Casanova JL. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr Opin Immunol 2016; 38:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang IC, Bailey CC, Weyer JL et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLOS Pathog 2011; 7:e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bailey CC, Huang IC, Kam C, Farzan M. IFITM3 limits the severity of acute influenza in mice. PLOS Pathog 2012; 8:e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLOS Pathog 2014; 10:e1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Everitt AR, Clare S, Pertel T et al. ; GenISIS Investigators; MOSAIC Investigators. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012; 484:519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang YH, Zhao Y, Li N et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun 2013; 4:1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z, Zhang A, Wan Y et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci USA 2014; 111:769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X, Tan B, Zhou X et al. Interferon-inducible transmembrane protein 3 genetic variant rs12252 and influenza susceptibility and severity: a meta-analysis. PLOS One 2015; 10:e0124985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu W, Zhang W, Duggan ES, Booth JL, Zou MH, Metcalf JP. RIG-I and TLR3 are both required for maximum interferon induction by influenza virus in human lung alveolar epithelial cells. Virology 2015; 482:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee N, Wong CK, Hui DS et al. Role of human Toll-like receptors in naturally occurring influenza A infections. Influenza Other Respir Viruses 2013; 7:666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Esposito S, Molteni CG, Giliani S et al. Toll-like receptor 3 gene polymorphisms and severity of pandemic A/H1N1/2009 influenza in otherwise healthy children. Virol J 2012; 9:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen C, Wang M, Zhu Z et al. Multiple gene mutations identified in patients infected with influenza A (H7N9) virus. Sci Rep 2016; 6:25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shirey KA, Lai W, Scott AJ et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature 2013; 497:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Awomoyi AA, Rallabhandi P, Pollin TI et al. Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J Immunol 2007; 179:3171–7. [DOI] [PubMed] [Google Scholar]
- 24. Zhou J, To KK, Dong H et al. A functional variation in CD55 increases the severity of 2009 pandemic H1N1 influenza A virus infection. J Infect Dis 2012; 206:495–503. [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Etxebarria K, Bracho MA, Galán JC et al. ; CIBERESP Cases and Controls in Pandemic Influenza Working Group. No major host genetic risk factor contributed to A(H1N1)2009 influenza severity. PLOS One 2015; 10:e0135983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mills TC, Rautanen A, Elliott KS et al. IFITM3 and susceptibility to respiratory viral infections in the community. J Infect Dis 2014; 209:1028–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gaio V, Nunes B, Pechirra P et al. Hospitalization risk due to respiratory illness associated with genetic variation at IFITM3 in patients with influenza A(H1N1)pdm09 infection: a case-control study. PLOS One 2016; 11:e0158181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu Z, Liu Y, Xu L et al. Extra-pulmonary viral shedding in H7N9 avian influenza patients. J Clin Virol 2015; 69:30–2. [DOI] [PubMed] [Google Scholar]
- 29. Ke C, Lu J, Wu J et al. Circulation of reassortant influenza A(H7N9) viruses in poultry and humans, Guangdong Province, China, 2013. Emerg Infect Dis 2014; 20:2034–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang C, Yu H, Horby PW et al. Comparison of patients hospitalized with influenza A subtypes H7N9, H5N1, and 2009 pandemic H1N1. Clin Infect Dis 2014; 58:1095–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bai L, Gu L, Cao B et al. Clinical features of pneumonia caused by 2009 influenza A(H1N1) virus in Beijing, China. Chest 2011; 139:1156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smieja M, Castriciano S, Carruthers S et al. Development and evaluation of a flocked nasal midturbinate swab for self-collection in respiratory virus infection diagnostic testing. J Clin Microbiol 2010; 48:3340–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee N, Chan PK, Hui DS et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 2009; 200:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee N, Hui DS, Zuo Z et al. A prospective intervention study on higher-dose oseltamivir treatment in adults hospitalized with influenza A and B infections. Clin Infect Dis 2013; 57:1511–9. [DOI] [PubMed] [Google Scholar]
- 36. Sladek R, Rocheleau G, Rung J et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445:881–5. [DOI] [PubMed] [Google Scholar]
- 37. Ma RC, Tam CH, Wang Y et al. Genetic variants of the protein kinase C-beta 1 gene and development of end-stage renal disease in patients with type 2 diabetes. JAMA 2010; 304:881–9. [DOI] [PubMed] [Google Scholar]
- 38. Assmann TS, Brondani Lde A, Bauer AC, Canani LH, Crispim D. Polymorphisms in the TLR3 gene are associated with risk for type 1 diabetes mellitus. Eur J Endocrinol 2014; 170:519–27. [DOI] [PubMed] [Google Scholar]
- 39. Le Goffic R, Balloy V, Lagranderie M et al. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLOS Pathog 2006; 2:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perales-Linares R, Navas-Martin S. Toll-like receptor 3 in viral pathogenesis: friend or foe? Immunology 2013; 140:153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salnikova LE, Smelaya TV, Moroz VV, Golubev AM, Rubanovich AV. Functional polymorphisms in the CYP1A1, ACE, and IL-6 genes contribute to susceptibility to community-acquired and nosocomial pneumonia. Int J Infect Dis 2013; 17:e433–42. [DOI] [PubMed] [Google Scholar]
- 42. Tu X, Chong WP, Zhai Y et al. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect 2015; 71:101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clemens EB, Grant EJ, Wang Z et al. Towards identification of immune and genetic correlates of severe influenza disease in Indigenous Australians. Immunol Cell Biol 2016; 94:367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zúñiga J, Buendía-Roldán I, Zhao Y et al. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J 2012; 39:604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simonsen L, Spreeuwenberg P, Lustig R et al. ; GLaMOR Collaborating Teams. Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: a modeling study. PLOS Med 2013; 10:e1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.