Abstract
Background
Virus infections result in a range of clinical outcomes for the host, from asymptomatic to severe or even lethal disease. Despite global efforts to prevent and treat virus infections to limit morbidity and mortality, the continued emergence and re-emergence of new outbreaks as well as common infections such as influenza persist as a health threat. Challenges to the prevention of severe disease after virus infection include both a paucity of protective vaccines as well as the early identification of individuals with the highest risk that may require supportive treatment.
Methods
We completed a screen of mice from the Collaborative Cross (CC) that we infected with influenza, severe acute respiratory syndrome-coronavirus, and West Nile virus.
Results
The CC mice exhibited a range of disease manifestations upon infections, and we used this natural variation to identify strains with mortality after infection and strains exhibiting no mortality. We then used comprehensive preinfection immunophenotyping to identify global baseline immune correlates of protection from mortality to virus infection.
Conclusions
These data suggest that immune phenotypes might be leveraged to identify humans at highest risk of adverse clinical outcomes upon infection, who may most benefit from intensive clinical interventions, in addition to providing insight for rational vaccine design.
Keywords: Collaborative Cross, immune correlates of mortality, RNA virus infection
We used a screen of genetically diverse mice from the Collaborative Cross infected with RNA viruses in combination with comprehensive preinfection immunophenotyping to identify baseline immune correlates of protection from mortality to virus infection.
Virus infections lead to significant morbidity and mortality worldwide. Although prophylactic vaccinations exist for some infections, we lack vaccines against many pathogens, and the continued emergence of new viral outbreaks drives a constant arms race to limit human disease associated with viruses. One barrier to the design of disease prevention strategies is a lack of knowledge of natural immune correlates of protection from infection and from disease and death. Without this information about the types of immune responses that need to be generated, the design of vaccines or other immune-modulating therapies will remain an empirical, unpredictable process. However, immune correlates are difficult to identify and require either a case-control clinical study with prospectively collected samples or a large, prolonged longitudinal study of immune responses in infected individuals pre- and postinfection. In addition, findings regarding immunity to one type of infection may not prove useful in application against another infection, and so understanding what types of immune responses might be universal in combatting many or all viruses would pave the way for a widespread effort to combat multiple viruses simultaneously.
Study of immunity in small animal models such as mice has provided a wealth of data on immunity to infection. Advantages of these animal models include (1) the ability to easily study immunity at pre- and postinfection timepoints as well as (2) a high degree of control over both genetics, infection route, and environmental factors that result in extremely reproducible datasets. However, the latter is also a disadvantage when a single strain of mice is used for infection studies, because inbred mice lack genetic diversity. Therefore, these inbred strains typically respond reproducibly to infection, thereby preventing the elucidation of immune correlates of protection for a genetically diverse population such as humans. To overcome these barriers, we used the Collaborative Cross (CC). The CC is a population of recombinant inbred mouse strains with high levels of standing genetic variation. The CC strains are derived from 8 founder mouse strains that include 5 classic inbred strains and 3 wild-derived strains [1–4]. We and others have previously shown that the CC can be used to model the diversity in human immune responses and reproduce human disease states not captured in standard inbred mouse models [5–14], and we additionally demonstrated that the CC better represents the large diversity in T-cell immunophenotypes represented in the human population [15]. Thus, the CC affords us with the ability to study immunity and clinical disease to infection in an animal model with diverse genotypes. In this study, we used a large screen of mice from CC F1 crosses (CC-RIX, CC recombinant intercross) infected with 3 distinct viruses—H1N1 influenza (flu), severe acute respiratory syndrome-coronavirus (SARS-CoV), and West Nile virus (WNV)—to reveal novel baseline immune correlates associated with protection from mortality after infection.
METHODS
Mice
The CC-RI mice were obtained from the Systems Genetics Core Facility at the University of North Carolina-Chapel Hill (UNC) [16]. Between 2012 and 2017, CC-F1 mice (CC-RIX) were generated for this research study at UNC in a specific pathogen-free (SPF) facility based on the following principles: (1) each CC-RI strain used in an F1 cross had to have been certified distributable [16]; (2) the UNC Systems Genetics Core Facility was able to provide us sufficient breeding animals for our program to generate N = 100 CC-F1 animals in a target 3-month window; (3) each CC-RIX had to have 1 parent with an H2Bb haplotype (from either the C57BL/6J or 129S1/SvImJ founders), and 1 parent with a haplotype from the other 6 CC founder strains; (4) each CC-RI had to be used at least 1× (preferably 2×) as a Dam, and 1× (preferably 2×) as a Sire in the relevant CC-RIX; (5) finally, we included 2 CC-RIX multiple times across the 5 years of this program to specifically assess and control for batch and seasonal effects. The use of CC-RIX allowed us to explore more lines than the more limited number of available RI strains, and, in addition, CC-RIX lines were bred to ensure that lines were heterozygous at the H-2b locus, having 1 copy of the H-2b haplotype and 1 copy of the other various haplotypes. This design was selected such that we could examine antigen-specific T-cell responses for our parallel studies of immunogenetics of virus infection, while concurrently maintaining genetic variation throughout the rest of the genome.
Six- to eight-week-old F1 hybrid male mice were transferred from UNC to the University of Washington (UW) and housed directly into a biosafety level (BSL)-2+ laboratory within an SPF barrier facility. Male 8- to 10-week-old mice were used for all baseline immune and WNV experiments, with 3–6 mice per experimental group. Concurrently, 6- to 8-week-old female F1 hybrid mice were transferred to either a BSL-3 (SARS-CoV) or a BSL-2 cubicle facility (influenza) at UNC, where mice were allowed to acclimate for 2 weeks before infection (at 8–10 weeks of age). All animal experiments were approved by the UW or UNC Institutional Animal Care and Use Committee. The Office of Laboratory Animal Welfare of National Institutes of Health approved the UNC (no. A3410-01) and the UW (no. A3464-01), and this study was carried out in strict compliance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Virus and Infection
West Nile virus TX-2002-HC (WN-TX) was propagated as previously described [17]. Mice were subcutaneously (s.c.) inoculated in the rear footpad with 100 plaque-forming units (PFU) WN-TX. Mice were monitored daily for morbidity (percentage of initial weight loss) and clinical disease scores [18]. Mouse-adapted SARS-CoV MA15 was propagated and tittered on Vero cells as previously described [11, 19]. Mice were intranasally infected with 105 PFU of SARS-CoV MA15 and measured daily for weight loss. Mice exhibiting extreme weight loss or signs of clinical disease were observed 3 times a day and euthanized if necessary based on humane endpoints. Influenza virus A/Ca/04/09 (H1N1) was generated from a reverse genetics system in MDCK cells [20]. Mice were inoculated intranasally with 5000 PFU of this virus in 50 μL phosphate-buffered saline. Mice were monitored daily for morbidity and mortality.
Each virus inoculum dose was selected based on doses that caused a range of susceptibility phenotypes in the 8 founder strains. Previous studies for all viruses were done on a C57BL/6 background, so this dose was then tested in the founder lines to ensure a range of susceptibility, mortality, and immune responses. For example, 30% of C57BL/6 mice succumb to 100 PFU WNV delivered s.c., with some other founder lines having greater than 50% mortality and others no mortality. We aimed to maximize phenotypic diversity while still maintaining sufficient survival such that we could assess immune phenotypes at various times postinfection.
Flow Cytometry
Spleens were prepared for flow cytometry staining as previously described [5, 6, 15, 18]. All antibodies were tested using cells from the 8 CC founder strains to confirm that antibody clones were compatible with the CC before being used for testing.
Statistical Analysis
When comparing groups, Mann-Whitney tests were conducted, with P < .05 considered significant. Error bars are ±standard error of the mean.
RESULTS
Altered Baseline T-Cell Ratio and Phenotypes Associated With Protection From Mortality After Virus Infections
Although many studies have sought to identify immune correlates of protection from single virus infections, we used multiple infection models to identify globally conserved immune correlates of protection from mortality after virus infection. Thus, we used infection of mice with 3 different viruses: flu and SARS-CoV, which infect the respiratory tract of the host, as well as WNV, which is a mosquito-borne neurotropic infection. We focused on mortality as an endpoint to indicate severe disease, because the unique clinical manifestations of each infection makes scoring difficult. In this screen, 18–28 mice each from over 100 different CC-RIX lines were infected with each of the 3 viruses, followed by monitoring for survival up to 28 days postinfection. For this analysis, we identified CC-RIX that had no mortality after infection with any of the 3 viruses (“No mortality”) and lines that had some degree of mortality to all 3 (“Mortality in all three”). This resulted in identification of 8 lines with no mortality and 11 lines with mortality to all 3, providing a high and low susceptibility cohort for downstream studies (Table 1). The other ~80 CC-RIX had intermediate mortality phenotypes, or lacked immune cell phenotypic data, and were not included in the subsequent analysis but are listed in Supplementary Table 1.
Table 1.
CC F1 Lines Grouped by Mortality Rates
| Strain | Flu | SARS-CoV | WNV |
|---|---|---|---|
| (CC003 × CC062)F1 | 0 | 0 | 0 |
| (CC041 × CC016)F1 | 0 | 0 | 0 |
| (CC029 × CC027)F1 | 0 | 0 | 0 |
| (CC046 × CC068)F1 | 0 | 0 | 0 |
| (CC012 × CC038)F1 | 0 | 0 | 0 |
| (CC013 × CC041)F1 | 0 | 0 | 0 |
| (CC030 × CC061)F1 | 0 | 0 | 0 |
| (CC038 × CC013)F1 | 0 | 0 | 0 |
| (CC001 × CC055)F1 | 8.7 | 8.0 | 50.0 |
| (CC019 × CC027)F1 | 9.1 | 13.3 | 8.3 |
| (CC040 × CC015)F1 | 58.3 | 3.8 | 33.3 |
| (CC074 × CC062)F1 | 23.8 | 83.3 | 50.0 |
| (CC074 × CC058)F1 | 16.7 | 54.5 | 33.3 |
| (CC015 × CC059)F1 | 31.8 | 4.8 | 50.0 |
| (CC075 × CC035)F1 | 8.7 | 11.5 | 41.7 |
| (CC018 × CC065)F1 | 8.3 | 16.7 | 55.6 |
| (CC035 × CC020)F1 | 7.7 | 5.6 | 66.7 |
| (CC043 × CC033)F1 | 7.4 | 4.0 | 33.3 |
| (CC052 × CC014)F1 | 8.3 | 10.0 | 33.3 |
Abbreviations: CC, Collaborative Cross; flu, H1N1 influenza; SARS-CoV, severe acute respiratory syndrome-coronavirus; WNV, West Nile virus.
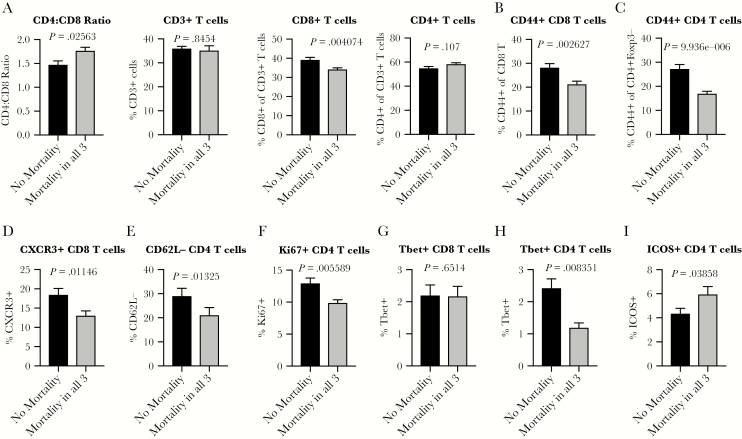
To determine baseline immune signatures that correlate with mortality, we then examined the frequency of different populations and phenotypes of T cells within the spleen at steady state by assessment of a second cohort of age-matched mice from each of these lines (Supplementary Figure S1). We found a statistically lower CD4:CD8 T-cell ratio preinfection in mice from lines that experienced no mortality after any of the infections (Figure 1A). There was no difference in the frequency of CD3+ T cells or the frequency of CD3+ T cells that were CD4+, but there was a significantly lower higher fraction of CD3+ T cells that were CD8+ in mice from lines that experienced no mortality after infection with WNV, SARS-CoV, or flu (Figure 1A). These data suggested that higher baseline CD8 T-cell levels compared to CD4 T cells correlated with reduced risk of mortality. Along with this difference in CD4 and CD8 T-cell ratio, there were significantly elevated frequencies of memory-phenotype-like, CD44+ CD8 and CD4 T cells in mice from lines with no mortality after infection with any of the 3 viruses (Figure 1B and C). In addition to an increased frequency of CD44+ T cells correlating with survival upon infection, there are also increased preinfection frequencies of activated CD4 and CD8 T cells present in mice from lines that survive a subsequent infection. In particular, there are increased frequencies of CXCR3+CD8 T cells (Figure 1D), CD62L−CD4 T cells (Figure 1E), and Ki67+CD4 T cells (Figure 1F). Thus, the presence of an increased fraction of activated T cells that have recently proliferated and/or are poised to respond to signals that direct them out of lymphoid tissue and into distant tissue sites where virus is actively replicating may also confer a survival advantage after virus infections. These data, taken together, suggest that mice resistant to infection-induced mortality maintain a skew towards higher CD8 T-cell levels with higher baseline T-cell activity than animals with lower survival.
Figure 1.
A distinct baseline T-cell ratio and activation signature associates with protection from mortality after virus infections. Age-matched male or female CC-RIX were infected with H1N1 influenza, severe acute respiratory syndrome-coronavirus, or West Nile virus and monitored for survival. Based on these data, they were grouped into “No Mortality” or “Mortality in all 3” categories as shown in Table 1. A second cohort of 3–6 age-matched male mice of the same CC-RIX were euthanized, and splenic cells were analyzed by flow cytometry staining to determine the CD4:CD8 T-cell ratio, the frequency of CD3+ T-cells, the frequency of CD3+ T cells that were CD8+ or CD4+ (A), and the frequency of CD44+CD8 T cells (B), CD44+CD4 T cells (C), CXCR3+CD8 T cells (D), CD62L−CD4 T cells (E), Ki67+CD4 T cells (F), Tbet+CD8 T cells (G), Tbet+CD4 T cells (H), and inducible costimulator (ICOS)+ CD4 T cells (I). Statistical significance was determined by Mann-Whitney test.
Building from these findings, we next assessed the frequency of baseline splenic T cells that expressed the transcription factor Tbet, which indicates a Th1 phenotype linked with secretion of interferon (IFN)γ, known to be a potent antiviral cytokine [21]. Although there is no difference in Tbet expression by CD8 T cells between no mortality versus mortality groups (Figure 1G), there is a statistically significant increase in Tbet+ CD4 T cells in lines with no mortality (Figure 1H). We hypothesize that this may predispose CD4 T cells in mice from these lines to rapidly participate in the antiviral response. In contrast, there is an increase in the frequency of inducible costimulator (ICOS)+CD4+ T cells in lines with mortality (Figure 1I). It has been demonstrated that ICOS plays a role in immunity to virus infections including flu, lymphocytic choriomeningitis virus, and vesicular stomatitis virus, at the level of antibody responses, germinal center formation, and CD4 T-cell responses [22], thereby suggesting that a steady-state reduction in ICOS expression by CD4+ T cells could result in impaired antiviral responses. However, it is also possible that attenuated immunity after infection could result in diminished immune-mediated pathology and could benefit the host in terms of survival. Indeed, although a robust antipathogen immune response can rapidly clear virus, it can also come at the cost of enhanced destruction of infected tissues. A previous study that tested treatment of flu-infected mice with anti-ICOS antibody found that it reduced pulmonary T-cell inflammation [23], thus indicating a potential mechanism whereby reduced ICOS expression could benefit host survival after virus infection through reduction of tissue-specific inflammation.
Increased Frequencies of Regulatory T Cells at Steady-State Is Associated With Protection From Death After Infection
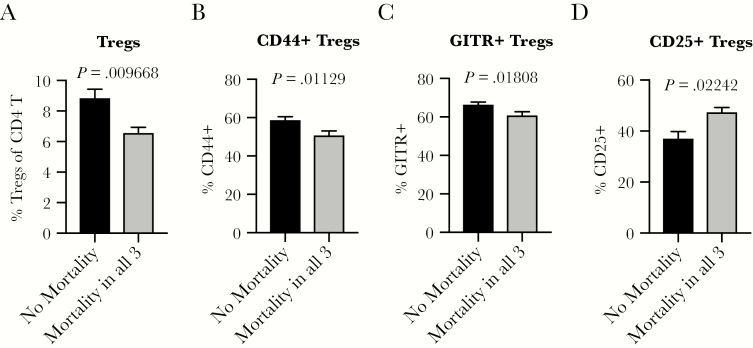
Based on our finding that distinct baseline T-cell activation states may play a role in protection from severe clinical outcomes such as mortality after viral infection (Figure 1), we next examined steady-state regulatory T-cell (Treg) frequency and phenotype in the spleen. A significantly higher baseline frequency of Foxp3+ Tregs (Figure 2A), as well as an increased frequency of Tregs that express activation markers CD44 and glucocorticoid-induced tumor necrosis factor (TNF) receptor (GITR) (Figure 2B and C), is present in mice from lines that are protected from death after infection. This is consistent with the notion that a homeostatic state of immune quiescence mediated at least in part by Tregs could play a role in protection from severe clinical outcomes after virus infection. It is interesting to note that we found a decreased frequency of Tregs expressing CD25 in mice from lines with no mortality after virus infections (Figure 2D and Supplementary Figure S2). CD25 expression affords Tregs the ability to be competitive consumers of interleukin (IL)-2, thereby depriving conventional CD4 T cells of this essential cytokine and thus restricting their expansion [24, 25]. Thus, it is possible that the reduced frequency of CD25+ Tregs indicates less and/or altered suppressive capacity by Tregs, which could be critical for maintaining a balance of robust immunity that can clear virus with limitation of immunopathology-driven disease, morbidity, and mortality.
Figure 2.
An increased frequency of regulatory T cells (Tregs) at steady-state predicts protection from death after subsequent virus infections. Age-matched male or female CC-RIX were infected with H1N1 influenza, severe acute respiratory syndrome-coronavirus, or West Nile virus and monitored for survival. Based on these data, they were grouped into “No Mortality” or “Mortality in all 3” categories as shown in Table 1. A second cohort of 3–6 age-matched male mice of the same CC-RIX were euthanized, and splenic cells were analyzed by flow cytometry staining to determine the frequency of Foxp3+ Tregs (A), CD44+ Tregs (B), glucocorticoid-induced tumor necrosis factor receptor (GITR)+ Tregs (C), and CD25+ Tregs (D). Statistical significance was determined by Mann-Whitney test.
Enhanced T-Cell Production of Proinflammatory Cytokines at Baseline Is Associated With Increased Risk of Death After Virus Infection
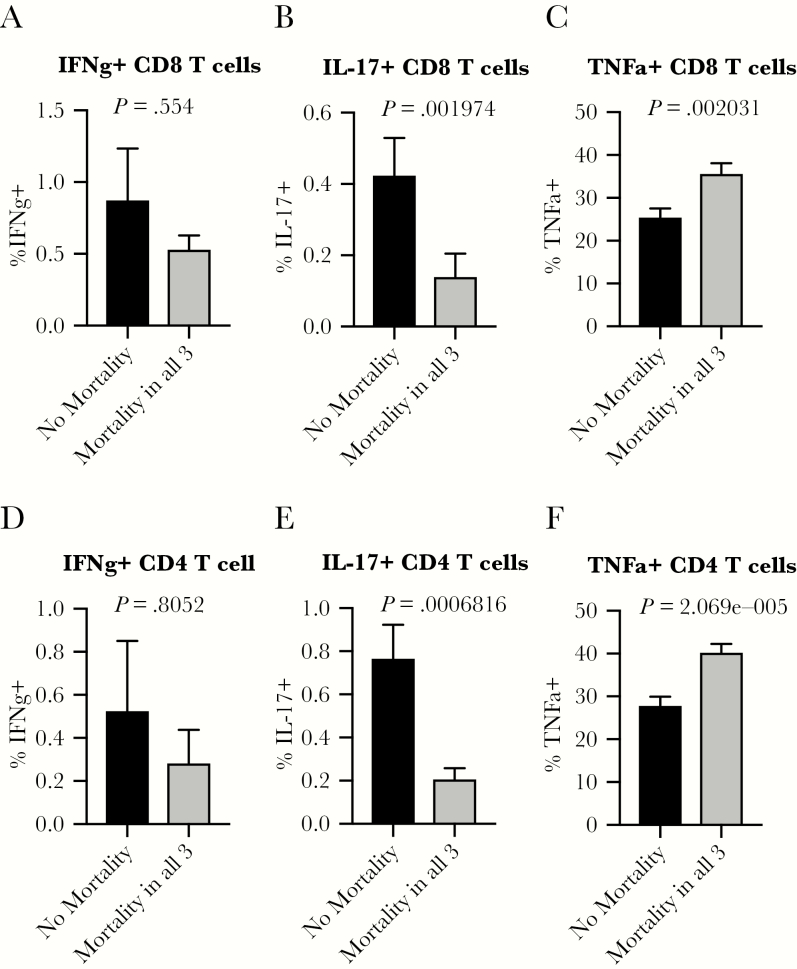
To further investigate our hypothesis of the need for a balanced state of appropriately tuned immune responses to survive viral infection, we examined T-cell capacity to produce cytokines at the steady state. There is no difference in CD8 T-cell or CD4 T-cell production of IFNγ at baseline in mice from lines with or without mortality (Figure 3A and D). Although the frequency of T cells producing IL-17 is very low at baseline, there is a significant, more than 3-fold increase in the fraction of CD8 and CD4 T cells producing IL-17 in mice from lines that were protected from death after viral challenge (Figure 3B and E and Supplementary Table 2). Th17 cells are best known for their roles in immunosurveillance of mucosal barrier surfaces and in many autoimmune and inflammatory diseases. The role of IL-17 during intracellular and viral infections remains unclear, because there is evidence both for protective and pathological roles [26, 27]. Finally, we observed that a reduced frequency of CD8 and CD4 T cells producing TNFα at baseline is associated with protection from mortality upon virus infections (Figure 3C and F). We hypothesize that a propensity to produce this proinflammatory cytokine could play a detrimental role in immune-mediated tissue damage upon infection, consistent with previous findings with SARS-CoV [28], and perhaps this could serve as a biomarker for individuals who may be at higher risk of severe clinical disease upon virus infections.
Figure 3.
Enhanced T cell-mediated proinflammatory cytokine potential at baseline is associated with risk of death after virus infections. Age-matched male or female CC-RIX were infected with H1N1 influenza, severe acute respiratory syndrome-coronavirus, or West Nile virus and monitored for survival. Based on these data, they were grouped into “No Mortality” or “Mortality in all 3” categories as shown in Table 1. A second cohort of 3–6 age-matched male mice of the same CC-RIX were euthanized, and splenic cells were treated with anti-CD3/CD28 for intracellular cytokine staining assessment of interferon (IFN)γ (A and D), interleukin (IL)-17 (B and E), and tumor necrosis factor (TNF)α (C and F) expression by CD8 (A–C) and CD4 (D–F) T cells. Statistical significance was determined by Mann-Whitney test.
DISCUSSION
We used CC mice in combination with a large screen of postinfection mortality after infection with flu, SARS-CoV, and WNV to identify immune correlates of protection from severe clinical outcome, as indicated by death. Through this study, we have identified several baseline immunological features that may contribute to protection from severe disease and clinical outcomes upon virus infections of many kinds. These include a lower CD4:CD8 T-cell ratio, thereby indicating that CD8 T cells may play a key role in mediating this balance between clearing pathogen while sparing collateral damage to host tissues. Moreover, an increased level of basal T-cell activation is associated with protection, and given that CD44+, a marker associated with T-cell memory, is one of the markers found in this signature (Figure 1B and C), it is possible that rapid bystander activity may play a critical role in this protective response. These CD44+ T cells may represent true memory T cells that have resulted from previous microbial exposures, because these mice were housed in specific pathogen-free (SPF) but not germ-free conditions. Alternatively, these cells could also represent unconventional memory phenotype cells that acquired a memory phenotype without exposure to cognate antigen [29–33]. In either case, however, it is possible that these cells could participate in early viral control through bystander-mediated action and thus potentially confer a survival advantage to the host through rapid viral containment before the generation of a virus-specific T-cell response [34]. Indeed, this would be consistent with previous findings that unconventional memory T cells proliferate rapidly in response to antigen [35] and can participate in protective immunity against pathogens such as Listeria monocytogenes and influenza virus [36–38]. In addition, we found that an increased frequency of Tregs with a unique suppressive profile correlated with protection (Figure 2), which supports the notion that balance between active immunity and suppression is likely critical to spare the host from severe disease after infection. We have previously found that Tregs can play a role in protection from human immunodeficiency virus infection through analysis of a case-control cohort [39], and, in addition, there is precedent for Tregs playing a role in protection from immunopathology after infection [40–42]. Furthermore, it has previously been shown that Treg activity is required during viral infections to allow for appropriate generation and migration of immune effector cells to the site of infection [43–45]. Thus, it is possible that this increased Treg abundance and expression of the suppressive marker GITR play a role in coordinating effective antiviral immunity. Alternatively, it is also possible that Tregs could assist in attenuating antiviral immunity upon viral clearance, thereby sparing the host additional collateral damage that could be associated with a prolonged active immune response. The results, taken together, argue that an augmented but targeted T-cell response plays a role in resistance to viral-induced mortality. Finally, a reduced steady-state T-cell capacity to produce the proinflammatory cytokine TNFα is correlated with protection, which is consistent with less risk of collateral damage upon T-cell receptor-mediated stimulation. We hypothesize that the CD44+ memory-phenotype CD8 T cells, found in greater steady-state abundance in CC-RIX that did not suffer from mortality upon infection, do not confer protection via a canonical, antigen-specific “true memory”-mediated mechanism such as cytokine production. Instead, we speculate that these CD44+CD8+ T cells become bystander-activated during inflammation associated with infection, and thus they acquire cytotoxic function that could be mediated by alternative pathways such as NKG2D-mediated target cell killing [34]. Altogether, these observations distinguish a T-cell profile associated with protection from viral-induced mortality.
Due to the nature of this large, collaborative study that used thousands of mice over the course of several years, there are some limitations to discuss. All mice for the project were obtained from the Systems Genetics Core Facility located at UNC. These mice were used for flu and SARS-CoV experiments at UNC or for WNV experiments, including flow cytometry-based immune phenotyping of naive and infected mice, at the UW. To reduce the amount of breeding as well as the production and use of mice, we used female mice for flu and SARS-CoV studies at UNC, and male mice were used for WNV studies at UW. Thus, although only males were used for baseline splenic immunophenotyping, and males (WNV) and females (influenza and SARS-CoV) were used for infection studies that yielded data on mortality, this ensured that any correlates of protection identified in this analysis were not sex-specific. Moreover, we examined immune cells from the spleen by flow cytometry, as a measure of preinfection immune phenotypes, but we did not examine blood or other viral target tissue such as the lung, using this same flow cytometry panel. Thus, we cannot speculate on preinfection lung immunophenotypes associated with viral infection outcomes. In addition, most human data comes from analysis of immune cells within the blood, and although our analysis of immune cells within the spleen can be used to predict what might be protective in humans, it is not the most common tissue type analyzed, and thus a direct blood-to-blood comparison is not possible from our study. In addition, the majority of immune phenotypes screened for represent T-cell phenotypes, and there are likely other innate and humoral immune states that could be associated with mortality, and this warrants further study. Finally, although the results presented herein demonstrate an association between a set of baseline T-cell immunophenotypes and protection from mortality upon infection with ribonucleic acid viruses, we have not tested these predictions through mechanistic study. However, we speculate that it is not 1 single immune phenotype that solely drives protection from mortality, and so we predict that blocking or augmenting a single immune phenotype alone would not significantly alter the degree of mortality upon infection.
CONCLUSIONS
Overall, our results define distinct baseline T-cell correlates associated with survival after viral challenge. This study not only identifies specific basal immune characteristics that are associated with protection from mortality upon virus infections, but it also underscores the need for a protective immune response to be balanced to protect the host not only from uncontrolled virus replication but also from disease associated with robust immunity. Although validation studies in humans are required to confirm these correlates of protection from severe clinical disease after virus infection, it is possible that this knowledge can advance efforts to design vaccines and other infection prevention strategies. Likewise, the T-cell profile can inform methods by which to screen for individuals at increased risk of severe clinical disease upon virus infections. More important, the approach highlights the utility of exploring multiple infections in the context of a genetically diverse populations.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank our collaborators in the Systems Immunogenetics Group for helpful discussions and generation of mice. In particular, we thank Ginger Shaw for generating the RIX mice used in this study.
Author contributions. D. R. M., S. K. M., M. T. F., F. P.-M., M. G., M. T. H., R. S. B., and J. M. L. designed the research studies; J. B. G., J. L. S., V. D. M., L. E. G., A. S., K. S. P., C. R. M., K. M. V., R. G., and M. T. F. conducted experiments and acquired and analyzed data; G. C., S. J., and M. A. M. performed data cleaning and integration; and J. B. G. and J. M. L. wrote the first draft of the manuscript. All authors read the manuscript and contributed editorial suggestions.
Financial support. Funding for this study was provided by National Institutes of Health Grant U19AI100625.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Churchill GA, Airey DC, Allayee H, et al. ; Complex Trait Consortium The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 2004; 36:1133–7. [DOI] [PubMed] [Google Scholar]
- 2. Collaborative Cross Consortium. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 2012; 190:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011; 477:289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome 2007; 18:473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Graham JB, Swarts JL, Wilkins C, et al. A mouse model of chronic West Nile virus disease. PLoS Pathog 2016; 12:e1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graham JB, Thomas S, Swarts J, et al. Genetic diversity in the Collaborative Cross model recapitulates human West Nile virus disease outcomes. MBio 2015; 6:e00493–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graham JB, Swarts JL, Thomas S, et al. Immune correlates of protection from West Nile virus neuroinvasion and disease. J Infect Dis 2019; 19:1162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brinkmeyer-Langford CL, Rech R, Amstalden K, et al. Host genetic background influences diverse neurological responses to viral infection in mice. Sci Rep 2017; 7:12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elbahesh H, Schughart K. Genetically diverse CC-founder mouse strains replicate the human influenza gene expression signature. Sci Rep 2016; 6:26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferris MT, Aylor DL, Bottomly D, et al. Modeling host genetic regulation of influenza pathogenesis in the Collaborative Cross. PLoS Pathog 2013; 9:e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gralinski LE, Ferris MT, Aylor DL, et al. Genome wide identification of SARS-CoV susceptibility loci using the Collaborative Cross. PLoS Genet 2015; 11:e1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kollmus H, Pilzner C, Leist SR, Heise M, Geffers R, Schughart K. Of mice and men: the host response to influenza virus infection. Mamm Genome 2018; 29:446–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leist SR, Baric RS. Giving the genes a shuffle: using natural variation to understand host genetic contributions to viral infections. Trends Genet 2018; 34:777–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rasmussen AL, Okumura A, Ferris MT, et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 2014; 346:987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graham JB, Swarts JL, Mooney M, et al. Extensive homeostatic T cell phenotypic variation within the Collaborative Cross. Cell Rep 2017; 21:2313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Welsh CE, Miller DR, Manly KF, et al. Status and access to the Collaborative Cross population. Mamm Genome 2012; 23:706–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suthar MS, Ma DY, Thomas S, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 2010; 6:e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Graham JB, Swarts JL, Lund JM. A mouse model of West Nile virus infection. Curr Protoc Mouse Biol 2017; 7:221–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gralinski LE, Sheahan TP, Morrison TE, et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. MBio 2018; 9:e01753–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 2009; 460:1021–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol 2013; 13:777–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertram EM, Tafuri A, Shahinian A, et al. Role of ICOS versus CD28 in antiviral immunity. Eur J Immunol 2002; 32:3376–85. [DOI] [PubMed] [Google Scholar]
- 23. Humphreys IR, Edwards L, Snelgrove RJ, Rae AJ, Coyle AJ, Hussell T. A critical role for ICOS co-stimulation in immune containment of pulmonary influenza virus infection. Eur J Immunol 2006; 36:2928–38. [DOI] [PubMed] [Google Scholar]
- 24. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 2012; 30:531–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol 2011; 11:119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das S, Khader S. Yin and yang of interleukin-17 in host immunity to infection. F1000Res 2017; 6:741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai CW, Blase JR, Zhang X, Eickhoff CS, Hoft DF. Th17 cells are more protective than Th1 cells against the intracellular parasite Trypanosoma cruzi. PLoS Pathog 2016; 12:e1005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDermott JE, Mitchell HD, Gralinski LE, et al. The effect of inhibition of PP1 and TNFα signaling on pathogenesis of SARS coronavirus. BMC Syst Biol 2016; 10:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jameson SC. T cell homeostasis: keeping useful T cells alive and live T cells useful. Semin Immunol 2005; 17:231–7. [DOI] [PubMed] [Google Scholar]
- 30. Le Campion A, Bourgeois C, Lambolez F, et al. Naive T cells proliferate strongly in neonatal mice in response to self-peptide/self-MHC complexes. Proc Natl Acad Sci U S A 2002; 99:4538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Min B, McHugh R, Sempowski GD, Mackall C, Foucras G, Paul WE. Neonates support lymphopenia-induced proliferation. Immunity 2003; 18:131–40. [DOI] [PubMed] [Google Scholar]
- 32. Schüler T, Hämmerling GJ, Arnold B. Cutting edge: IL-7-dependent homeostatic proliferation of CD8+ T cells in neonatal mice allows the generation of long-lived natural memory T cells. J Immunol 2004; 172:15–9. [DOI] [PubMed] [Google Scholar]
- 33. Surh CD, Sprent J. Regulation of mature T cell homeostasis. Semin Immunol 2005; 17:183–91. [DOI] [PubMed] [Google Scholar]
- 34. Chu T, Tyznik AJ, Roepke S, et al. Bystander-activated memory CD8 T cells control early pathogen load in an innate-like, NKG2D-dependent manner. Cell Rep 2013; 3:701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haluszczak C, Akue AD, Hamilton SE, et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med 2009; 206:435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee JY, Hamilton SE, Akue AD, Hogquist KA, Jameson SC. Virtual memory CD8 T cells display unique functional properties. Proc Natl Acad Sci U S A 2013; 110:13498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sosinowski T, White JT, Cross EW, et al. CD8α+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J Immunol 2013; 190:1936–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lanzer KG, Cookenham T, Reiley WW, Blackman MA. Virtual memory cells make a major contribution to the response of aged influenza-naïve mice to influenza virus infection. Immun Ageing 2018; 15:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pattacini L, Baeten JM, Thomas KK, et al. ; Partners PrEP Study Team Regulatory T-cell activity but not conventional HIV-specific T-cell responses are associated with protection from HIV-1 infection. J Acquir Immune Defic Syndr 2016; 72:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*). Annu Rev Immunol 2009; 27:551–89. [DOI] [PubMed] [Google Scholar]
- 41. Richert-Spuhler LE, Lund JM. The immune fulcrum: regulatory T cells tip the balance between pro- and anti-inflammatory outcomes upon infection. Prog Mol Biol Transl Sci 2015; 136:217–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smigiel KS, Srivastava S, Stolley JM, Campbell DJ. Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol Rev 2014; 259:40–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lund JM, Hsing L, Pham TT, Rudensky AY. Coordination of early protective immunity to viral infection by regulatory T cells. Science 2008; 320:1220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ruckwardt TJ, Bonaparte KL, Nason MC, Graham BS. Regulatory T cells promote early influx of CD8+ T cells in the lungs of respiratory syncytial virus-infected mice and diminish immunodominance disparities. J Virol 2009; 83:3019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soerens AG, Da Costa A, Lund JM. Regulatory T cells are essential to promote proper CD4 T-cell priming upon mucosal infection. Mucosal Immunol 2016; 9:1395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.