We describe the MERS-CoV-neutralizing antibody repertoires from a survivor and characterized 13 ultrapotent mAbs. The mAbs, MERS-GD27 and MERS-GD33, exhibited the strongest neutralizing activities. Both targeted receptor-binding domain via different contacts. The epitopes of MERS-GD27 overlapped with DPP4-binding sites.
Keywords: crystallographic analysis, human monoclonal antibody, MERS-CoV, neutralizing antibody repertoires
Abstract
Background
The Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe respiratory infection with a high (~35%) mortality rate. Neutralizing antibodies targeting the spike of MERS-CoV have been shown to be a therapeutic option for treatment of lethal disease.
Methods
We describe the germline diversity and neutralizing activity of 13 potent human monoclonal antibodies (mAbs) that target the MERS-CoV spike (S) protein. Biological functions were assessed by live MERS-CoV, pseudotype particle and its variants, and structural basis was also determined by crystallographic analysis.
Results
Of the 13 mAbs displaying strong neutralizing activity against MERS-CoV, two with the immunoglobulin heavy-chain variable region (IGHV)1-69-derived heavy chain (named MERS-GD27 and MERS-GD33) showed the most potent neutralizing activity against pseudotyped and live MERS-CoV in vitro. Mutagenesis analysis suggested that MERS-GD27 and MERS-GD33 recognized distinct regions in S glycoproteins, and the combination of 2 mAbs demonstrated a synergistic effect in neutralization against pseudotyped MERS-CoV. The structural basis of MERS-GD27 neutralization and recognition revealed that its epitope almost completely overlapped with the receptor-binding site.
Conclusions
Our data provide new insights into the specific antibody repertoires and the molecular determinants of neutralization during natural MERS-CoV infection in humans. This finding supports additional efforts to design and develop novel therapies to combat MERS-CoV infections in humans.
The Middle East respiratory syndrome coronavirus (MERS-CoV) is a novel, zoonotic pathogen first recognized in September 2012 in Saudi Arabia. The MERS-CoV causes an acute and severe respiratory illness with a high mortality rate in humans [1], which is similar to the severe acute respiratory syndrome CoV that appeared in China in 2002 [2]. Since September 2012, MERS cases have been reported in more than 27 countries with over 2123 laboratory-confirmed cases. A ~35% fatality rate has been reported to the World Health Organization as of December 2017 (http://www.who.int/emergencies/mers-cov/en/). On May 2015, a Korean man, exposed in the Guangdong province, was identified as the first case (GD01) of MERS-CoV infection imported to China [3].
The genome sequence of MERS-CoV has similarities to bat CoVs (BtCoV HKU4 and HKU5) [4, 5]. The CoV spike (S) protein, a class I transmembrane protein, is the major envelope protein on the surface of CoVs. It presents as a trimer and mediates receptor binding, membrane fusion, and virus entry [6]. The receptor-binding domain (RBD) located in the S1 domain of the MERS-CoV S protein is responsible for binding to the cellular receptor identified as dipeptidyl peptidase 4 ([DPP4], CD26) and is critical for the binding and entry of the virus [6, 7]. Therefore, neutralizing antibodies (Abs) capable of blocking such an interaction could be promising preventive and/or therapeutic candidates [8, 9].
Several research groups have been identifying human polyclonal or monoclonal antibodies (mAbs) capable of neutralizing MERS-CoV since 2014 [10–14]. These Abs have been isolated from (1) human Ab libraries, (2) transgenic “humanized” mice, (3) transchromosomic bovines, or (4) naive B cells of an infected individual [2, 10, 15–19]. Several of these mAbs also showed therapeutic efficacy in an animal model (mouse, rhesus monkey, or common marmoset) with MERS-CoV infection [2, 14, 19–21]. Antibodies elicited from acute infections have a relatively low number of mutations coinciding with germline Abs in the early stages of infection, but they are still capable of potent inhibitory activity [22]. However, to our knowledge, human neutralizing mAbs from patients with natural MERS-CoV infections have not provided relevant detailed information due to the limited panning strategies and numbers of mAbs obtained.
In this study, we describe the isolation and characterization of 13 human mAbs from the B cells of an infected patient. Sequencing data revealed germline features of specific fully human mAbs targeting the MERS-CoV S protein. The in vitro neutralization activity for two of the most potent mAbs (MERS-GD27 and MERS-GD33) was selected to be determined. Furthermore, the structural basis for neutralization of MERS-GD27 against MERS-CoV was also explored.
MATERIALS AND METHODS
The Recovered Middle East Respiratory Syndrome Coronavirus Patient
A Korean businessman exposed to the virus in the Guangdong province on May 28, 2015 was identified as the first imported case of MERS infection in China. Diagnosis was established by reverse-transcription polymerase chain reaction detection and complete genome sequence analysis, which was defined as ChinaGD01 [3]. Convalescent sera and peripheral blood mononuclear cells were collected and isolated from the patient on June 20, 2015. The serum Abs displayed high reactivity to MERS-CoV S protein (Sino Biological Inc., Beijing, China) and neutralizing activity as determined by an enzyme-linked immunosorbent assay (ELISA) and MERS-CoV pseudovirus production, neutralization assay [23].
Ethics Statement
The study from which human samples were obtained was approved by the Chinese Center for Disease Control and Prevention Institutional Review Board, and all participants provided written informed consent.
Monoclonal Antibody Production
The production of Abs was based on the specific steps in the Supplementary Materials and the previous methods [24].
Enzyme-Linked Immunosorbent Assay, Neutralization Assays, and Combination Effects of the 13 Neutralizing Monoclonal Antibodies
All experiments were performed as described previously [23].
BioLayer Interferometry Analysis
The interaction of purified Abs with MERS-CoV S protein was monitored using BioLayer Interferometry (BLI) carried out at 25°C in single-cycle mode. The anti-hIgG-Fc was immobilized with the 2-fold serially dilution Abs on the 96-well microplate according to manufacturer’s protocol. The S protein was injected and S-binding responses were measured. The apparent equilibrium dissociation constants (KD) for each S-Ab interaction were calculated using Octet RED96 Software (Pall ForteBio).
Competition Binding Assay
The MERS-GD27 and MERS-GD33 were first labeled with biotin. Afterward, each Ab was 5-fold serially diluted, and the half-maximal effective concentration (EC50) was selected to incubate with the EC50 of MERS-GD27-biotin and MERS-GD33-biotin. The responses of the two mAbs binding to the S protein were compared.
Viral Infections, Isolation, and Titration
All infectious MERS-CoVs were conducted within an approved animal biosafety level-3 laboratory. Experimental designs and methods involving challenge with live MERS-CoV, isolation, and titration were described previously [23].
Crystallization, Data Collection, and Structure Determination
Diffraction data were collected on the BL17U beamline at Shanghai Synchrotron Research Facility [25] and processed with HKL2000 [26]. The structure was determined by molecular replacement with the crystallographic software PHASER [27]. Iterative refinement with the program PHENIX and model building with the program COOT were performed to complete the structure refinement [28, 29]. Structure validation was performed with the program PROCHECK [30], and all structural figures were generated with PYMOL (47).
Statistical Analyses
ImMunoGeneTics information system (IMGT) was used for immunogenetics analyses, and sequence alignments were made with ClustalW. The half maximal inhibitory concentration (IC50) was calculated for each Ab using the dose-response inhibition model in GraphPad Prism 5 software. Data were expressed by means ± standard deviations or means ± standard errors, and normality for all data sets was tested using the Anderson Darling normality test. Synergistic, additive, and antagonistic interplay between Abs for virus neutralization was evaluated by the median effect analysis method with the CompuSyn software (ComboSyn Inc.).
RESULTS
Isolation and Germline Analysis of Neutralizing Antibodies From a Recovered Patient
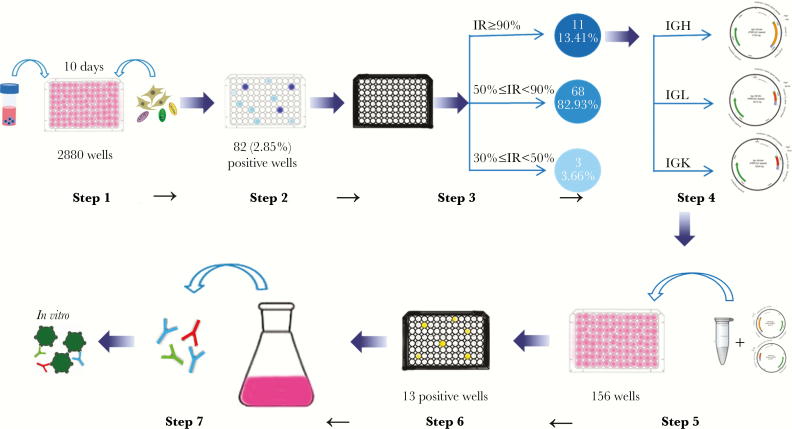
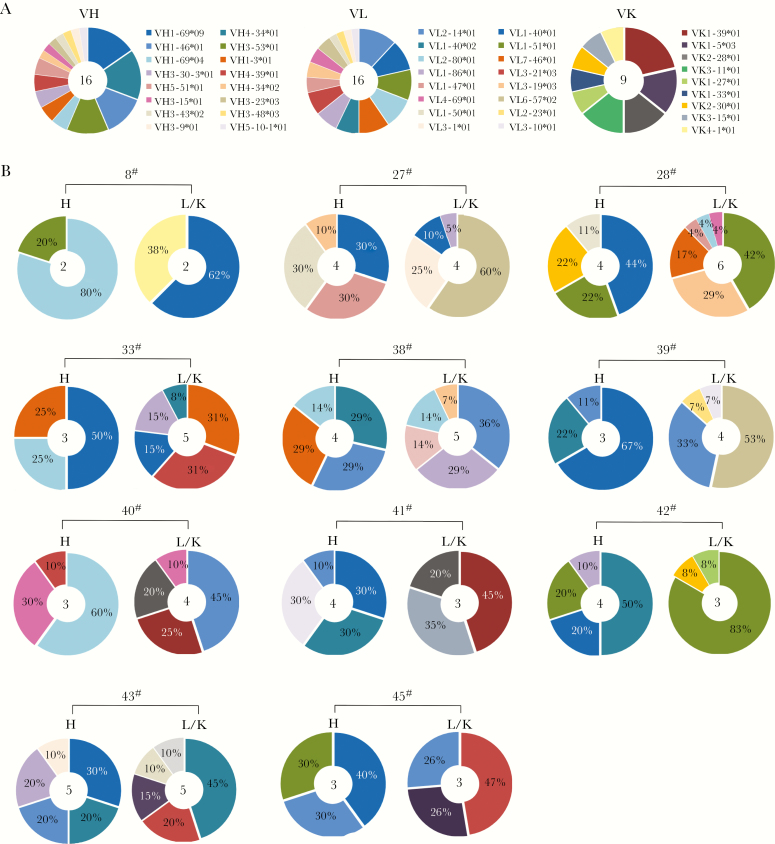
To provide a more comprehensive view for human mAbs against MERS-CoV, we sought to isolate and identify human mAbs targeting the MERS-CoV S protein. We were interested in determining the germline origin genes of the most potent neutralizing Abs in a recovered patient with natural MERS-CoV infection. Workflow for generation of the human mAbs was shown in Figure 1. Eighty-two B-cell cultures of ∼2880 wells were positive (2.85%) against the S protein by ELISA, and 11 of 82 B-cell culture wells targeting the MERS-CoV S protein showed the most potent neutralizing activity (inhibition rate [IR], ≥90%) against the MERS-CoV S pseudovirus (Figure 1). The variable regions encoding the heavy and light chains (VH and VL/VK) were amplified from the above 11 exceptionally potent B-cell cultures and cloned into expression vectors (PBR322 based) for sequencing and immunogenetics analysis. To expand the screening of Ab genes, we sequenced and analyzed 110 VH plasmid sequences and found 39 distinct VH sequences originating from 16 germline genes (Figure 2A, Supplementary Table S1). We sequenced and analyzed 220 VL plasmid sequences and found 56 distinct VL lambda and kappa sequences originating from 16 and 9 different germline genes (Figure 2A and Supplementary Table S1), respectively. We noted that the 4 major families of the immunoglobulin heavy-chain variable region (IGHV) germline were IGHV1-69*04/09, IGHV4-34*01/02, IGHV1-46*01, and IGHV3-53*01, with percentages of 18.6%, 16.28%, 11.63%, and 11.63%, respectively. The major family of the immunoglobulin light-chain/kappa-chain variable germline were IGLV1-40*01/02, IGLV2-14*01, IGKV1-39*01, IGKV1-5*03, IGKV2-28*01, and IGKV3-11*01, with percentages of 16.66%, 11.90%, 21.43%, 14.29%, 14.29%, and 14.29%, respectively. Among neutralizing active mAbs, the VH1-69 gene usage was dominated in the heavy chains, whereas the V1-40 and V1-39 genes of the lambda and kappa chains were used with the highest frequencies in the light chains of the human IgG repertoire (Figure 2A).
Figure 1.
Workflow for generation of human monoclonal antibodies (mAbs) by cloning antibody genes from primary human B cells. Step 1. B cells were isolated from peripheral blood mononuclear cells of a recovered Middle East respiratory syndrome coronavirus (MERS-CoV) patient and then cultured in the 96-well plates in the presence of 3T3 cells, human interleukin (hIL)-2, CpG2006, and hIL-21 for 10 days. Step 2. Culture supernatants were used to detect MERS-CoV spike (MERS-CoV S) for binding activities using enzyme-linked immunosorbent assay. Step 3. Positive wells were used to detect neutralization activities against MERS-CoV S pseudoviruses. Step 4. The variable regions were cloned into expression vectors and analyzed by sequencing technology. Step 5. The selected VH and VL/VK clonal genes were transiently cotransfected into HEK-293T cells from the same well. Step 6. The culture supernatants were detected against MERS-CoV S pseudoviruses for neutralization activities. Step 7. The neutralizing mAbs were purified, and the immunological function was validated. Abbreviations: IGH, immunoglobulin heavy-chain; IGL, immunoglobulin light chain; IGK, immunoglobulin ; IR, inhibition rate.
Figure 2.
The germline characteristics of the anti-Middle East respiratory syndrome coronavirus spike monoclonal antibodies (mAbs). (A) Clonal diversity of B cells and (B) the mAbs of 11 wells with exceptionally potent neutralizing activity. The VH, VL, and VK repertoires were shown as a pie chart, with each slice representing a unique VH, VL, and VK clone. The percentage of each slice for A was shown in Supplementary Table S1. The total number of sequences was indicated by the number at the center of each pie chart.
Various germline genes and frequencies were found for 11 of the individual B-cell wells with robust neutralizing activities against MERS-CoV pseudoviruses (Figure 2B). Remarkably, distinct VH sequences of the neutralizing Ab gene were originated from germline genes VH1-69, and distinct VL sequences of the neutralizing Ab gene were originated from 5 different germline genes (Table 1, Supplementary Table S2). The VH segments showed very low levels of somatic hypermutation (SHM), ranging from 0 to only 3 amino acid substitutions. The lengths of the VH complementarity-determining region 3 (CDR3) for this VH vary from 16 to 18 amino acids. However, the VL genes were more diverse. One VL uses kappa chain, and 6 use lambda chains (Supplementary Table S2). The SHM ranged from 0 to 11 amino acid substitutions.
Table 1.
The MERS-CoV-Neutralizing Human Monoclonal Antibodies Repertoires From B Cells
| mAb ID | Secondary IDa | Strategy | HV Gene | HD Gene | HJ Gene | HCDR 3 | SHM | V-Region Identity % (nt) | LV/K Gene | LJ Gene | LCDR 3 | SHM | V-Region Identity % (nt) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ab8 | H8-7/L8-15 | B cells of patient | 1-69*04 | 3-10*01 | 3*02 | 18 | 0 | 100.00 | 1-40*01 | 2*01 | 11 | 1 | 98.96 |
| Ab27 | H27-8/L27-17 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 2 | 97.92 | 1-40*01 | 2*01 | 11 | 1 | 98.96 |
| Ab28 | H28-6/L28-13 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 3 | 98.26 | 1-51*01 | 2*01/3*01 | 11 | 3 | 97.89 |
| Ab33-1 | H33-1/L33-7 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 0 | 99.65 | 1-40*01 | 2*01 | 11 | 1 | 99.86 |
| Ab33-2 | H33-1/L33-12 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 0 | 99.65 | 1-40*02 | 2*01 | 12 | 0 | 97.22 |
| Ab38 | H38-7/L38-20 | B cells of patient | 1-69*04 | 3-10*01 | 3*02 | 18 | 0 | 100.00 | 2-14*01 | 2*01 | 10 | 11 | 90.46 |
| Ab39-1 | H39-9/L39-12 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 1 | 98.61 | 1-40*02 | 2*01 | 12 | 0 | 97.22 |
| Ab39-2 | H39-9/L39-13 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 1 | 98.61 | 1-50*01 | 1*01 | 11 | 4 | 93.75 |
| Ab40 | H40-2/K40-2 | B cells of patient | 1-69*04 | 4-17*01 | 6*02 | 16 | 0 | 99.31 | 2-28*01 | 4*01 | 9 | 0 | 99.66 |
| Ab41 | H41-5/K41-6 | B cells of patient | 1-69*09 | 4-17*01 | 6*02 | 16 | 0 | 99.65 | 2-28*01 | 4*01 | 9 | 0 | 99.87 |
| Ab42 | H42-6/L42-8 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 3 | 98.26 | 1-51*01 | 2*01/3*01 | 11 | 3 | 97.54 |
| Ab43 | H43-9/L43-2 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 1 | 98.61 | 1-40*02 | 2*01 | 12 | 0 | 97.22 |
| Ab45 | H45-8/L45-13 | B cells of patient | 1-69*09 | 3-10*01 | 3*02 | 18 | 0 | 99.65 | 2-14*01 | 2*01 | 10 | 11 | 90.52 |
| m336 | 922664705-10 | Phage display library | 1-69*06 | 2-2*03 | 3*02 | 18 | 4 | NA | 1-17*01 | 4*01 | 9 | 2 | NA |
| MCA1 | 1200169277-9 | Phage display of patient’s PBMC | 1-69 | 2-2*01 | 3*02 | 17 | 0 | NA | 3-20*01 | 5*01 | 9 | 0 | NA |
| LCA60 | NA | B cells of patient | 3-15*01 | NA | NA | 18 | 17 | NA | 2-23*01 | NA | NA | 12 | NA |
| MERS-4 | NA | scFv-yeast display library | 3-30-3*01 | 6-19*01 | 4*02 | 7 | NA | 94.5 | 1-47*01 | 17*01 | 13 | NA | 98.0 |
| MERS-27 | NA | scFv-yeast display library | 3-11*03 | 3-3*01 | 1*01 | 16 | NA | 93.9 | 1D-33*01 | 5*01 | 10 | NA | 93.0 |
| Seven nAbs | NA | Phage display library | 1-69*06/09, 3-30*03, 1-3*01 | 2-2*01, 3-10*01, 3-10*02,4-17*01, 4-23*01 | 3*02, 4*02 | 11, 13, 14, 16, 18, 19 | 0–10 | NA | 3-1*01, 1-47*01, 3-15*01, 4-1*01, 3-20*01, 10-45*01 | 1*01, 2*01, 3*01, 3*02, 4*01 | 9, 11 | 2–9 | NA |
| 4C2b | 939186871/4 | Immunized mice | 5-6-4*01 | NA | 2*01 | 9 | NA | NA | 10–96*01 | 1*01 | 9 | NA | NA |
Abbreviations: CoV, coronavirus; ID, identification; mAb, monoclonal antibody; MERS, Middle East respiratory syndrome; NA, not available; nAbs, neutralizing antibodies; nt, nucleotide; PBMC, peripheral blood mononuclear cells; SHM, somatic hypermutation.
aThe origin of light and heavy chains.
bHumanized mAb.
Functional Characterization of Antibodies With Potent Neutralizing Activity Against Middle East Respiratory Syndrome Coronavirus In Vitro
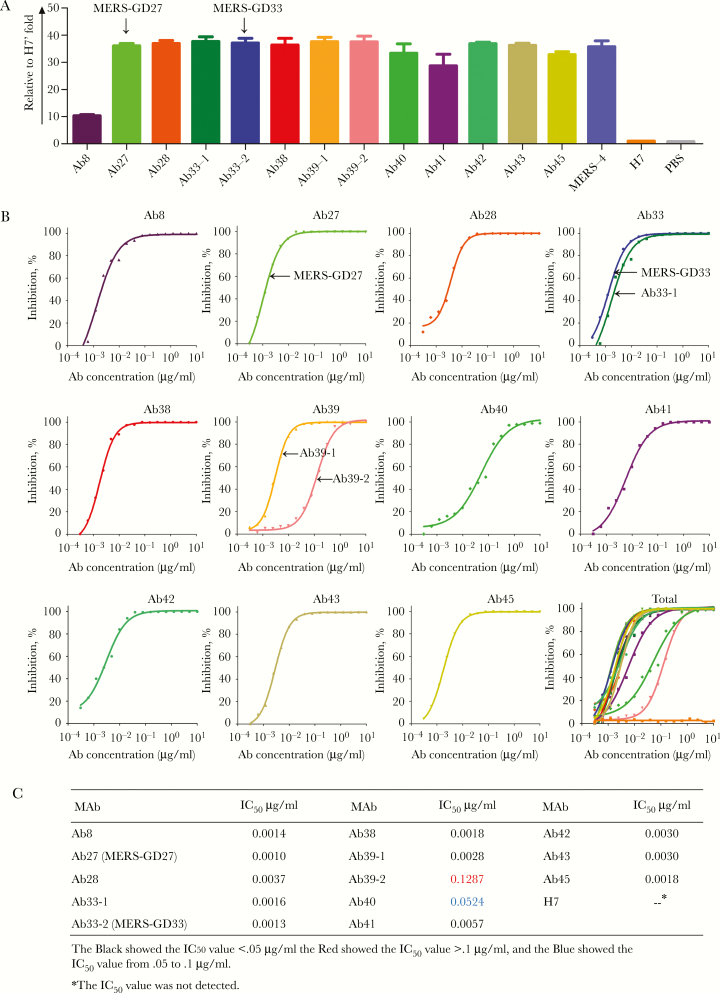
To characterize the function of human mAbs targeting the MERS-CoV S protein, we produced 13 mAbs from 11 cell cultures in large quantities from different combinations of VH and VL/VK genes in the same well by a double gene expression vector transiently transfected into human embryonic kidney (HEK) 293FS cells. All of the Abs target the S protein from the endoplasmic reticulum membrane protein complex (EMC) strain, as determined by ELISA, whereas the H7 (a mAb against hemagglutination of the influenza virus) was used as an irrelevant Ab control and phosphate-buffered saline was used as a blank control (Figure 3A). Among the 13 mAbs, 11 (Ab27, Ab28, Ab33-1, Ab33-2, Ab38, Ab39-1, Ab39-2, Ab40, Ab42, Ab43, and Ab45) displayed strong binding to the S protein (>30-fold of H7), 1 mAb (Ab41) ranged from 15- to 30-fold of H7, and 1 mAb (Ab8) showed weak binding (<15-fold of H7) (Figure 3A).
Figure 3.
Binding activity and neutralizing activity of 13 monoclonal antibodies (mAbs). (A) Binding characterization determined from enzyme-linked immunosorbent assay. Middle East respiratory syndrome coronavirus spike (MERS-CoV S) protein was coated on the 96-well plate 24 hours before the binding test. The mAbs and goat antihuman immunoglobulin (Ig)G Ab/horseradish peroxidase (HRP) antibodies were added sequentially. The absorbance at 450 nm was recorded, and the data presented here was normalized to the value of an irrelevant antibody (H7). The bar chart was depicted by GraphPad Prism 5 software. Data are depicted as the means ± standard deviations from 3 repeats. (B) Neutralization of 13 mAbs against MERS-CoV pseudovirus. Pseudotyped virus was incubated with mAbs before infection of DPP4-expressing Huh-7.5 cells. Luciferase activities were measured, and percentage of neutralization was calculated for 2-fold serial dilutions of each antibody in concentrations from 100 μg/mL to 3 ng/mL. (C) Summary of the half maximal inhibitory concentration (IC50) values of the 13 mAbs.
The neutralizing activity of these mAbs was then tested on Huh7.5 cells against MERS-CoV pseudovirus. Various levels of neutralizing activity were detected for individual mAbs (Figure 3B and C). The neutralizing activity (indicated as IC50 value) of 13 mAbs developed in this study was summarized in Figure 3C. Eleven of 13 mAbs displayed strong neutralizing activity against the MERS-CoV pseudovirus (IC50 value, <0.05 μg/mL), whereas one (Ab39-2) showed a weak neutralizing activity against the MERS-CoV pseudovirus (IC50 value, >0.1 μg/mL). The Abs Ab27 and Ab33-2 (named MERS-GD27 and MERS-GD33, respectively) demonstrated the most potent neutralizing activities, with IC50 values of 0.0010 and 0.0013 μg/mL, respectively, against the MERS-CoV pseudovirus. Some mAbs shared the same VH genes, but their neutralizing activity was rather different. For example, Ab39-1 and Ab39-2 were derived from the same well, and the VH germline gene (H39-9) showed various levels of neutralizing activity when paired with different VLs.
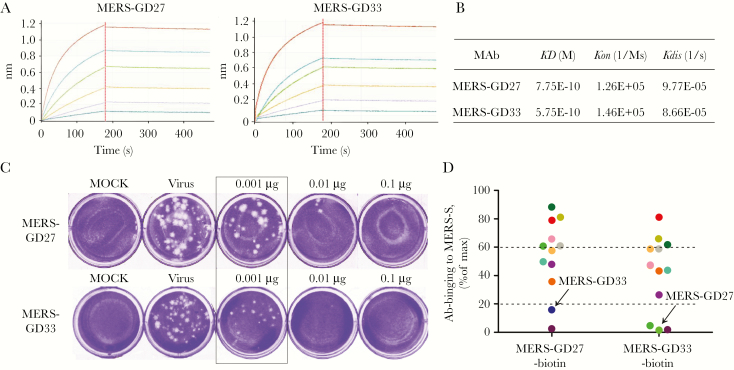
MERS-GD27 and MERS-GD33 Showed the Most Potent Neutralizing Activity Against the Live Middle East Respiratory Syndrome Coronavirus
MERS-GD27- and MERS-GD33-like Abs were naturally present in the patient’s serum (Supplementary Figure S1). We characterized the binding activity of MERS-GD27 and MERS-GD33 by BLI (Figure 4A), and the neutralizing activities against live MERS-CoV were also studied (Figure 4C). MERS-GD27 and MERS-GD33 showed subnanomolar affinity for the MERS-CoV S protein (KD equivalent to 0.775 and 0.575 nM, respectively), which was consistent with MERS-4 (KD equivalent to 0.978 nM) [15]. When neutralizing activity was tested by plaque reduction neutralization test using the live MERS-CoV stock (hCoV-EMC), MERS-GD27 and MERS-GD33 demonstrated potent inhibitory activity against MERS-CoV infection with an IC50 of 0.001 μg/mL of MERS-GD27 and MERS-GD33, respectively (Figure 4C).
Figure 4.
The functional verification of Middle East respiratory syndrome (MERS)-GD27 and MERS-GD33 in vitro. (A) Specific interaction between MERS-CoV S protein and MERS-GD27 and MERS-GD33 characterized by biolayer interferometry. The monoclonal antibodies (mAbs) were captured on the 96-well microplate immobilized with anti-hIgG-Fc and tested for binding with gradient concentrations of MERS-coronavirus spike (CoV S) protein. (B) The apparent dissociation constants were calculated and summarized. (C) Neutralizing activities of MERS-GD27 and MERS-GD33 against live MERS-CoV and against plaque formation of Vero-E6 cells by plaque reduction neutralization test. Three different concentrations of Abs (0.001, 0.01, and 0.1 μg/well) were incubated with 30 plaque-forming units/well live MERS-CoV. Cells were stained with crystal violet at the end of treatment, and the plaques were determined. The inhibitory activity is over 50% in the box. (D) Competition studies among MERS-GD27 and MERS-GD33 with other mAbs. The 96-well plate was first coated with MERS-CoV S 24 hours before the competitive binding test. The mixture of Ab-biotins and other antibodies was incubated in the concentration for 50% of maximal effect. Phosphate-buffered saline was used as a blank control. According to the different competitive binding, 3 groups were created (see Supplementary Table S3).
To determine whether these mAbs recognize different epitopes, we labeled two Abs (MERS-GD27 and MERS-GD33) with biotin, which had no influence on the affinity of the Abs (Supplementary Figure S2), and tested the epitopes between mAb biotin and the other Abs by competitive ELISA (Figure 4D and Supplementary Table S3). Thirteen mAbs could be divided into different groups based on their binding activities (high, medium, and low) of competing ability with MERS-GD27 or MERS-GD33. We found that MERS-GD27 and MERS-GD33 did not bind to the same epitopes of S protein and therefore had a low level of competing activity.
Epitope Mapping and Cooperativity of MERS-GD27 and MERS-GD33
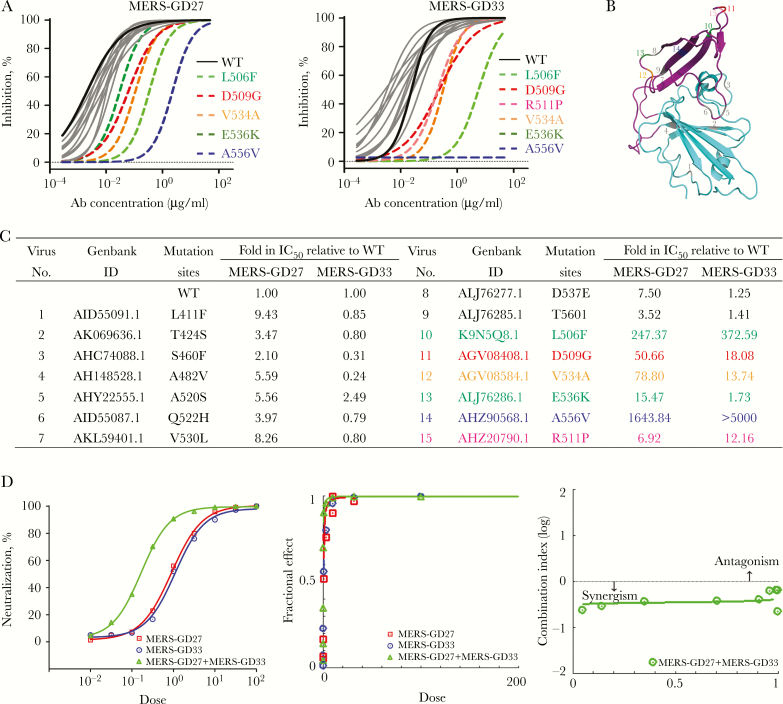
To test the epitopes and the range of neutralizing activity of MERS-GD27 and MERS-GD33 against MERS-CoV, we generated a total of 15 mutant MERS-CoV S glycoproteins with single- or multiple-residue substitutions, which represent most known variant residues at the interface between RBD and DPP4 (Figure 5A). Pseudotyped viruses bearing 10 of 15 mutant MERS-CoV S glycoproteins could be neutralized by either MERS-GD27 or MERS-GD33. MERS-GD27 showed effective neutralizing activity against MERS-CoV, as indicated by the robust inhibition on infection by all but 5 pseudotyped viruses bearing the mutant S glycoprotein (Figure 5A), and 5 mutations (L506F, D509G, V534A, E536K, and A556V) resulted in substantial increases in resistance against neutralization of MERS-GD27. MERS-GD33 differed from MERS-GD27 based on the neutralizing activity against the MERS-CoV pseudoviruses (Figure 5A and C). MERS-GD33 could effectively neutralize the virus with the E536K mutation but could not neutralize the virus with the R511P mutation.
Figure 5.
Epitope mapping by mutagenesis of pseudotyped Middle East respiratory syndrome coronavirus (MERS-CoV) and combination effects in neutralizing pseudotyped MERS-CoV for MERS-GD27 and MERS-GD33. (A) Neutralizing analysis of MERS-GD27 and MERS-GD33 against MERS-CoV wild-type (WT) and its variant mutants; site-directed mutagenesis was introduced into the WT receptor-binding domain (RBD) sequence to create 15 mutant RBDs of other strains. Discrepant residues significantly reducing the neutralizing activities were indicated by colored and dashed lines, respectively. (B) The spatial relationship of the critical residues. Six highlighted positions and 9 gray positions on the crystal structure of RBD. (C) Summary of inhibition on infection by MERS-GD27 and MERS-GD33 against all pseudotyped viruses bearing the mutant S glycoprotein relative to WT. (D, left) Percentage of neutralization was calculated for serial 3-fold dilutions of each antibody alone and in combination at constant ratios in a range of concentrations from 81 times to 1/81 of half maximal inhibitory concentrations (IC50s). On the x-axis, a dose of 1 was at the IC50 concentration. (Middle) Fractional effect (FA) plots generated by the CompuSyn program. (Right) Median effect plot of calculated combination index (CI) values (logarithmic) versus FA values, in which a log CI of <0 is synergism and a log CI of >0 is antagonism.
Because MERS-GD27 and MERS-GD33 bound to different epitopes of S protein (Figure 4D) and showed different epitope mapping on mutant MERS-CoV S glycoproteins (Figure 5A), we went further to determine whether the combination of the two mAbs in virus neutralization was synergistic, additive, or antagonistic. As shown in Figure 5D, a constant ratio between MERS-GD27 and MERS-GD33 was set by their respective IC50 concentrations, as determined above in Figure 3B and C. Dose-dependent neutralization activity for MERS-GD27, MERS-GD33, or the combination was then evaluated by serial 3-fold dilutions in concentrations from 81 times of the IC50 to 1/81 of the IC50. Figure 5D showed the percentage of neutralization (Figure 5D, left), fractional effect (Figure 5D, middle), and the combination index (CI) (Figure 5D, right) using the CompuSyn program. Percentage of neutralization for combination was approximately 0.499-fold and 6.05-fold reduction in IC50 compared with that for MERS-GD27 or MERS-GD33 alone. Furthermore, the CI values of combination at fractional effect values of effective dose 50%, 75%, 90%, and 95% (ED50, ED75, ED90, and ED95) were 0.36, 0.37, 0.39 and 0.40, respectively. Because a CI value of 1 indicates an additive effect, <1 indicates synergism, and >1 indicates antagonism, the combination was clearly in strong synergism (0.36 to 0.40) at ED50, ED75, ED90, and ED95 [31, 32]. Figure 5D, right, showed a logarithmic CI scale, and the combination demonstrated strong synergistic effect at lower and higher Ab concentration.
Structural Basis for Neutralization by MERS-GD27
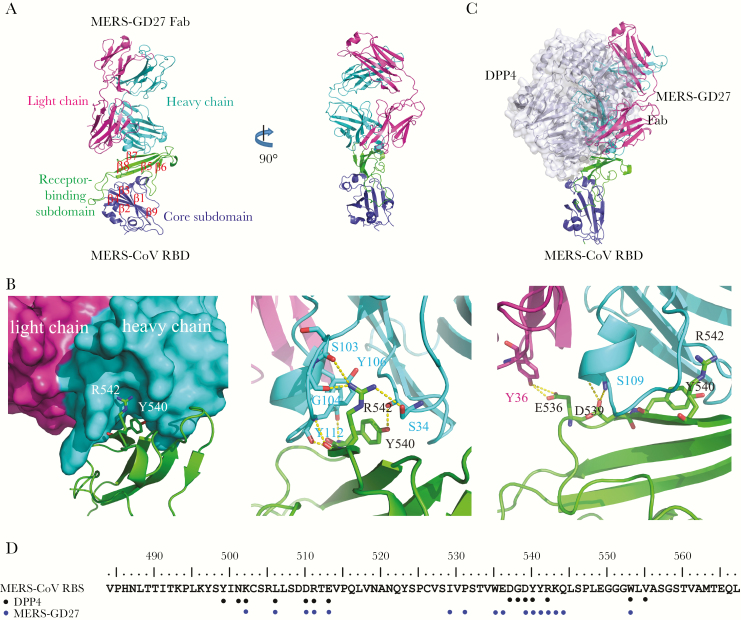
To elucidate the molecular mechanism of MERS-CoV neutralization by MERS-GD27, we reconstituted the complex of the RBD of MERS-CoV S glycoprotein with the Fab fragment of MERS-GD27 and determined its crystal structure at a resolution of 3.0 Å (Figure 6A). As previously defined, the MERS-CoV RBD has two subdomains: the core subdomain comprising a 5-stranded antiparallel sheet (β1, β2, β3, β4, and β9) and the receptor-binding subdomain comprising a 4-stranded antiparallel sheet (β5, β6, β7, and β8) located between the β4 and β7 of the core subdomain. The MERS-GD27 binds the receptor-binding subdomain (Figure 6A), using a total of 20 residues from the 3 heavy-chain complementarity determining region (HCDR) loops and light-chain complementarity determining region (LCDR)1 to interact with 16 residues from the β5 strand, β5-β6 loop, β6-β7 loop, β7 strand, and β7-β8 loop of the receptor-binding subdomain (Supplementary Table S4). The buried surface contributed by the heavy chain at the interface was approximately 770 Å2, which was much larger than the approximately 100 Å2 buried surface by the light chain. The core at the binding interface was formed by the RBD β7 strand and the 3 HCDR loops (Figure 6A), in which RBD Tyr540 interacted with Ab Ser34 and Tyr112, and RBD Arg542 interacts with Ab Ser34, Ser103, Gly104, and Tyr106. Around the binding core, RBD Glu536 and Asp539 formed hydrogen-bonding interactions with Ab light chain Tyr36 and heavy chain Ser109, respectively (Figure 6B).
Figure 6.
The overall structure of Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain (RBD) in complex with neutralizing antibody MERS-GD27 and the binding interface. (A) A ribbon diagram of the complex in which the RBD core subdomain, RBD receptor binding subdomain, MERS-GD27 heavy chain, and MERS-GD27 light chain are colored blue, green, cyan, and purple, respectively. (B) At the binding interface, the β7 strand of RBD interacts with the MERS-GD27 heavy chain. (C) The structural superimpositions of RBD/MERS-GD27 and RBD/DPP4 complexes. (D) The binding sites of MERS-GD27 and DPP4 on receptor binding subdomain.
The superimposition of the MERS-GD27/RBD and DPP4/RBD complex structures showed that the MERS-GD27 would directly compete with the binding of the receptor DPP4 (Figure 6C and D). By comparing the footprints of the MERS-GD27 and DPP4 on the RBD, we also found that most RBD residues recognized by the MERS-GD27, such as Trp535, Glu536, Leu506, Asp510, Glu513, Ser539, Try540, and Trp553, had been previously shown to be critical for DPP4 binding. Hence, MERS-GD27 bound to an epitope that likely would block viral attachment and entry.
DISCUSSION
In this study, we aimed to comprehensively profile the human B-cell response to MERS-CoV infection. We used recombinant S protein to screen B cells and then cloned an extensive panel of anti-S mAbs from the peripheral B cells of a recovered MERS-CoV patient who was infected during the 2015 Korea outbreak. We successfully demonstrated the isolation and verification of 13 specific human mAbs from an individual who was naturally infected with MERS-CoV. We found that antiviral Abs targeting the S protein of MERS-CoV used diversity germline V genes as the origins (Figure 2A, Supplementary Table S1). The 13 mAbs demonstrated distinct binding activity and neutralizing activity. Of the 11 mAbs displaying strong neutralizing activity against MERS-CoV, 2 of the mAbs with an IGHV1-69-encoded heavy chain (MERS-GD27 and MERS-GD33) that bound different epitopes on the S protein showed the most potency and largest range of neutralizing activity against the MERS-CoV in vitro. Furthermore, we identified the structural basic of MERS-GD27 neutralization and recognition by crystallographic analysis, which revealed that its epitope almost completely overlapped with the receptor-binding site.
Compared with the previous reports of human mAbs targeting the MERS-CoV S protein (Table 1), our results showed a high diversity in the germline origin genes of mAbs in a recovered patient with a natural MERS-CoV infection; these covered all the VH germline genes except for two mAbs (MERS-27 was from the scFv-yeast display library and 4C2 was from immune mice). We also found that several VH germline genes were never reported in previous human mAbs against MERS-CoV. It is notable that various genetically distinct germline-encoded neutralizing mAbs existed in the MERS-CoV-recovered patient’s immune repertoires. Another noticeable feature of our MERS-CoV natural infection-derived immune repertoires was the high frequency of IGHV1-69 allele usage, which suggested that this germline gene was a common precursor of MERS-CoV Abs and might recognize similar epitopes. The IGHV1-69 gene was also preferentially used by other antiviral Abs, including Abs against human immunodeficiency virus (HIV)-1 [33], influenza virus [34], and hepatitis C virus [35]. Accordingly, the VH genes of 13 mAbs originated from the VH1-69*04/09 with low SHM, suggesting that the clones arose from naive B cells, and each of them paired with the lambda (VL1-40*01/02, VL1-50*01, VL1-51*01, and VL2-14*01) and kappa (VK2-28*01) genes, respectively (Supplementary Table S2). The germline nature of these Abs implied that Abs could be elicited relatively fast because of the low number of SHM required for affinity maturation. This was in contrast to some Abs against HIV-1 and likely other chronic infections that could be highly divergent from their putative germline predecessors, and they may require extensive and complex affinity maturation pathways to obtain breadth and potency [36, 37].
Coronaviruses could mutate, especially during cross-species transmission, and were important for virus adaptation to new host receptors, which was similar to the other ribonucleic acid viruses [38, 39]. Natural variation in F506 had been reported for England-1 MERS-CoV, which was isolated from the second MERS patient from Qatar [40]. Given viral infection and neutralization escape, we termed “divergent combination immunotherapy” [41] and suggested investigating the cooperation of the more potent neutralizing mAbs with the cross-neutralized protective mAbs. Our results indicated that 13 mAbs present diverse binding phenotypes of competing ability with MERS-GD27 or MERS-GD33, which suggests divergent epitopes. Mutagenesis analysis suggested that MERS-GD27 and MERS-GD33 recognized distinct regions in S glycoproteins, but more work would be required for the fine mapping of epitopes in the future. The epitope mapping of MERS-GD27 and MERS-GD33 could also be helpful for identifying residues critical for neutralization and for investigating virus evolution under immune pressure, which was a force selecting for virus mutation. In addition, synergistic effect was also found when MERS-GD27 and MERS-GD33 were used in combination for neutralization against pseudotyped MERS-CoV, providing a theoretical basis for the combined use in animal and clinical testing in the future.
All published human MERS-CoV-neutralizing Abs targeted the DPP4-binding site on the RBD in spite of identifying them from different Ab libraries and panning strategies, which demonstrated that the RBD was dominant in the selection systems. Previously determined complex structures included the RBD bound by Abs m336, MCA1, MERS-27, 4C2, and D12, respectively. Structural comparisons of them with the MERS-GD27/RBD complex structure showed 2 major orientations of the Ab when binding the RBD (Supplementary Figure S3). Antibodies m336, MCA1, and MERS-GD27 adopted the first orientation by having a large amount of steric clashes with the DPP4 receptor in the structural superimposition (Supplementary Figure S3A–C), whereas the other Abs MERS-27, 4C2, and D12 tilted from the first orientation and had much less steric clashes with the DPP4 receptor (Supplementary Figure S3D–F). MERS-GD27 and m336 had the largest overlap in their epitopes with DPP4 (Supplementary Figure S3C), and the pseudovirus IC50 values for m336 (0.003 μg/mL) [10, 11] and MERS-GD27 (0.001 μg/mL) were lower than those of MERS-27 (63.96 nM) [15], 4C2 (0.71 μg/mL) [18], MCA1 (0.39 μg/mL) [20], and D12 (0.013 μg/mL) [42]. It seemed the extent of overlapping with the DPP4-binding site was critical for the neutralization capability of these Abs.
CONCLUSIONS
Overall, our results on ultrapotent human neutralizing Ab repertoires provided new insight into the MERS-CoV-specific Abs induced by natural infection at the serological and clonal levels. Based on this study, effective and potent neutralizing Abs can be designed and developed for the treatment of human cases of MERS-CoV infection.
Supplementary Data
Refer to Web version on PubMed Central for supplementary material
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Dr. Jincun Zhao (State Key Laboratory of Respiratory Disease, First Affiliated Hospital of Guangzhou Medical University, China) for helpful discussions and Drs. Bart L. Haagmans and Ron A. M. Fouchier (Erasmus MC, Rotterdam, the Netherlands) for providing the Middle East respiratory syndrome coronavirus (isolate hCoV-EMC/2012).
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was funded by the National Key Research and Development Program of China (2016YFD0500300 [to W. T. and X. W.] and 2016YFC1200901 and 2016YFC1200200 [to B. H. and J. Z.]) and the Megaproject for Infectious Disease Research of China (2016ZX10004001-003; to W. T.).
Potential conflicts of interest. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–20. [DOI] [PubMed] [Google Scholar]
- 2. Corti D, Zhao J, Pedotti M, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci U S A 2015; 112:10473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Liu D, Shi W, et al. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in china: phylogenetics and coalescence analysis. MBio 2015; 6:e01280–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 2013; 19:1819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ithete NL, Stoffberg S, Corman VM, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis 2013; 19:1697–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013; 495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013; 500:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crowe JE., Jr Principles of broad and potent antiviral human antibodies: insights for vaccine design. Cell Host Microbe 2017; 22:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Magnani DM, Silveira CT, Ricciardi MJ, et al. Potent plasmablast-derived antibodies elicited by the national institutes of health dengue vaccine. J Virol 2017; 91:pii:e00867-17. doi:10.1128/JVI.00867-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ying T, Du L, Ju TW, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol 2014; 88:7796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ying T, Prabakaran P, Du L, et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun 2015; 6:8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corti D, Passini N, Lanzavecchia A, Zambon M. Rapid generation of a human monoclonal antibody to combat Middle East respiratory syndrome. J Infect Public Health 2016; 9:231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pascal KE, Coleman CM, Mujica AO, et al. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A 2015; 112:8738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson RF, Bagci U, Keith L, et al. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology 2016; 490:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang L, Wang N, Zuo T, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med 2014; 6:234ra59. [DOI] [PubMed] [Google Scholar]
- 16. Du L, Zhao G, Yang Y, et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol 2014; 88:7045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang XC, Agnihothram SS, Jiao Y, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A 2014; 111:E2018–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Wan Y, Liu P, et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res 2015; 25:1237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luke T, Wu H, Zhao J, et al. Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS-CoV in vivo. Sci Transl Med 2016; 8:326ra21. [DOI] [PubMed] [Google Scholar]
- 20. Chen Z, Bao L, Chen C, et al. Human neutralizing monoclonal antibody inhibition of Middle East respiratory syndrome coronavirus replication in the common marmoset. J Infect Dis 2017; 215:1807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Doremalen N, Falzarano D, Ying T, et al. Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antiviral Res 2017; 143:30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. MAbs 2010; 2:347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang W, Wang H, Deng Y, et al. Characterization of anti-MERS-CoV antibodies against various recombinant structural antigens of MERS-CoV in an imported case in China. Emerg Microbes Infect 2016; 5:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith K, Garman L, Wrammert J, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 2009; 4:372–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang QS, Yu F, Huang S, et al. The macromolecular crystallography beamline of SSRF. Nucl Sci Tech 2015; 26:12–7. [Google Scholar]
- 26. Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–26. [DOI] [PubMed] [Google Scholar]
- 27. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–54. [DOI] [PubMed] [Google Scholar]
- 29. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–32. [DOI] [PubMed] [Google Scholar]
- 30. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 1993; 26:283–91. [Google Scholar]
- 31. Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010; 70:440–6. [DOI] [PubMed] [Google Scholar]
- 32. Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984; 22:27–55. [DOI] [PubMed] [Google Scholar]
- 33. Prabakaran P, Zhu Z, Chen W, et al. Origin, diversity, and maturation of human antiviral antibodies analyzed by high-throughput sequencing. Front Microbiol 2012; 3:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 2011; 85:11048–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan CH, Hadlock KG, Foung SK, Levy S. V(H)1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood 2001; 97:1023–6. [DOI] [PubMed] [Google Scholar]
- 36. Yu F, Song H, Wu Y, et al. A potent germline-like human monoclonal antibody targets a pH-sensitive epitope on H7N9 influenza hemagglutinin. Cell Host Microbe 2017; 22:471–83 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou T, Zhu J, Wu X, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 2013; 39:245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cotten M, Watson SJ, Kellam P, et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 2013; 382:1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li W, Zhang C, Sui J, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 2005; 24:1634–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bermingham A, Chand MA, Brown CS, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 2012; 17:20290. [PubMed] [Google Scholar]
- 41. Tang XC, Marasco WA. Human neutralizing antibodies against MERS coronavirus: implications for future immunotherapy. Immunotherapy 2015; 7:591–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang L, Shi W, Joyce MG, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun 2015; 6:7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.