Investigations in pigs and porcine differentiated airway epithelial cells (PDAECs) reflect a loss in virulence of A(H1N1)pdm09 viruses isolated from humans and swine (2009–2015). PDAEC may be an in vitro test system for virulence of influenza A viruses.
Keywords: pandemic H1N1 2009 influenza A viruses, differentiated airway epithelial cells, evolution, adaptation
Abstract
We analyzed the virulence of pandemic H1N1 2009 influenza A viruses in vivo and in vitro. Selected viruses isolated in 2009, 2010, 2014, and 2015 were assessed using an aerosol-mediated high-dose infection model for pigs as well as air-liquid interface cultures of differentiated airway epithelial cells. Using a dyspnea score, rectal temperature, lung lesions, and viral load in the lung as parameters, the strains from 2014–2015 were significantly less virulent than the strains isolated in 2009–2010. In vitro, the viruses from 2009–2010 also differed from the 2014–2015 viruses by increased release of infectious virus, a more pronounced loss of ciliated cells, and a reduced thickness of the epithelial cell layer. Our in vivo and in vitro results reveal an evolution of A(H1N1)pdm09 viruses toward lower virulence. Our in vitro culture system can be used to predict the virulence of influenza viruses.
The influenza pandemic of 2009 started in Mexico and was caused by an influenza A virus of the H1N1 subtype [1, 2]. The virus most likely originated from influenza A viruses circulating in swine and was a reassortant virus [1, 2]. Its 8 RNA genome segments were derived from shift of genes of Eurasian swine influenza virus (NA, M) and North American triple reassortant virus (HA, NS, genes of the ribonucleoprotein complex) [1, 2]. In the first year of the pandemic, infections resulted in >100 000 deaths worldwide [2–4]. Increased lethality was particularly observed in patients with diabetes and obesity, but also in pregnant women [5–7]. The majority of infections, however, had a less severe course of disease [8]. The virus replicated well in the new host and established a new lineage in the human population, replacing the previous lineage of seasonal H1N1 influenza A viruses. At present, viruses originating from the pandemic virus are co-circulating together with viruses of the A(H3N2) subtype and influenza B viruses of 2 lineages [9]. Establishment of stable lineages within new hosts is only possible after adaptation to the new host cells [10, 11].
When influenza A viruses enter the respiratory tract, they encounter a layer of epithelial cells that function as a primary barrier to infection. To prevent infection, airway epithelial cells are equipped with a mucociliary clearance system based on mucus-producing cells and ciliated cells [12]. Ciliated cells transport mucus and the entrapped foreign substances out of the airways and thus prevent their detrimental effect. In addition to mucociliary clearance, the epithelial cells also have a barrier function. Tight junctions connecting the cells in the epithelial layer prevent the paracellular entry of foreign substances including pathogenic microorganisms [12, 13]. Air-liquid interface (ALI) cultures of respiratory epithelial cells from different species—that is, humans, ferrets, pigs, mice, and cattle [14–18]—have been used to analyze infections by respiratory viruses. Recently it has been shown that late during infection, the epithelial cell layer gets thinner because of a substantial loss of ciliated cells due to apoptosis; nevertheless, it retains its barrier function [18]. This is achieved by the basal cells that differentiate into specialized cells and compensate for the loss of infected cells. These effects of the influenza virus infection correspond to the changes observed in pathological studies of samples from influenza virus–infected airways in humans [19, 20].
In the present study, 1 porcine (SC14) and 4 human (HA09, JE09, JE10, KI15) A(H1N1)pdm09 isolates collected between 2009 and 2015 were analyzed for differences in their virulence for pigs and their effects on differentiated swine airway epithelial cells. The virulence of the viruses from 2014 and 2015 in pigs was decreased compared with viruses from 2009 and 2010. The effects of the viruses on the animals were paralleled by their effects on cultured airway epithelial cells. Infection of porcine ALI cultures by virulent influenza viruses resulted in a more extensive loss of ciliated cells, a more pronounced reduction of the epithelial thickness, and a higher titer of virus released into the supernatant compared with the less virulent isolates.
MATERIALS AND METHODS
Cells and Viruses
Virus cultivation and titration was performed in Madin–Darby canine kidney (MDCK, CRL-2936, American Type Culture Collection [ATCC]) cells as described previously [21]. Five A(H1N1)pdm09 viruses were randomly selected from the collections of viruses available of that time: A/Germany/1580/2009 (HA09), A/Germany/5258/2009 (JE09), A/Germany/2688/2010 (JE10), A/Germany/18909686/2015 (KI15), and A/sw/Germany/19989/2014 (SC14). The viruses belong to A(H1N1)pdm09 influenza virus clades 1 (JE10), 2 (HA09, JE09), and 6 (SC14, KI15). HA09 virus was isolated in April 2009, JE09 in July 2009, JE10 in March 2010, SC14 in April 2014, and KI15 in January 2015.
The GenBank accession numbers of viral genes are as follows: HA09: GU480807, HQ104924–HQ104929, and HM598305; JE09: KJ549775–KJ549782; JE10: MK159113–MK159120; SC14: KX013010–KX013017; KI15: MK159105–MK159112.
Infection Trials
Ethics Statement
The animal trials were approved by the Landesverwaltungsamt Sachsen-Anhalt (reference numbers AZ 42502-3-401, AZ 42502-3-642Ä, AZ 42502-3-743, and AZ 45502-3-579).
All trial procedures and animal care activities were conducted in accordance with the guidelines and under approval of Good Clinical Practice (VICH GL9, CVMP/VICH/595/98), the Directive 2001/82/EC on the community code relating to veterinary medicinal products, and German Animal Protection Law.
Pigs
Four different trials (Supplementary Table 1) were carried out using 11- to 16-week-old pigs (Large White × German Landrace) of the same pig farm. The pig herd was monitored regularly for the freedom of pathogens (Supplementary Materials and Methods) by investigation of blood samples and nasal swabs (every 6 months).
Treatments
All pigs were treated with antibiotics before challenge to reduce the influence of bacterial coinfection (Tulathromycin-Draxxin 10% ad usum veterinarium, Zoetis GmbH—1 mL per pig in the first week after birth and then every 14 days).
Study Procedures
Infection was carried out under high-dose aerosol nebulization conditions as described before [21, 22]. The investigations were done in the frameworks of the FluResearchNet and European Surveillance Network for Influenza in Pigs 3 networks under Biosafety Level 2 conditions in infection units with high-efficiency particulate air H13 filters by one of the network partners. For a detailed description of the study procedures, see the Supplementary Materials and Methods.
Studies in Porcine Airway Epithelial Cells
Preparation of ALI Cultures of Porcine Airway Epithelial Cells
Porcine lungs used in this study were obtained from a local slaughterhouse. ALI cultures of porcine airway epithelial cells were generated as previously described [23, 24].
Virus Infection
Well-differentiated porcine bronchial epithelial cells (PBECs) were washed 3 times with phosphate-buffered saline (PBS) and then infected by 1 of the 5 A(H1N1)pdm09 viruses at an infectious dose of 104 tissue culture infectious dose 50 per filter in 100 μL ALI medium. After incubation at 37°C for 1 hour, the infectious medium was removed and cells were washed 3 times with PBS to remove unbound virions. Finally, 600 μL ALI medium was added to the basal compartment to maintain the cells. To determine the virus release, 100 µL ALI medium was added at the respective time points to the apical compartment and incubated at 37°C. After 30 minutes, the apical medium was collected and used to determine the viral infectivity by endpoint dilution titration on MDCK cells as described previously [25].
Immunofluorescence Microscopy
All infected and mock-infected samples were washed with PBS 3 times and fixed with 3% paraformaldehyde (PFA) for 20 minutes. PFA was removed and 0.1 M glycine was added for 5 minutes. Samples were permeabilized with 0.2% Triton X-100, washed 3 times with PBS, and treated with 1% bovine serum albumin (BSA) for 30 minutes to block nonspecific binding sites. Primary and secondary antibodies were diluted with 1% BSA in PBS and incubated with the samples for 1 hour each. After washing with PBS, samples were incubated with DAPI (4′,6-diamidino-2-phenylindole) for 20 minutes. Finally, transwell filters were taken out by using a scalpel and embedded in Prolong Gold Antifade Reagent (Life Technologies, Darmstadt, Germany), and stored at 4°C for further analysis.
Antibodies against influenza A virus nucleoprotein (NP) (1:750, AbDSeroTec, Kidlingtom, United Kingdom), against mucin-5 AC (1:250, Santa Cruz Biotechnology, Dallas, Texas), and against zonula occludens-1(1:250, Life Technologies, Darmstadt, Germany) and Cy3-labeled antibody against β-tubulin (1:500, Sigma, St Louis, Missouri) were used as primary antibodies. Green fluorescence and red fluorescence (Alexa Fluor 488 and 568) conjugated antibodies (1:1000, Life Technologies, Darmstadt, Germany) were used as secondary antibodies. Samples were analyzed by using an inverse immunofluorescence microscope Nikon Eclipse Ti-S (Nikon) and true confocal system Spektraldetektor 5 (TCS SP5) confocal laser scanning microscope (Leica). Images were analyzed by using NIS-Elements Viewer 4.20 software (Nikon), LAS AF Lite software (Leica), and ImageJ/Fuji software.
Measurement of Cilia Coverage and Thickness of Cultures
To measure the cilia coverage of samples, images were taken by an inverse immunofluorescence microscope Nikon Eclipse Ti-S (Nikon) as TIF version and put in ImageJ/Fuji software to measure the relative cilia coverage area on the basis of the procedure of the software. Finally, data were statistically analyzed by using GraphPad Prism 5 software. To measure the thickness of cultures, images were taken by a TCS SP5 confocal laser scanning microscope (Leica) as TIF version (XZY) and measured for thickness by using LAS AF Lite sofware (Leica). Finally, data were statistically analyzed by using GraphPad Prism 5 software.
Statistical Analyses
Mann–Whitney U test was performed to evaluate statistical significances for data obtained in the animal experiments by using the software program SPSS 15.0. All in vitro experiments were performed at least 3 times and data were analyzed with Tukey multiple comparison test by using GraphPad Prism 5 software. Results were shown as means with standard deviations. A P value < .05 was considered significant. For further information, see the Supplementary Materials and Methods.
RESULTS
Virulence of A(H1N1)pdm09 Viruses in Pigs
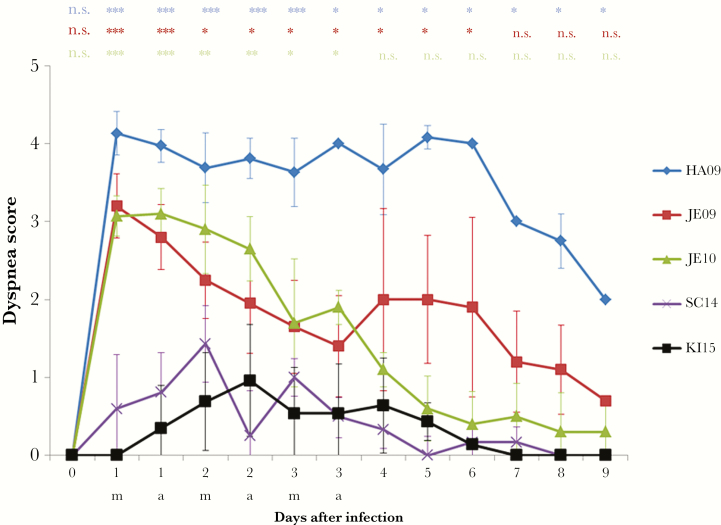
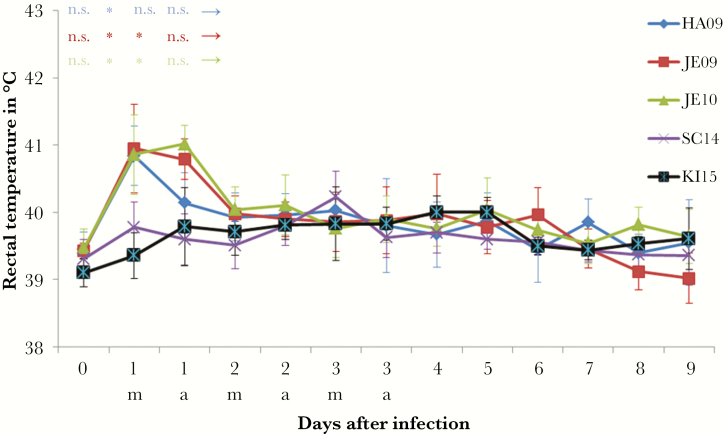
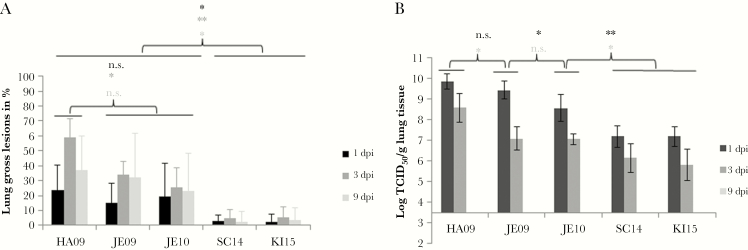
The virulence of the 5 A(H1N1)pdm09 viruses (HA09, JE09, JE10, SC14, and KI15) was analyzed in experimental infections of animals by determining the dyspnea score, the rectal temperature, the lung lesions, and the viral load in the lung. The viruses differed remarkably in their capability to induce signs of disease in pigs. The viruses isolated in 2009 and 2010 induced severe dyspnea in pigs, whereas viruses isolated in 2014 and 2015 failed to do so (Figure 1). With respect to the dyspnea score, the HA09 strain was most virulent inducing strong dyspnea in all pigs even 6 days after infection (Figure 1). HA09 was also the only virus that caused the death of some animals; 3 of 17 pigs died within the first 2–6 days after infection, indicating a mortality of about 18%. Two dyspnea peaks (1 dpi and 4–6 dpi) were observed after infection with JE09. The JE10 A(H1N1)pdm09 induced strong dyspnea 1 dpi that gradually decreased the following days. Low virulence was determined for the strains SC14 and KI15, which showed only slightly increased values. A difference between the viruses was also observed when the rectal temperature was measured. The viruses isolated in 2009–2010 induced fever, that is, >40°C. A short duration of fever is typical for influenza A virus infections of pigs. Strains SC14 or KI15 did not affect the rectal temperature (Figure 2).The pigs infected with 2009–2010 viruses displayed significantly more severe macroscopic lung lesions in comparison to pigs infected with viruses isolated in 2014–2015 (Figure 3A). The lesions were visible at least up to 9 dpi. Only marginal lesions were detectable in pigs infected by strains SC14 or KI15 (Figure 3A). In all animals, virus was detectable in the lungs at 1 and 3 dpi, but not at 9 dpi. The viral lung load after infection differed between the strains: Viruses isolated in 2009 induced significantly increased viral lung loads on 1 dpi when compared to virus isolated in 2010 (P ≤ .006); the latter induced a significantly higher viral lung load on 1 dpi in comparison to 2014 and 2015 A(H1N1)pdm09 viruses (P < .001; Figure 3B).
Figure 1.
Dyspnea in pigs infected by A(H1N1)pdm09 viruses. Dyspnea score (arithmetic mean), after infection of approximately 11- to 16-week-old pigs by different A(H1N1)pdm09 viruses isolated from 2009 to 2015. Statistical probability (Mann–Whitney U test): *P ≤ .05, **P ≤ .01, ***P ≤ .001. There were no significant differences between viruses SC14 and KI15; therefore, all statistical calculations were done with the viruses isolated in 2009–2010 (HA09, JE09, JE10) in comparison to KI15. Significant differences were also found within the group of virulent viruses: HA09 > JE09 (P < .001 at days 1–3 and P < .01 at days 5–9) > JE10 (P < .019 at day 5). Number of pigs: HA09, 17; JE09, 15; JE10, 15; SC14, 13; KI15, 19. Animals that died from influenza (group HA09) were provided with the highest score (4) in the calculations. Abbreviations: a, afternoon; m, morning; n.s., not significant.
Figure 2.
Rectal temperatures (°C, arithmetic mean) were determined after infection of approximately 11- to 16-week-old pigs by different A(H1N1)pdm09 viruses isolated from 2009–2015. Statistical probability (Mann–Whitney U test): *P ≤ .05, **P ≤ .01, ***P ≤ .001. There were no significant differences between viruses SC14 and KI15; therefore, all statistical calculations were done with the viruses isolated in 2009–2010 (HA09, JE09, JE10) in comparison to KI15. Number of pigs: HA09, 17; JE09, 15; JE10, 15; SC14, 13; KI15, 19. Abbreviations: a, afternoon; m, morning; n.s., not significant.
Figure 3.
Lung lesions (A) and viral lung load (B). Gross lung lesions (%, arithmetic mean with standard deviation) and the viral lung load (log TCID50, geometric mean with standard deviation) were determined after aerosol infection of approximately 11- to 16-week-old pigs by A(H1N1)pdm09 viruses isolated from 2009–2015. Statistical probability (Mann–Whitney U test): *P ≤ .05, **P ≤ .01, ***P ≤ .001. There were no significant differences between viruses SC14 and KI15; therefore, all statistical calculations were done with viruses HA09, JE09, and JE10 in comparison to KI15; 3.5 = detection limit due to predilutions of the samples. Significant differences were also found within the group of virulent viruses; lung lesions: HA09 > JE09/JE10 (P ≤ .021 at day 3); viral load: HA09 > JE09 (P = .001 at day 3) > JE10 (P = .006 at day 1). Number of pigs: HA09, 17; JE09, 15; JE10, 15; SC14, 13; KI15, 19. Abbreviations: dpi, days postinfection; n.s., not significant; TCID50, tissue culture infectious dose 50.
There was no difference between the virus isolated from pigs (SC14) and the human virus from 2015 (KI15) with respect to their virulence (Figures 1–3).
Taken together, the animal infections revealed that the virulence for pigs was significantly lower in animals infected by strains SC14 or KI15 compared with strains HA09, JE09, and JE10.
Infection of Porcine Differentiated Airway Epithelial Cells With A(H1N1)pdm09 Viruses
Cell Tropism and Transepithelial Electrical Resistance
Recently we have reported about the infection of well-differentiated PBECs cultured under ALI conditions. Both 2 swine influenza viruses and 2 human laboratory strains were found to infect mainly ciliated cells. Though infection resulted in the loss of ciliated cells, the viruses did not induce a reduction of the transepithelial electrical resistance (TEER). With respect to these 2 parameters, strains HA09, JE09, JE10, SC14, and KI15 did not differ from each other. They affected mainly ciliated cells and did not affect the epithelial barrier function (data not shown).
Replication Kinetics of A(H1N1)pdm09 Viruses in Porcine ALI
Well-differentiated PBECs were infected with either of the 5 viruses. Supernatants were collected at 24, 48, 72, 96, and 120 hours postinfection, and used to determine the amount of infectious virus released at the respective time point. The kinetics of virus release is shown in Figure 4. The viruses from 2009–2010 grew to higher titers than the 2 viruses from 2014–2015. At 48 and 72 hours postinfection, the differences were >10-fold and highly significant (P < .0001; Figure 4).
Figure 4.
Replication kinetics of A(H1N1)pdm09 viruses in porcine bronchial epithelial cells (PBECs). PBEC cultures were apically infected with pdmH1N1 isolates at infection dose of 104 TCID50/filter, at 37°C. After 1 hour, samples were washed 3 times with phosphate-buffered saline to remove free virions. Supernatants were collected at 24, 48, 72, 96, and 120 hours postinfection and analyzed for infectivity with TCID50 assay. At 48 and 72 hours postinfection, the differences were >10-fold and highly significant between viruses from 2009–2010 and viruses from 2014–2015. The results were shown as mean ± standard deviation of samples of 3 independent experiments, each with 3 replicates. Significance (***P < .001) was analyzed with Tukey multiple comparison test by using GraphPad Prism 5 software. Abbreviation: TCID50, tissue culture infectious dose 50.
Effect of A(H1N1)pdm09 Viruses on Ciliated Cells
Porcine ALI cultures of well-differentiated PBECs were analyzed for the loss of ciliated cells after infection by either of the 5 A(H1N1)pdm09 viruses. On 8 dpi, PBECs were subjected to β-tubulin staining to visualize cilia.
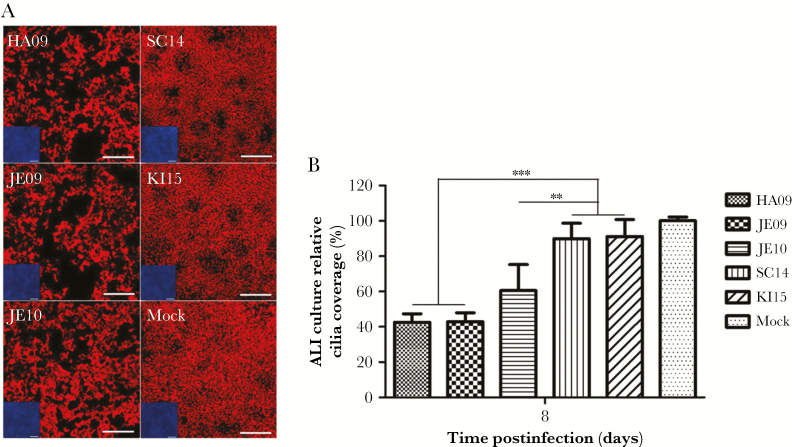
In mock-infected samples, the majority of cells were ciliated cells (Figure 5A). This high proportion was slightly decreased in ALI cultures infected by either of the 2014–2015 strains. A marked difference was observed when these cultures were compared to samples infected by either of the 2009–2010 strains (Figure 5A). A quantification of the area covered by cilia staining revealed a reduction of about 40%–50% between these 2 groups of viruses and the difference was significant (P ≤ .0049; Figure 5B). We also analyzed the thickness of the epithelial layer. For this purpose, ALI cultures were subjected to DAPI staining to visualize nuclei and to immunostaining with anti-β-tubulin antibodies to visualize cilia. As shown in Figure 6A, in vertical sections, the thickness of the samples infected by SC14 or KI15 was somewhat reduced compared to uninfected cultures. A stronger reduction was observed in ALI cultures infected by JE10. An even more pronounced reduction of the epithelial thickness was detectable after infection by HA09 or JE09. The differences in the thickness between the 2014–2015 strains and the 2009–2010 strains were significant (P ≤ .0363; Figure 6B). Taken together, infection of swine ALI cultures by different A(H1N1)pdm09 viruses results in virus release over a long period of time, loss of ciliated cells, and a reduction of the thickness of the epithelial layer. These effects are more pronounced in infections by the 2009–2010 viruses than they are in cultures infected by the strains SC14 or KI15. Results obtained with this in vitro culture system parallel those obtained in infection studies with pigs.
Figure 5.
Cilia coverage of porcine bronchial epithelial cells (PBEC) on 8 days postinfection. PBEC cultures were apically infected with A(H1N1)pdm09 viruses at infection dose of 104 tissue culture infectious dose50/filter. A, After 8 days postinfection, PBEC cultures were stained with Cy3-labelled β-tubulin to detect cilia (red). Scale bar = 100 μm. B, Quantitative analysis of area covered by cilia at 8 days postinfection. The air-liquid interface (ALI) culture relative cilia coverage (%) is shown as mean ± standard deviation compared to mock-infected cultures. Nine samples from 3 independent experiments were determined, and 3 fields from 1 sample were evaluated. Significance (**P < .01, ***P < .001) was analyzed with Tukey multiple-comparisons test by using GraphPad Prism 5 software.
Figure 6.
Thickness of porcine bronchial epithelial cells (PBECs) on day 8 postinfection. PBEC cultures were apically infected with A(H1N1)pdm09 isolates at infection dose of 104 tissue culture infectious dose50/filter. After day 8 postinfection, cilia (red) and nuclei (blue) of PBEC cultures were stained with Cy3-labelled β-tubulin and DAPI. A, The thickness of PBEC cultures infected with A(H1N1)pdm09 viruses at day 8 postinfection. For accurate determination of the thickness, vertical image stacks of 5 planes were merged. Scale bar = 50 μm. B, Quantitative analysis of the thickness of PBEC culture at day 8 postinfection. The air-liquid interface (ALI) culture relative thickness (%) is shown as mean ± standard deviation compared to mock-infected cultures. Nine samples from 3 independent experiments were determined, and 3 fields from 1 sample were evaluated. Significance (*P < .05, **P < .01) was analyzed with Tukey multiple-comparisons test by using GraphPad Prism 5 software.
DISCUSSION
Pigs are susceptible to infection by human and avian influenza A viruses [26]. They can be infected with human A(H1N1)pdm09 viruses and—in contrast to humans—their influenza A virus infections are not prone to be influenced by seasonal factors [27]. Moreover, signs of disease and course of swine influenza are comparable with that in humans [21]. This makes pigs an interesting infection model system for A(H1N1)pdm09 viruses [28, 29]. We investigated A(H1N1)pdm09 viruses isolated at different times from human patients (HA09, JE09, JE10, and KI15) or from a pig (SC14) under experimental high-dose aerosol-mediated infection conditions [21, 22].
Our results demonstrate that the porcine and human A(H1N1)pdm09 viruses from 2014 and 2015 are significantly less virulent for pigs than the 3 viruses from 2009–2010. The KI15 as well as the SC14 virus were less virulent than the others. Pigs from an age of 6 months onward usually develop less pronounced signs of disease after influenza A virus infection (R. Dürrwald, unpublished data). The difference of 4–5 weeks in age of pigs of the KI15 group in comparison to the other groups can be neglected, especially since a similar virus (SC14) showed similar characteristics. Despite the fact that the JE09 and the JE10 viruses induced disease in pigs, a comparison of the virulence parameters (dyspnea, lung lesions, virus load) indicated a gradual decrease in the virulence already in these early isolates of A(H1N1)pdm09 viruses: HA09 > JE09 > JE10 (see P values in the Results and the figure legends). The data support the notion that there is a gradual adaptation of A(H1N1)pdm09 viruses to their new hosts associated with a reduction in their capacity to induce severe disease. This process took place in humans and was shared by the virus isolated from a pig. The results obtained with our pig model point to an evolution of the A(H1N1)pdm09 virus toward lower virulence.
In vitro effects of infection with virulent viruses were evident by different parameters: virus release, loss of ciliated cells, and reduction of the thickness of the epithelial cell layer. Despite these effects, the barrier function of the epithelial cells was maintained as demonstrated by TEER. These results indicate that a regeneration process has occurred that compensated for the loss of ciliated cells. Our findings are consistent with the clinical symptoms observed in pigs. Reduced mucociliary clearance efficiency may contribute to the severity of dyspnea. Virus release over an extended time period and release of apoptotic cells may lead to an obstruction of bronchioli by cell debris. The quick recovery of most pigs may be favored by the sustained barrier functions and by the regeneration process of the airway epithelium.
Influenza viruses are generally well adapted to replication in their reservoir host. In the case of spillover infections, the virus has to modify its replication properties to propagate in the new host [30]. Since the first isolation of a human influenza virus, 3 different subtypes, H1N1, H2N2, and H3N2, had been successful in establishing virus lineages in the human population. Viruses of other subtypes (eg, H5N1, H7N9) occasionally infect humans and may cause severe or fatal disease in infected individuals; however, these viruses were so far not able to spread the infection among the population, which is a sign of insufficient adaptation to the spillover host [27, 31, 32]. The A(H1N1)pdm09 virus was able to replace the seasonal H1N1 lineage. The adaptation of the virus to the human host was accompanied—as demonstrated by our data—by a reduction of virulence for pigs. The A(H1N1)pdm09 virus is easily transmitted between humans and pigs but transmission between humans seems to be more easily accomplished than that between pigs because the virus did only rarely establish stable lineages after primary incursion into the pig population and preferred to acquire new genes (ie, N2) before establishing stable propagation in pigs [33]. Thus, the original A(H1N1)pdm09 virus was most probably not yet well adapted to mammalian species, including pigs and humans, despite its ability to replicate efficiently in the latter. This may be the reason for the severe course of disease in infection experiments with HA09 and JE09 virus.
Strains JE09 and JE10 differ by a number of amino acid exchanges from the HA09 strain. However, none of these mutations were found in both viruses. Therefore, it is not possible to assign the somewhat reduced virulence of JE09 and JE10 to a certain amino acid. Similar findings were reported by others that compared the virulence of early pandemic H1N1 viruses in ferrets and mice [34, 35]. A different picture was obtained when the virulent viruses were compared with the low-virulent viruses. Among the different influenza virus genes, a total of 20 amino acid changes were found that distinguished these 2 groups of viruses (Supplementary Table 2). The successful establishment of the new virus lineage is evident from the fact that all 84 sequences of human A(H1N1)pdm09 viruses that have been isolated in different parts of Germany and deposited in the Global Initiative on Sharing All Influenza Data repository share the 20 amino acid changes mentioned above. None of these amino acids is known to be a marker for virulence. Thus, these mutations may reflect adaptations to human cells.
The 2014 virus was isolated from a pig. We do not know when this virus was transmitted from humans to pigs and how many passages in pigs it underwent. The relatively large number of mutations in comparison to KI15 hints to a longer evolution in pigs. Despite this, its ancestor most probably was not directly derived from a 2009 or 2010 A(H1N1)pdm09 virus. Whatever the passage history of this swine virus SC14 is, it resembles the KI15 virus in its virulence properties. Most probably, this low virulence reflects properties that the virus acquired during evolution in the human population.
To determine how the different amino acid changes cooperate to confer the phenotype of reduced virulence would require an enormous number of animal experiments. In this context, it is intriguing that the results of our in vitro infections with filter-grown differentiated airway epithelial cells reflected the virulence differences between the viruses as determined in pig experiments. Our observations with ALI cultures are consistent with pathological changes reported for samples derived from diseased patients or animals [20, 35, 36]. The affected epithelium frequently shows only a single layer of flattened basilar epithelial cells covering the basement membrane. Viruses with different virulence differ in the size of the foci of infection that are characterized by the loss of cilia. With this in vitro culture system available that—upon infection by influenza virus—reflects the virulence characteristics of the respective virus, it will be much easier to analyze the importance of individual mutations or combinations of amino acid exchanges or epigenetic mechanisms with respect to virulence in pigs. In future studies, it will be interesting to find out whether the porcine in vitro cell culture model can also be applied to viruses of other species, for example, avian influenza viruses, and to determine whether the virulence-related differences between human influenza viruses are also observed upon infection of human differentiated airway epithelial cells.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. This work was performed by Y. F. in partial fulfillment of the requirements for doctoral degrees from the University of Veterinary Medicine Hannover. Part of this work was contributed by F. M. in partial fulfillment of the requirements for the Doctor of Veterinary Medicine degree from the University of Veterinary Medicine Hannover. We thank Dr Sigrid Baumgarte, Institut für Hygiene und Umwelt, Hamburg (current position Laborärztliche Arbeitsgemeinschaft für Diagnostik und Rationalisierung GmbH, Medizinisches Zentrum Dr Kramer & Kollegen, Geesthacht), for providing virus HA09.
Financial support. The animal trials of this study were funded to R. D. by Bundesministerium für Bildung und Forschung (grant numbers 01KI 07143) within the framework of the FluResearchNet and by the European Union FP7-INFLUENZA-2010 ESNIP3 program (grant number 259949) within FP7 Health. The work of G. H. was supported by grants from the Deutsche Forschungsgemeinschaft (grant numbers He1168/15-1 and He1168/19-1). This work was further supported in part by grants from the German Federal Ministry of Education and Research (grant numbers 01KI7142 and 01KI1006J) awarded to R. D., R. Z. and M. S.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009; 459:931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization. WHO global alert and response: pandemic (H1N1) 2009—update 112. Geneva, Switzerland: WHO, 2010. [Google Scholar]
- 4. Dawood FS, Iuliano AD, Reed C, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 2012; 12:687–95. [DOI] [PubMed] [Google Scholar]
- 5. Jiménez-García R, Hernández-Barrera V, Rodríguez-Rieiro C, et al. Hospitalizations from pandemic influenza [A(H1N1)pdm09] infections among type 1 and 2 diabetes patients in Spain. Influenza Other Respir Viruses 2013; 7:439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun Y, Wang Q, Yang G, Lin C, Zhang Y, Yang P. Weight and prognosis for influenza A(H1N1)pdm09 infection during the pandemic period between 2009 and 2011: a systematic review of observational studies with meta-analysis. Infect Dis (Lond) 2016; 48:813–22. [DOI] [PubMed] [Google Scholar]
- 7. Creanga AA, Johnson TF, Graitcer SB, et al. Severity of 2009 pandemic influenza A (H1N1) virus infection in pregnant women. Obstet Gynecol 2010; 115:717–26. [DOI] [PubMed] [Google Scholar]
- 8. Dawood FS, Jain S, et al. ; Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in human. N Engl J Med 2009; 360:2605–15. [DOI] [PubMed] [Google Scholar]
- 9. Bedford T, Riley S, Barr IG, et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 2015; 523:217–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 2010; 7:427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watanabe T, Kawaoka Y. Influenza virus-host interactomes as a basis for antiviral drug development. Curr Opin Virol 2015; 14:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ganesan S, Comstock AT, Sajjan US. Barrier function of airway tract epithelium. Tissue Barriers 2013; 1:e24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tilley AE, Walters MS, Shaykhiev R, Crystal RG. Cilia dysfunction in lung disease. Annu Rev Physiol 2015; 77:379–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirchhoff J, Uhlenbruck S, Goris K, Keil GM, Herrler G. Three viruses of the bovine respiratory disease complex apply different strategies to initiate infection. Vet Res 2014; 45:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dijkman R, Jebbink MF, Koekkoek SM, et al. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J Virol 2013; 87:6081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam E, Ramke M, Groos S, Warnecke G, Heim A. A differentiated porcine bronchial epithelial cell culture model for studying human adenovirus tropism and virulence. J Virol Methods 2011; 178:117–23. [DOI] [PubMed] [Google Scholar]
- 17. Liu X, Luo M, Zhang L, Ding W, Yan Z, Engelhardt JF. Bioelectric properties of chloride channels in human, pig, ferret, and mouse airway epithelia. Am J Respir Cell Mol Biol 2007; 36:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu NH, Yang W, Beineke A, et al. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Sci Rep 2016; 6:39668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu WC, Chan RW, Wang J, et al. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. J Virol 2011; 85:6844–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weinheimer VK, Becher A, Tönnies M, et al. Influenza A viruses target type II pneumocytes in the human lung. J Infect Dis 2012; 206:1685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duerrwald R, Schlegel M, Bauer K, Vissiennon T, Wutzler P, Schmidtke M. Efficacy of influenza vaccination and Tamiflu treatment—comparative studies with Eurasian swine influenza viruses in pigs. PLoS One 2013; 8:e61597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dürrwald R, Selbitz H-J. Swine influenza control by vaccination. Pig progress. Spec Resp Dis 2001; 20:11–4. [Google Scholar]
- 23. Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 2005; 107:183–206. [DOI] [PubMed] [Google Scholar]
- 24. Meng F, Wu NH, Seitz M, Herrler G, Valentin-Weigand P. Efficient suilysin-mediated invasion and apoptosis in porcine respiratory epithelial cells after streptococcal infection under air-liquid interface conditions. Sci Rep 2016; 6:26748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang W, Punyadarsaniya D, Lambertz RL, et al. Mutations during the adaptation of H9N2 avian influenza virus to the respiratory epithelium of pigs enhance sialic acid binding activity and virulence in mice. J Virol 2017; 91 pii:e02125-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010; 7:440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simon G, Larsen LE, Dürrwald R, et al. ; ESNIP3 Consortium. European surveillance network for influenza in pigs: surveillance programs, diagnostic tools and swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS One 2014; 9:e115815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajao DS, Vincent AL. Swine as a model for influenza A virus infection and immunity. ILAR J 2015; 56:44–52. [DOI] [PubMed] [Google Scholar]
- 29. Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol 2012; 20:50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cotter CR, Jin H, Chen Z. A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLoS Pathog 2014; 10:e1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Potter CW. A history of influenza. J Appl Microbiol 2001; 91:572–9. [DOI] [PubMed] [Google Scholar]
- 32. Neumann G, Kawaoka Y. Transmission of influenza A viruses. Virology 2015; 479–480:234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lange J, Groth M, Schlegel M, et al. Reassortants of the pandemic (H1N1) 2009 virus and establishment of a novel porcine H1N2 influenza virus, lineage in Germany. Vet Microbiol 2013; 167:345–56. [DOI] [PubMed] [Google Scholar]
- 34. Meunier I, Embury-Hyatt C, Stebner S, et al. Virulence differences of closely related pandemic 2009 H1N1 isolates correlate with increased inflammatory responses in ferrets. Virology 2012; 422:125–31. [DOI] [PubMed] [Google Scholar]
- 35. Elderfield RA, Watson SJ, Godlee A, et al. ; MOSAIC Investigators. Accumulation of human-adapting mutations during circulation of A(H1N1)pdm09 influenza virus in humans in the United Kingdom. J Virol 2014; 88:13269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meng F, Punyadarsaniya D, Uhlenbruck S, et al. Replication characteristics of swine influenza viruses in precision-cut lung slices reflect the virulence properties of the viruses. Vet Res 2013; 44:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.