Abstract
Avian influenza virus (AIV) H9N2 emerged in the 1990s as an economically important disease in poultry and occasionally infects humans and other mammals. The aim of this study was to evaluate the acquisition and retention of H9N2 AIV on and within the house fly, Musca domestica (Linnaeus 1758), under laboratory conditions. In first experiment, 100 adult house flies were divided into control and treatment groups equally. Treatment group was fed with a meal containing H9N2 virus, while control group was supplied with an identical meal without virus. Fifteen minutes after exposure in each group, flies were washed twice to remove surface particles, disinfected and then homogenized for testing. The two external body surface washes and the homogenate samples were tested for H9N2 to distinguish exterior from interior viral load. Second experiment was performed likewise but five flies from each group were taken at 0, 6, 24, 48, 72, 96, and 120 h post-exposure. All samples were subjected to real-time reverse-transcription polymerase chain reaction (RRT-PCR) for detecting H9-Specific viral RNA. Results of the first experiment showed that viral RNA was detectable in both of external surface and homogenates samples. Second experiment revealed that persistence of H9N2 AIVs on external body surface and within the body of M. domestica were 24 and 96 h, respectively. Moreover, viral RNAs concentration declined during the time after exposure to AIV H9N2 either outside or within house flies. Overall, house fly was able to acquire and preserve H9N2 AIV experimentally, which may contribute the spread of virus among poultry farms.
Keywords: avian influenza, house fly, H9N2, acquisition, retention
Low-pathogenic avian influenza virus (LPAIV H9N2) has circulated in domestic poultry since 1994 after emerging in China (Kwon et al. 2006). Two major issues arose with LPAIV H9N2 1) due to persistent outbreaks and widespread vaccination programs it served as a barrier to poultry market economics and 2) from 1998 on, 15 reports of human infections were published suggesting an increasing likelihood of a pandemic (Cox et al. 2017). Frequency of AI infections in different countries is affected by the structure of poultry sector, including density of poultry production, the infection status, biosecurity practices, and environmental conditions (Paul et al. 2011, Sims et al. 2017). LPAIV infections have been reported with varying mortality and morbidity in different developing countries. Since the first Iranian outbreak of H9N2 in 1998, mortality between 20 and 65% has been reported and endemic H9N2 AI virus has imposed a huge strain on the economy of poultry industry (Nili and Asasi 2002).
AIV, although relatively unstable in the environment, remains viable on condition that organic materials such as nasal secretions or feces as well as cool and moisture increase resistance of the virus to inactivation. AIV remained viable up to 105 d in liquid manure during winter, 30–35 d at 4°C in feces and 7 d at 20°C, and therefore it stands a better chance of dispersal (Sims et al. 2017). In addition, transmission of AIV seems to occur through viruses shed from nares, mouth, conjunctiva, and cloaca, either by direct bird-to-bird contact or indirectly via aerosol droplets, contaminated food, water, fomites, and people. Therefore, synanthropic animals such as rodents, wild terrestrial birds, and insects like flies may be possible vectors of AIV (Velkers et al. 2017). Previously in areas with AI outbreak history, H5 highly pathogenic avian influenza (HPAI) viruses were isolated from house flies, blow flies, black garbage flies, and small dung flies (Brugh and Johnson 1986, Sawabe et al. 2006).
The house fly (Musca domestica L.) is a ubiquitous synanthropic insect frequently associated with poultry waste. House flies undergo holometabolous metamorphosis with an approximate 10-day life cycle and 1–3 km flight range. Transmission of pathogens occurs via mouth parts, vomit droplets, feces and dislodgement from exoskeleton, especially legs, through the ovipositing and feeding behavior (Wanaratana et al. 2013).
More recently, transmission of influenza virus by flies arouses the interest of scientists. Sawabe et al. (2006), not only isolated H5N1 from internal organs in two species of blow flies in the vicinity of infected poultry farm but also detected viable H5N1 virus in the blow flies up to 48 h post-exposure (PE) to virus experimentally (Sawabe et al. 2009). Wanaratana et al. (2011) showed that infective H5N1 virus could be detected at least 72 h PE from house flies while viral RNA was still found by real-time reverse-transcription polymerase chain reaction (RRT-PCR) 96 h PE. Furthermore, their following study showed that sufficient amount of H5N1 virus for spreading disease to healthy chickens can be carried by house flies (Wanaratana et al. 2013). Nielsen et al. (2011) revealed the potential of house fly as a carrier of HPAI virus after investigating persistence of H5N7 and H7N1 virus in the digestive tract of infected house flies. The aim of this study was to investigate the acquisition of H9N2 AIV by the house fly under laboratory conditions and to determine virus persistence in external surface and within house fly using quantitative reverse-transcription PCR.
Materials and Methods
Virus
The influenza virus, A/chicken/Iran/SR110/2007 (H9N2) (NCBI GenBank Accession Number EU157927.33) used in this study was propagated in the allantoic cavity of 10-d-old embryonated chicken eggs according to the OIE guidelines (OIE 2015). After the third passage, allantoic fluid of inoculated embryonated chicken eggs was stored at −80°C until being used in virus exposure experiments. The virus titer was determined as 108 50% chicken embryo infective dose per milliliter (EID50/ml).
House Flies
House flies were captured by insect aerial net from rotten fish baits at a dairy farm. After transferring flies to the laboratory, the flies were identified based on morphological criteria outlined by Carvalho and Mello-Patiu under a stereomicroscope (Carvalho and Mello-Patiu 2008). The mixed-sex adult house flies were placed and kept as a source in specially designed fly cages constructed from six 50 × 50 cm transparent acrylic glass boards. Four sides of the cages were finely drilled. Two extraholes (15 cm in diameter) were intended as entry and exit or to allow manipulation. All through the incubation periods, house flies were kept at constant room temperature with 25–40% humidity and a 12:12 (L:D) h cycle, in the entomology laboratory, veterinary school of Ferdowsi University of Mashhad. Prior to commencing the experiments, ten flies were randomly examined to confirm the absence of AI H9N2 viruses by RRT-PCR. All samples were free of the AI H9N2 virus.
Experiment 1 (Ability of House Fly to Acquire the AIV H9N2)
One hundred adult house flies were randomly divided and housed in two sterile fly cages as control (n = 50) and treatment (n = 50) groups. Flies in both groups were deprived of food and water for 24 h. The control group was then supplied with a cotton pad soaked with 10% sucrose diluted in the ultra-high-temperature (UHT) processed milk (Wanaratana et al. 2011), while the treatment group was fed with 13.5 ml of the same meal plus 1.5 ml of allantoic fluid containing H9N2 virus doses equivalent to 108 EID50 (Nielsen et al. 2011). Water was also provided ad libitum using a soaked cotton pad in each cage. Fifteen minutes PE, all flies in each group were immobilized at −20 ◦C for 2 min and then transferred into two 50-ml Falcon conical tubes (SPL Life Sciences Co., Ltd., Pocheon, South Korea). The external body surface of flies was washed up with 10 ml of sterile phosphate buffered saline (PBS; 135 mmol/liter NaCl, 3 mmol/liter Na2HPO4 12 H2O, and 13 mmol/liter NaH2PO4 2H2O, pH 7.2). Then the first wash fluid (S1) was transferred into the 1.5-ml microtubes. Once again, the external body surface of flies was washed by vortexing with 10 ml of 70% ethanol and rinsed by 10 ml of distilled water after discarding the ethanol. Thereafter, fly bodies were transferred to a new falcon tube by sterile forceps and again body surface of flies was washed up with 10 ml of sterile PBS. Second body surface specimen (S2) from the last PBS washing was collected in microtubes. The washed bodies of all 50 flies were homogenized together by chilled and sterile mortar and pestle while 10 ml of PBS was gently added. The homogenate was taken in sterile microtubes and centrifuged at 3000×g for 5 min at 4°C (Eppendorf Centrifuge 5417R, Hamburg, Germany). The supernatant was collected in sterile microtubes (H1). S1, S2, and H1 specimens were refrigerated at −80°C to be subjected to RRT-PCR for detecting the H9-Specific viral RNA. The whole experiment 1 was repeated five times.
Experiment 2 (Persistency Period of AIV H9N2 in/on House Fly)
All the steps were identically performed as described in the first experiment unless otherwise mentioned. House flies were randomly divided into control (n = 60) and treatment (n = 60) groups. Following 15-min exposure of the treatment group to AI H9N2 virus, water, and food cotton pads were removed from both cages. For the rest of the experiment, flies consumed clean (virus-free) diet; 10% sucrose in UHT milk, and water. Five flies from each group were randomly taken using an aspirator at each time point; 0, 6, 24, 48, 72, 96, and 120 h PE. After immobilization at −20°C for 2 min, two specimens were obtained by washing external body surface of flies with 1.5 ml of PBS inside falcons; S3 (first outer surface wash fluid before ethanol washing) and S4 (Second wash fluid after disinfecting outer surface). The third specimen (H2) was also obtained after whole body homogenizing of house flies as described in first experiment, but with 1.5 ml of PBS. All samples were labeled with group, sample name, and time point, then were frozen until being subjected to RRT-PCR. The experiment 2 also had five replicates.
RNA Extraction
Total RNAs of specimens were extracted using High Pure Viral RNA Kit according to the manufacturer’s instructions (Roche, Mannheim, Germany). Briefly, 200 µl of wash fluids or homogenate was mixed with 400 µl of Binding Buffer supplemented with Poly (A). The mixture was transferred to High Pure Filter tube assembly and centrifuged for 15 s at 8000×g. Then, 500 µl of Inhibitor Removal Buffer was added to the upper reservoir and centrifuged for 1 min. In the next step, 450 µl of wash buffer was added twice to the filter tube and centrifuged for 1 min after each washing. After each centrifugation, flow-through and the collection tube were discarded. Finally, the filter tube was placed in a 1.5-ml microtube, 50 µl of Elution Buffer was added to the upper reservoir and centrifuged. Extracted RNA was stored at −80°C.
Standard Curve Plotting
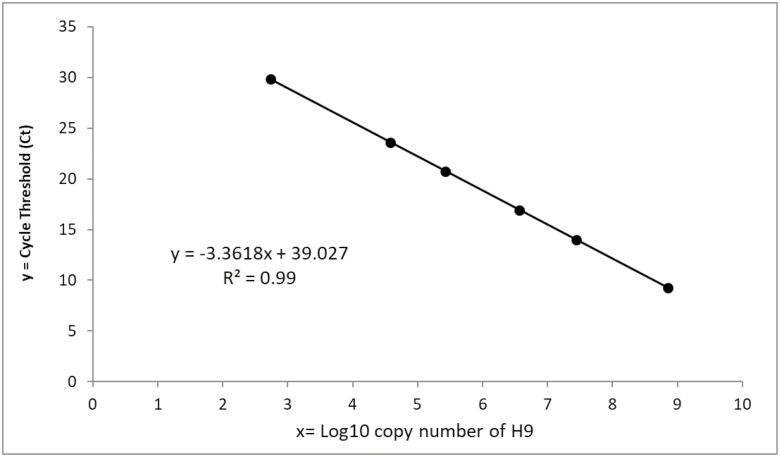
In order to perform an absolute quantitation by RRT-PCR assay, a standard curve was plotted using the PCR product of H9 gene with determined copy number. Firstly, the same primers of those used in real-time RT-PCR experiments were recruited in a conventional PCR amplification procedure. Then PCR product was electrophoresed on the agarose gel to purify the H9 gene band. Finally, its concentration was determined by Epoch microplate spectrophotometer (BioTek instrument, United States) as nanogram per microliter. The copy number of H9 gene in the standard was calculated according to the following formula: Number of copies (molecules) = (X ng × 6.0221 × 1023 molecules/mole)/ [(N × 660 g/mole) × 1 × 109 ng/g], where X is amount of amplicon (ng), N is length of dsDNA amplicon and 660 g/mole = average mass of 1 bp dsDNA. Standard curve was generated using a decimal serial dilution of the PCR product of H9 gene and plotting the threshold cycle values (Ct value) against the log concentrations of copy numbers (Fig. 1).
Fig. 1.
Standard curve (plot of the Ct values against the log10 concentration of a decimal serial dilution of H9 antigen) indicating the linearity of real-time PCR. The curve is linear over all 6 order of magnitude with R2 value of 0.999, slope of −3.36 and amplification efficiency of 98%.
RRT-PCR Assay
RRT-PCR was performed on RNA samples in duplicate according to the procedure developed by Monne et al. (2008) with some modifications, using the forward primer 5′-ATGGGGTTTGCTGCC-3′, reverse primer 5′-TTATATACAAATGTTGCAC(T)CTG-3′ and probe FAM-5′-TTCTGGGCCATGTCCAATGG-3′-TAMRA. Oasig lyophilized OneStep qRT-PCR Mastermix (Primerdesign Ltd, United Kingdom) and RotorGene 6000 thermocycler (Corbett, Australia) were utilized for RRT-PCR Test. Amplification protocol was 10 min at 55°C, 3 min at 95°C and 40 cycles of 95°C for 10 s and 57°C for 1 min. After optimization of the reaction, the regression coefficient (R2) was 0.99. Finally, the copy numbers in the samples were calculated by the standard curve method.
Results
Experiment 1
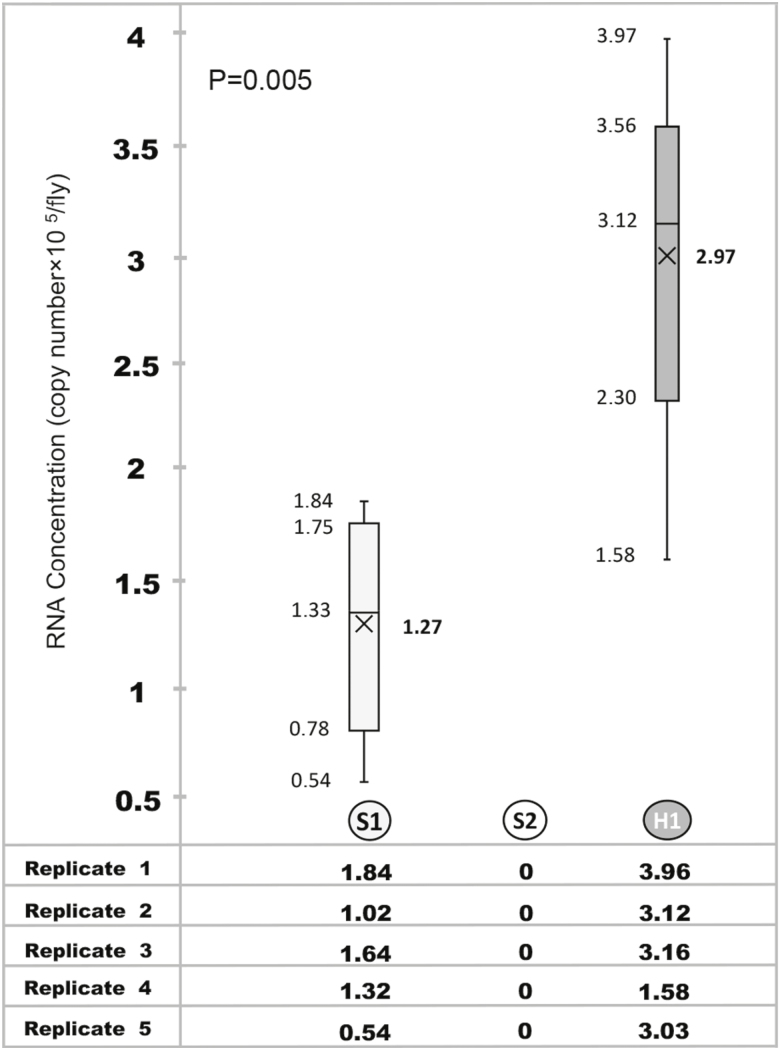
Viral RNA was detected at 1.27 ± 0.51 (mean ± SD) copies×105/fly in S1 samples which was significantly lower than 2.97 ± 0.86 copies × 105/fly in H1 samples (P = 0.005; Fig. 2). No viral RNA was found in S2 samples of treatment group. All samples of the control group were negative for AI.
Fig. 2.
Concentration of H9-Specific RNA in external body surface (S1 and S2) and homogenate (H1) of house fly after exposure to H9N2 AIV at concentration of 108 EID50/ml. Data are presented as box and whisker plots, where the bottom and top of the box are Q1 and Q3, horizontal lines represent median values, and the ends of the whiskers are the lowest and highest datum. (×) represents mean value.
Experiment 2
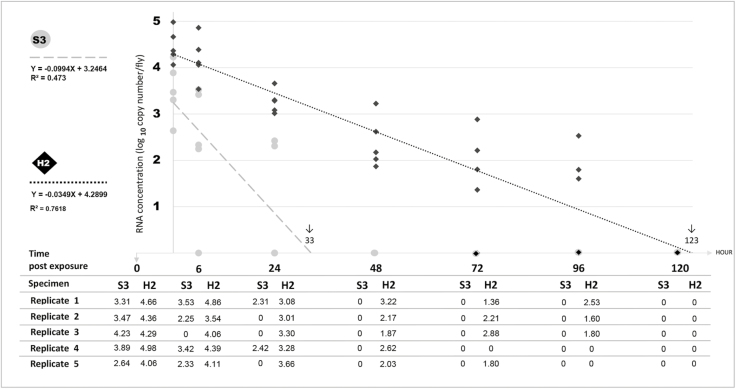
RNA Concentration of both S3 and H2 samples was regressed on time using a simple linear regression model (Fig. 3). S3 samples were positive only during first day after exposure at 0 (100%), 6 (80%) and 24 (40%) h PE with the average detection rate of 3.51, 2.3 and 0.95 Log10 copies/fly, respectively. The viral load measurement of H2 samples showed a time-dependent descending trend over 96 h following the logistic model (Y = −0.0349X + 4.2899, R2 = 0.7618). RRT-PCR results showed that AI H9N2 virus was detectable at 4.47, 4.19, 3.27, 2.38, 1.65, and 1.19 Log10 copies/fly in 0, 6, 24, 48, and 96 h but not at 120 h PE. The AI virus was not detected in both S4 and samples of negative control group.
Fig. 3.
Scatter plot showing the relationship between time and RNA concentration, expressed in terms of log10 copies per fly. A time-dependent descending trend over 96 h and the estimated persistence (↓) of H9N2 after fed to flies at a virus concentration of 108 EID50/ml are shown in external body surface (S3) and homogenate of house fly (H2).
Discussion
H9N2 avian influenza has spread worldwide and OIE has put it forward as one of the leading candidates for future influenza pandemics (OIE 2015). Notwithstanding the official designation as low-pathogenic, H9N2 subtype can cause a broad range of signs, varying from subclinical infection to 100% mortality in poultry farms (Nili et al. 2013) and poultry industry of countries such as Iran suffer huge economic losses via reduction of body weight gain, vaccination costs, and sometimes high mortality in poultry farms. In order to reduce the risk and problems posed to both public health and poultry sector, dissemination routes of this subtype need to be clarified.
Insects, wild birds, humans, other mammals, and also inanimate objects like fomites are believed to be able to transmit AI virus (Brugh and Johnson 1986). Among them, house fly has been considered as a vector of many pathogenic agents including bacteria, viruses, fungi, protozoan, parasitic oocysts, and eggs (Wanaratana et al. 2011, Junqueira et al. 2017). The predominance of optimal conditions for population growth of house flies made them the most important insect in poultry farms (Tyasasmaya et al. 2016). In this study, the possibility of acquisition and retention time period of this virus in/on house fly were investigated.
The results revealed that LPAIV could accommodate at external body surface of house fly and its persistency period was up to 24 h PE. These results are in accordance with those of Wanaratana et al. (2011) and Nazni et al. (2013) who respectively showed that H5N1 and H1N1 could be detectable up to 24 h on the outer body surface of house flies. Our findings appear to be well substantiated by three facts about house fly: the habitat, the behavior, and the body structures. House flies inhabit manure, spoiled foods, carcasses, and synanthropic environments (Sawabe et al. 2006, Junqueira et al. 2017). Additionally, surfaces are easily contaminated with virus particles due to fly foraging habits, regurgitation, and defecation (Sawabe et al. 2009), depositing eggs (Calibeo-Hayes et al. 2003) and also grooming activities (Tan et al. 1997). Anatomically, the fly’s body is covered with hairs, the legs are bristled, the foot pads adhesive, and the proboscis are all well suited to being contaminated (Tyasasmaya et al. 2016, Junqueira et al. 2017).
Our important finding was detection of H9-specific RNA within house flies up to 96 h, which was not detected any longer at 120 h after the exposure. These results are consistent with those of Wanaratana et al. (2011) who observed the same length of time for positive RRT-PCR results of whole homogenate of house fly bodies PE to AIV H5N1. More recently, in 2016, a viral exposure experiment also showed the existence of AIV H5N1 for at least 24 h in gastrointestinal tracts of house flies (Tyasasmaya et al. 2016). In crop of house flies, live Newcastle disease virus (NDV) was detected up to 96 h after initial exposure to NDV while it remained viable only 24 h in midgut and hindgut (Watson et al. 2007). However, the H5N1 AIV was isolated from the feces of blowflies Calliphora nigribarbis (Vollenhoven 1863) (Diptera: Calliphoridae) at 48 h after exposure (Sawabe et al. 2009). Under experimental conditions, sugar was used in feeding meal and high sugar concentration correlates with meal storage in crop, even for several days (Greenberg 1973). Thus, dissemination of virus through defecation and specially regurgitation seems more feasible. Flies were not dissected in the current study; however, in order to elucidate different probable routes of virus transmission and spread by flies, we are currently in the process of discovering virus retention sites in each dissected body fractures comparatively.
Our result also showed that the outer surface of house fly during initial 24 h could carry less load of viral RNAs than interior parts of flies which seems directly in line with the destructive effect of temperature and ultraviolet light on labile AI viruses (Chumpolbanchorn et al. 2006), but the most important clinically relevant finding was the decline in concentration of viral RNAs during the time after exposure to AIV H9N2 either outside or within house flies. The virus did not replicate inside the house fly, supporting the dominant paradigm of mechanical but not biological transmission of avian viruses; rotavirus, turkey coronavirus, reticuloendotheliosis virus, and NDV (Tan et al. 1997, Calibeo-Hayes et al. 2003, Davidson and Braverman 2005, Barin et al. 2010). Although this is the first experimental study elucidating the dynamic of H9N2 AIV retention in flies, it did not evaluate the viability of virus post-exposure. But generally, the RNA load probably is a high estimate of the viable virus that could be present and that when the samples are negative for RNA, there should not be any viable virus present, however, particularly HPAI H5N1 was shown to remain viable up to 72 h in a similar experiment (Wanaratana et al. 2011). Further works, which take viability of the viruses into account, is intended to be undertaken using subsequent inoculation to culture media or chickens. The answer to the question ‘if the number of viral particles suffice to be transmitted and to infect a chicken in different critical routes?’ is deferred to our future work in controlled field conditions.
In summary, the results of this study demonstrate the persistence of H9-specific RNA up to 24 h on the surface and up to 96 h within the house fly post-experimental feeding of H9N2 AIV. In endemic countries, especially those with low quality of biosecurity practices, flies might be involved in intrafarm or close transmission which needs practical implications like reducing flies population as well as boosting biosecurity measures.
Acknowledgments
We thank Ferdowsi University of Mashhad, Iran, for its financial support of this project under Grant number 2/30730. We also wish to thank Dr. Abbas Barin for providing the H9N2 influenza virus as well as Dr. Hesam Dehghani and Mr. Ali Zavari for their technical support.
References Cited
- Barin A., Arabkhazaeli F., Rahbari S., and Madani S. A.. . 2010. The housefly, Musca domestica, as a possible mechanical vector of Newcastle disease virus in the laboratory and field. Med. Vet. Entomol. 24: 88–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugh M., and Johnson D. C.. 1986. Epidemiology of avian influenza in domestic poultry, pp. 177–186. In V. A. Richmond (ed.), 2nd International Symposium on Avian Influenza, Athens, GA. Omnipress, United States Animal Health Association, Madison, WI. [Google Scholar]
- Calibeo-Hayes D., Denning S. S., Stringham S. M., Guy J. S., Smith L. G., and Watson D. W.. . 2003. Mechanical transmission of turkey coronavirus by domestic houseflies (Musca domestica Linnaeaus). Avian Dis. 47: 149–153. [DOI] [PubMed] [Google Scholar]
- Carvalho C. J. B. D., and Mello-Patiu C. A. D.. . 2008. Key to the adults of the most common forensic species of Diptera in South America. Rev. Bras. Entomol. 52: 390–406. [Google Scholar]
- Chumpolbanchorn K., Suemanotham N., Siripara N., Puyati B., and Chaichoune K.. . 2006. The effect of temperature and UV light on infectivity of avian influenza virus (H5N1, Thai field strain) in chicken fecal manure. Southeast Asian J. Trop. Med. Public Health 37: 102–105. [PubMed] [Google Scholar]
- Cox N. J., Trock S. C., and Uyeki T. M.. . 2017. Public health implications of animal influenza viruses, pp. 92–132. InSwayne D. E. (ed.), Animal influenza. Blackwell Publishing, Ames, IA. [Google Scholar]
- Davidson I., and Braverman Y.. . 2005. Insect contribution to horizontal transmission of Reticuloendotheliosis virus. J. Med. Entomol. 42: 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg B. 1973. Flies and disease, pp. 90–275. InGreenberg B. (ed.), Biology and disease transmission. Princeton University Press, Princeton, NJ. [Google Scholar]
- Junqueira A. C. M., Ratan A., Acerbi E., Drautz-Moses D. I., Premkrishnan B. N. V., Costea P. I., Linz B., Purbojati R. W., Paulo D. F., Gaultier N. E., . et al. 2017. The microbiomes of blowflies and houseflies as bacterial transmission reservoirs. Sci. Rep. 7: 16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H. J., Cho S. H., Kim M. C., Ahn Y. J., and Kim S. J.. . 2006. Molecular epizootiology of recurrent low pathogenic avian influenza by H9N2 subtype virus in Korea. Avian Pathol. 35: 309–315. [DOI] [PubMed] [Google Scholar]
- Monne I., Ormelli S., Salviato A., De Battisti C., Bettini F., Salomoni A., Drago A., Zecchin B., Capua I., and Cattoli G.. . 2008. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J. Clin. Microbiol. 46: 1769–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazni W. A., Apandi M. Y., Eugene M., Azahari A. H., Shahar M. K., Zainah S., Vthylingam I., and Lee H. L.. . 2013. The potential of house fly, Musca Domestica (L.) in the mechanical transmission of influenza A subtype H1N1 virus under laboratory conditions. J. Gen. Mol. Virol. 5: 22–28. [Google Scholar]
- Nielsen A. A., Skovgård H., Stockmarr A., Handberg K. J., and Jørgensen P. H.. . 2011. Persistence of low-pathogenic avian influenza H5N7 and H7N1 subtypes in house flies (Diptera: Muscidae). J. Med. Entomol. 48: 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nili H., and Asasi K.. . 2002. Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathol. 31: 247–252. [DOI] [PubMed] [Google Scholar]
- Nili H., Mohammadi A., Habibi H., and Firouzi S.. . 2013. Pathogenesis of H9N2 virus in Chukar partridges. Avian Pathol. 42: 230–234. [DOI] [PubMed] [Google Scholar]
- Paul M., Wongnarkpet S., Gasqui P., Poolkhet C., Thongratsakul S., Ducrot C., and Roger F.. . 2011. Risk factors for highly pathogenic avian influenza (HPAI) H5N1 infection in backyard chicken farms, Thailand. Acta Trop. 118: 209–216. [DOI] [PubMed] [Google Scholar]
- Sawabe K., Hoshino K., Isawa H., Sasaki T., Hayashi T., Tsuda Y., Kurahashi H., Tanabayashi K., Hotta A., Saito T., . et al. 2006. Detection and isolation of highly pathogenic H5N1 avian influenza A viruses from blow flies collected in the vicinity of an infected poultry farm in Kyoto, Japan, 2004. Am. J. Trop. Med. Hyg. 75: 327–332. [PubMed] [Google Scholar]
- Sawabe K., Tanabayashi K., Hotta A., Hoshino K., Isawa H., Sasaki T., Yamada A., Kurahashi H., Shudo C., and Kobayashi M.. . 2009. Survival of avian H5N1 influenza A viruses in Calliphora nigribarbis (Diptera: Calliphoridae). J. Med. Entomol. 46: 852–855. [DOI] [PubMed] [Google Scholar]
- Sims L. D., Weaver J., and Swayne D. E.. . 2017. Epidemiology of avian influenza in agricultural and other man-made systems, pp. 302–336. InSwayne D. E. (ed.), Animal influenza. Blackwell Publishing, Ames, IA. [Google Scholar]
- Tan S. W., Yap K. L., and Lee H. L.. . 1997. Mechanical transport of rotavirus by the legs and wings of Musca domestica (Diptera: Muscidae). J. Med. Entomol. 34: 527–531. [DOI] [PubMed] [Google Scholar]
- Tyasasmaya T., Wuryastuty H., and Sievert K.. . 2016. Avian influenza virus H5N1 remained exist in gastrointestinal tracts of house flies 24 hours post-infection. J. Veteriner. 17: 205–210. [Google Scholar]
- Velkers F. C., Blokhuis S. J., Veldhuis Kroeze E. J. B., and Burt S. A.. . 2017. The role of rodents in avian influenza outbreaks in poultry farms: a review. Vet. Q 37: 182–194. [DOI] [PubMed] [Google Scholar]
- Wanaratana S., Panyim S., and Pakpinyo S.. . 2011. The potential of house flies to act as a vector of avian influenza subtype H5N1 under experimental conditions. Med. Vet. Entomol. 25: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanaratana S., Amonsin A., Chaisingh A., Panyim S., Sasipreeyajan J., and Pakpinyo S.. . 2013. Experimental assessment of houseflies as vectors in avian influenza subtype H5N1 transmission in chickens. Avian Dis. 57: 266–272. [DOI] [PubMed] [Google Scholar]
- Watson D. W., Niño E. L., Rochon K., Denning S., Smith L., and Guy J. S.. . 2007. Experimental evaluation of Musca domestica (Diptera: Muscidae) as a vector of Newcastle disease virus. J. Med. Entomol. 44: 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation for Animal Health (OIE) 2015. Chapter 2.3.4. Avian influenza. InManual of diagnostic tests and vaccines for terrestrial animals. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf. [Google Scholar]