Abstract
Avian influenza caused by avian influenza virus (AIV) has a negative impact on poultry production. Low-pathogenic AIV (LPAIV) is naturally present in wild birds, and the introduction of the virus into domestic poultry is assumed to occur through contact with wild birds and by human activity, including the movement of live and dead poultry, and fomites such as clothing and vehicles. At present, the possible role of insects in the spread of AIV is dubious. The objective of the present work was to investigate the potential transmission of LPAIV by persistence of the virus in the alimentary tract of house flies, Musca domestica L. (Diptera: Muscidae). Flies were fed three virus concentrations of two AIV strains and then incubated at different temperatures for up to 24 h. The persistence of the two virus strains in the flies declined with increasing incubation temperatures and incubation periods. Similarly, increased virus uptake by the flies increased the persistence of virus. Persistence of infective AIV in flies differed significantly between the two virus strains. The laboratory experiments of the present study indicate that the house fly can be a potential carrier of AIV.
Keywords: avian influenza virus, insect carrier, transmission, house fly, Musca domestica
Avian influenza virus (AIV) is an enveloped RNA virus that belongs to the genus Influenzavirus A of the family Orthomyxoviridae. Influenza A viruses are classified into subtypes by the combination of their two surface glycoproteins hemagglutinin and neuraminidase, of which 16 (H1–H16) and nine (N1–N9) antigens, respectively, have been recognized (reviewed in Pantin-Jackwood and Swayne 2009). AIV of H5 and H7 subtypes occurs as low-pathogenic (LPAIV) and high-pathogenic (HPAIV) strains, based on their virulence in chickens. The natural reservoir of avian influenza virus is wild aquatic birds within the orders Anseriformes and Charadriiformes (Stallknecht 2003), and AIV circulates between wild birds and domestic poultry as well as within poultry flocks through complex routes that are not fully understood (Alexander 2008). In domestic poultry, most AIV infections are subclinical or give rise to mild clinical disease. Low-pathogenic isolates of the H5 and H7 subtypes may mutate into highly pathogenic (HP) variants after introduction into fowl, which induce severe, systemic disease with high mortality (Swayne and King 2003, Swayne 2007, Pantin-Jackwood and Swayne 2009).
During outbreaks of HP avian influenza (AI), spread of AIV between poultry flocks is assisted by human activity, but spread by contact with wild birds also may contribute to dissemination (Alexander 2008). Although the role of insects in transmission of AIV has been considered, few studies have been published. AIV was detected in 25 of 72 pools of house flies, Musca domestica L. (Diptera: Muscidae), collected within a 4-mo period from poultry flocks in Pennsylvania and Maryland, infected with H5N2 AIV (Wilson et al. 1986). Also, HPAIV H5N1 has been isolated from two blow fly species, Aldrichina grahami (Aldrich) and Calliphora nigribarbis Snellen van Vollenhoven (Diptera: Calliphoridae), collected in the vicinity of infected poultry at a farm in Kyoto, Japan (Sawabe et al. 2006). The persistence of HPAIV H5N1 in house flies and blow flies as well as Newcastle disease virus (family Paramyxoviridae, genus avulavirus, NDV), turkey coronavirus (family Coronaviridae, genus Coronavirus, TCV), and reticuloendotheliosis virus (family Retroviridae, genus Gammaretrovirus, REV) has been studied in the laboratory (Calibeo-Hayes et al. 2003; Davidson and Braverman 2005; Chakrabarti et al. 2007, 2008; Watson et al. 2007; Sawabe et al. 2009; Barin et al. 2010; Wanaratana et al. 2011). In all studies, flies fed with the viruses became transiently contaminated and the persistence of the virus in crop or alimentary canal declined with prolonged incubation period of the flies. Importantly, a recent study by Wanaratana et al. (2011) demonstrated that H5N1 HPAIV can persist in the viscera of house flies up to 72 h postexposure in the laboratory. Watson et al. (2007) and Calibeo-Hayes et al. (2003) further examined the transmission of NDV (Roakin strain) and TCV, respectively, to susceptible poultry by house flies. Based on these studies, Watson et al. (2007) concluded the house fly to be a poor vector of NDV (Roakin). In contrast, Calibeo-Hayes et al. (2003) successfully infected turkeys by placing them in contact with TCV-exposed flies.
The current study investigated the persistence of LPAIV in the alimentary tract of house flies that were fed various concentrations of subtypes H5N7 and H7N1 viruses and incubated at different temperatures for up to 24 h. Flies were subsequently tested for the persistence of infective virus.
Materials and Methods
Rearing of House Flies.
Adult house flies were obtained from a colony bred for >250 generations at the Department of Integrated Pest Management, Aarhus University, Slagelse, Denmark. Newly emerged flies were kept in cages and supplied sucrose, powdered milk and tap water ad libitum until used in the experiments at 4–6 d of age. Except for one experiment, female flies were used preoviposition.
Virus Strains.
Low-pathogenic (H7N1) A/African Starling/England/983/79 supplied by VLA, Weybridge, United Kingdom, and (H5N7) A/Mallard/Denmark/75.64.650/03 AIV strains were propagated in specific-pathogen-free (SPF) embryonated chicken eggs (ECEs) (Lohmann, Cuxhaven, Germany). Infective allantoic fluid (AF) was stored at −80°C until use. The titers of H5N7 and H7N1 were 107.9 and 108.5 50% of egg lethal dose (ELD50/ml), respectively, when titrated in 9–11-d-old SPF embryos by 10-fold dilutions. One batch of each virus was used throughout the entire experiment. Two-fold virus dilutions were prepared in a sterile 10 mM phosphate-buffered saline (PBS) solution for use in the experiments.
AIV Exposure of House Flies.
In total of 1,195 flies were virus-fed, sham-fed, or negative controls. The two AIV subtypes were used in separate experiments, and three independent experiments were performed with each virus strain. After feeding, the flies were kept for 6, 12, or 24 h at 15, 25, or 35°C in humidified incubators with 40–70% RH and 24-h light. Before experiments, flies were deprived of water and feed overnight, and kept chilled in the dark. Before feeding with the AIV suspensions, the flies were anesthetized with CO2 and placed individually in disposable sterile 1,000-μl pipette tips cut to size for the fly's head and lapping proboscis to protrude. From this position, each fly dined on 1-μl volume of sterile or virus-positive AF diluted in PBS supplied by micropipette. Flies were fed two-fold diluted virus solutions (20, 2−2, or 2−4); hence, each fly was fed H5N7 virus doses equivalent to 104.9 ELD50, 104.3 ELD50, or 103.7 ELD50, or H7N1 virus in doses of 105.5 ELD50, 104.9 ELD50, or 104.3 ELD50. Flies that stopped or declined eating the full volume of virus solution were discarded. Fed flies were transferred to closed cages in groups of five for each combination of virus dilution, temperature, and incubation period. Fed flies were stored at room temperature until all 60 flies included in each experimental group were fed, after which the cages were placed in incubators and the incubation period of 6, 12, or 24 h was started. Importantly, the order with which flies were fed with the different virus dilutions was consistent among all experiments. Flies had unlimited access to whole-fat, pasteurized and homogenized milk during incubation. Sham-fed flies dined on equal volumes of sterile AF and were incubated as described above. Also, 0-h experiments were included, in which groups of five flies were fed 1-μl volume of either sterile AF or the three virus solutions for each virus strain. These flies were subsequently kept at room temperature (24°C) for 10 min without access to milk before termination. Likewise, for each experimental round another five flies were tested for virus as described below without any feeding or incubation.
Virus Isolation From Flies.
All flies were killed by freezing at −20°C for at least 30 min. Subsequently, body fluids, crop and intestinal organs from the abdomen were immediately harvested aseptically as described below. Alternatively, flies were kept at −80°C up to 48 h before harvest. While holding the fly with a sterile forceps in a sterile petri dish, the area surrounding the fly's cloaca was sterilized with a heated scalpel and punctured by a sterile needle. The content of the abdomen was harvested by a micropipette with a sterile disposable tip. The contents from each group of five flies were pooled in 0.5 ml of medium [PBS (8 mM) with penicillin (8,100 U/ml), streptomycin (8,100 μg/ml), gentamicin sulphate (200 μg/ml), nystatin (8,100 U/ml), fetal bovine serum (4.5% vol:vol), and phenol red 2‰ (0.4% vol:vol)]. To detect the presence of infectious virus, undiluted 200-μl aliquots of each pooled sample supernatant were inoculated into two SPF ECEs within 12 h of preparation by following a modified version of the EU Directive 2005/94/EC (http://vla.defra.gov.uk/science/sci_ai_reflab_man.htm). Samples were homogenized before inoculation. The individual groups of flies were scored as virus-positive if any of the SPF ECEs were infected (dead embryo and hemagglutinin-positive allantoic fluid) by primary inoculation or after one blind passage. Similarity between inoculated and isolated virus was confirmed by sequencing of reverse transcription-polymerase chain reaction (RT-PCR) amplicons. For each subtype of AIV, samples representing different combination of fed virus concentration, temperature, and incubation period were examined separately.
Persistence of Virus Dilutions.
After each experiment, the three two-fold diluted aliquots of the AIV stocks were incubated at 15, 25, and 35°C for 12 and 24 h. After incubation, selected virus-dilutions (the 2−4 dilution incubated at 35°C, the 2−2 dilution incubated at 25°C, and the 20 dilution incubated at 15°C) were titrated. Titrations were performed by 10-fold dilutions in SPF ECEs.
Extraction of Viral RNA.
Viral RNA was extracted from AF from selected eggs with virus isolates using the RNeasy mini kit (QIAGEN, Hilden, Germany) as described previously (Slomka et al. 2007). RNA was stored at −18°C.
RT-PCR and Sequencing.
H5 and H7 subtype specific PCRs were performed by following the H5 conventional PCR protocol and H7 cleavage-site real-time PCR protocol described in Slomka et al. (2007) and the European Union protocol (http://vla.defra.gov.uk/science/docs/sci_ai_vi536.pdf), respectively. Both strands of the amplicons were sequenced using the PCR primers at DNA Technology A/S (Risskov, Denmark).
Statistical Analysis.
The statistical analysis was carried out by logistic regression, using the General Linear model (GLM) procedure in SPlus (Insightful Corp. 2002). The dependent variable, AIV-positiveness of samples, was dichotomous and attained values of 0 or 1 only.
The probability p that an observation X describing virus status for an individual pool of five flies takes the value one or 0 (i.e., the fly pool is virus-positive [X = 1] or the fly pool is virus-negative [X = 0]) is modeled as follows, where logit is the logistic transformation logit(p) = log(p/(1 − p)):
logit (ρ) = αS + βT*T + βI*I + βC*C [1]
so that logit(p) is regressed on temperature (T; room temperature [24°C], 15, 25, and 35°C), incubation period postfeeding of the flies (I; 0, 6, 12, and 24 h), and AIV virus concentration (C; H5N7: 106.7, 107.3, and 107.9 ELD50/ml; H7N1: 107.3, 107.9, and 108.5 ELD50/ml); the fed virus concentrations were log10 transformed to stabilize the variance. The constant α was allowed to depend on virus strain (S), and interactions of up to third order between the four dependent variables T, I, C, and S was investigated using a step-down procedure where the interaction terms were added simultaneously to the model 1 and subsequently deleted to reduce this full model. This was done as stepwise reduction of the full model, including interactions, excluding nonsignificant parameters (P > 0.05) from the model, by using likelihood ratio tests for model reduction.
Because the analyzed pools of five flies are independent samples, the probability p for containing a virus-positive fly pool after a given incubation period may be translated as an estimate for the frequency of fly pools to be positive for virus. Hence, with q denoting logit(p), the probability p is found from q as
ρ = loagit(q)1 = exp(q)/(1+(exp(q))) [2]
Results
Overall, only two flies of the total 1,195 flies died during the incubation period. Both of these represented H7N1-fed individuals kept at 35°C for 24 h. The flies' level of activity and foraging at the three incubation temperatures was strongly affected by their poikilothermic nature. As expected, the pools of five flies selected randomly as negative controls before each experimental setup were all negative for virus, confirming the flies be bred under virus-free conditions. Similarly, all groups of flies fed with sterile allantoic fluid were negative for virus (Table 1). The persistence of infectious virus of the H5N7 and H7N1 AIV subtypes in the fly pools are given in Table 1. Correspondence between fed virus subtype and harvested samples from the flies was tested on randomly selected samples by RT-PCR and sequencing of the amplicons; identical similarity was confirmed in all cases. Virus persistence in the three virus dilutions for both virus strains was tested by incubating aliquots at 15, 25, and 35°C for 12 and 24 h. Subsequent titration of selected aliquots demonstrated the titers of the virus to be stable (data not shown).
Table 1.
Feeding with H5N7 and H7N1 subtypes
As seen in Table 1, the isolation of infectious virus from house flies strongly depended on T, fed C, and I. The importance of the four covariates T, C, I, and S was statistically tested by the logistic regression presented in model 1. Stepwise reduction of the full model gave rise to the final model described in model 3:
logit (ρ) = αS + βT*T + βI*I + βT.I*T*I + βS.C*C [3]
which included the main effects of T and I and in addition the interactions between temperature and incubation period T:I and between virus strain and fed virus concentration S:C. The estimated parameters in the reduced model 3 are listed in Table 2, and the constant terms αS and the parameter βS:C are virus strain specific. The logistic regression transformation was based on the experimental replicates presented in Table 1. The room temperature was ≈24°C, and the effect of this approximation on the statistical analysis was tested by a sensitivity analysis in which the room temperature was varied from 20 to 28°C. Stepwise reduction from model 1 including all interaction terms was repeated until no significant factors could be removed, and in all situations, the stepwise reduction gave rise to the reduced regression model 3, indicating the approximation did not affect the model.
Table 2.
Estimated parameters in logistic regression model 3
Due to the method used to feed the flies, there was uncertainty about the determination of the time covariate T; however, a sensitivity analysis demonstrated that this did not affect the reduced regression model 3. Likewise, a sensitivity analysis confirmed that the inclusion of the 0 h experimental data did not alter model 3. Statistical analyses confirmed that the persistence of infective virus from virus-fed fly pools decreased as a function of virus concentration, incubation period postfeeding, and temperature (Table 2). The coefficients for T and I cannot be regarded on their own, because of the significant T:I interaction effect (Table 2). Thus, for a temperature of 15°C, the effect of incubation period is given by the slope βI + 15βT:I, which Table 2 estimates as 0.206 −15·0.0165 = −0.0415. Obviously, the slope is lower for higher temperatures. Similarly, the effect of temperature at an incubation period of 6 h is βT + 6βT:I, which is estimated as 0.0328 − 6·0.0165 = −0.0662. In the same way, the coefficients for S and C must be combined with the corresponding interaction coefficient, to represent the effects of strain and virus concentration, respectively. As examples, the estimated persistence of virus-positive fly pools for each employed AIV subtype fed with a virus concentration of 107.3 ELD50/ml are shown in Fig. 1. The estimated incubation periods at which 50% of fly pools fed with either of the virus concentrations would be virus-positive are illustrated in Fig. 2.
Fig. 1.
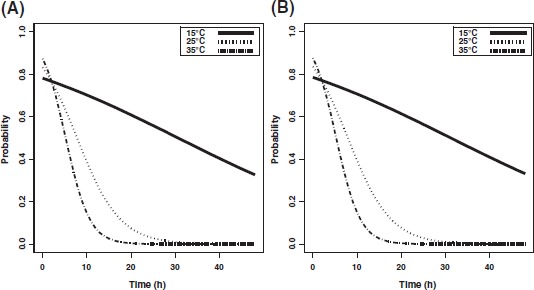
Persistence of H5N7 and H7N1 AIVs. The estimated persistence of H5N7 (A) and H7N1 (B) after fed to flies at a virus concentration of 107.3 ELD50/ml.
Fig. 2.
Estimated incubation period postfeeding as a function of temperature at which 50% of fed fly pools were virus positive. The incubation periods postfeeding at which 50% of the fly pools are positive for H5N7 (A) or H7N1 (B) are shown. Time points were calculated by using the reduced model 3 and the estimates listed in Table 2.
Discussion
Our results demonstrated the decline of virus at all concentrations and temperatures over time. In fact, only one group of flies was found virus positive out of 36 tested (3%) after 24 h at 25°C and 35°C, indicating that the two LPAIV strains did not replicate in the fly's alimentary tract. This indicates that the house fly may be a potential passive carrier of LPAIV, in agreement with previous observations on the survival of HPAIV, NDV, TCV, and REV in house flies and blow flies (Calibeo-Hayes et al. 2003, Davidson and Braverman 2005, Watson et al. 2007, Chakrabarti et al. 2008, Sawabe et al. 2009, Barin et al. 2010, Wanaratana et al. 2011).
The statistical analyses demonstrated the persistence of infectious LPAIV in the alimentary tract to be virus strain dependent. Although it cannot be excluded that the primary, secondary, and tertiary structural differences in the projecting surface molecules on the two LPAIV strains used may be correlated with different inactivation kinetics and excretion rate, this contribution was assumed to be questionable. The observed differences may reflect strain-related adaption of the two virus strains to cultivation in ECEs, strain-related pathogenicity and virus doses. The difference in persistence is especially prominent for the most diluted virus solutions. For example, one of the 0-h experiments with the most diluted virus solution of H5N7 was virus-negative, although the flies were fed only 10 min before termination. However, the reason for this strain difference is at present unknown, and further experiments including a variety of different LPAIV and HPAIV strains are required to address this issue.
Our study was based on flies individually fed known virus concentrations, and only those flies that ingested all of the provided volume of virus solution were included in the study. Although this procedure was time-consuming and caused some time delay before incubation, it was considered essential to ensure that all virus-exposed flies had ingested the required volume of virus. The house fly frequently regurgitates and defecates during foraging (Greenberg 1973), so the ingested doses of virus cannot be controlled by using ad libitum access to infective allantoic fluid. The focus of the current study was on the persistence of AIV in the alimentary tract; therefore, virus particles present on the fly's exoskeleton had to be excluded in the recovery procedures for infectious virus in the experiments. To obtain this, a method was developed for aseptic harvest of the content of the fly's posterior abdomen that involved thermal inactivation of external virus particles. The house fly's irregular integument surface may complicate external disinfection by solutions, a potential obstacle which we solved by thermal inactivation. This approach also made it possible to subsequently process the harvested pooled fly samples in small volumes convenient for virus isolation. However, it cannot be excluded that the method may have led to some thermal inactivation of infective virions located within the alimentary canal.
Excretion of virus particles during regurgitation and defecation in addition to inactivation of virions in the alimentary tract due to exposure to digestive enzymes, low pH levels, and bacterial flora probably contributed to the decrease in the number of virus positive-pools over time (Sinha 1976, Espinoza-Fuentes and Terra 1987, Terra et al. 1988, Nazni et al. 2005b). Despite our using preoviposit females in the study, egg development was pronounced after 24 h of incubation, which occasionally decreased the volume of harvested samples; an observation that might have affected the detection of AIV in these samples upon cultivation in ECEs.
In the presented work, the use of LPAIVs was based on the common occurrence of these viruses in wild birds and domestic poultry and the presumed transfer between these groups of hosts. Also, LPAIV of H5 and H7 subtypes have the potential to mutate to HPAIV in poultry, causing devastating outbreaks of disease. Because of their high morbidity and mortality, HPAIV do not circulate between wild birds and domestic poultry as does the LP variants. Furthermore, LPAIVs were used in our study for safety reasons because it was not feasible to contain house flies exposed to HPAIV in the laboratory. LPAIV may not represent a complete model for persistence of HPAIV in house flies. HPAIV H5N1 was recently shown to persist in the viscera of house flies up to 72-h postexposure in the laboratory (Wanaratana et al. 2011), whereas only one of the 36 virus-fed fly pools incubated at 25 or 35°C for 24 h was positive for virus in our study. Similarly, a velogenic exotic NDV strain was demonstrated to persist in house flies for a longer period of time than the less virulent mesogenic NDV (Roakin) strain (Watson et al. 2007, Chakrabarti et al. 2008). However, differences in the sensitivity of laboratory detection of viruses related to their pathogenicity may influence these results.
The synanthropic and omnivorous house fly is a very common and abundant livestock farm pest, which is attracted by moist manure and other fermenting organic waste that serves as a food source as well as developing environment of the fly (Axtell 1999, Malik et al. 2007, Hald et al. 2008). Because chickens capture and eat flying insects (Hald et al. 1998, Hald et al. 2008), flies foraging on virus-containing feces represent a risk in the spread of disease. Chickens infected with AIV shed virus in secretions and excretions, and although HPAIV typically are found in both feces and respiratory secretions, the primary route of shedding of LPAIV depends on the virus strain (Spickler et al. 2008). Viral quantities of 103.2–107.9 and 105.5–108.1 EID50/ml have been reported in cloacal and fecal samples, respectively, from chickens infected in the laboratory with LPAIV H1, H4, and H6 subtypes (Morales et al. 2009). Shed titers up to 104.5 and 104.3 EID50/ml have been observed in cloacal swabs from chickens infected with LPAIV H5N1 and H7N2 subtypes, respectively (Swayne and Beck 2005, Spackman et al. 2007). In our experiments, high virus doses were used to be able to monitor the persistence over time. Each fly ingested a minimum dose of 103.7 ELD50 (H5N7) and the pool of five flies a minimum total of 104.4 ELD50, which was sufficient to infect ECEs. Under natural conditions, the dose of virus that a house fly may ingest when foraging on virus-containing feces will depend on the amount and persistence of shed virus, the fly's hunger state as well as the size of the fly. Large flies are expected to uptake more food when foraging than smaller individuals and thus ingest more virus. A female house fly can ingest a volume to 2–4 μl (Kobayashi et al. 1999), and the frequency of foraging depends on factors such as temperature, level of stress and the food source. Although an individual fly might not ingest a sufficient virus dose to infect a chicken, the uptake of several infected flies may be critical and lead to infection. In this respect, Calibeo-Hayes et al. (2003) demonstrated that under controlled conditions the number of turkeys infected with TCV increased when placed in contact with higher numbers of TCV-exposed flies per bird.
In general, adult house flies remain within a few kilometers of their breeding sites for their entire life (Nazni et al. 2005a, Boase 2007, Stafford 2008). Therefore, house flies represent potential carriers of AIV among chickens placed in the same building or farm or between poultry located on premises in proximity.
Our study demonstrated that infective low-pathogenic avian influenza virus of the H7N1 and H5N7 subtypes can be isolated from the alimentary tract of house flies for at least 24 h postfeeding and that factors such as temperature, incubation period postfeeding, and load of ingested virus play an important role in the persistence of infective virus. Because house flies commonly occur on premises with poultry production, the data indicate that house flies can be a potential risk in the dissemination of avian influenza virus. However, studies that address whether flies and in particular house flies and blow flies are able to cause infection after ingestion of AIV in poultry should have high priority. Similarly, studies on the possibilities of flies to ingest chicken infective doses of AIV in poultry production environments and in nature are needed.
Acknowledgments
The technical assistance of Hanne Røigaard-Petersen, Susanne Nordby Stubbe, Søs Hinge Siem, and Gitte Jensen is highly acknowledged. The employed A/African Starling/England/983/79 (H7N1) AIV was kindly provided by the Veterinary Laboratories Agency, Surrey, United Kingdom. The presented work was funded by grant 3304-FVFP-07-754-01 from the Ministry of Food, Agriculture and Fisheries, Denmark.
References Cited
- Alexander D. J. 2008. Orthomyxoviridae—avian influenza, pp. 317–332. InPattison M., McMullin P. F., Bradbury J. M., Alexander D. J. (eds.), Poultry diseases, 6 ed. W. B. Saunders, Philadelphia, PA. [Google Scholar]
- Axtell R. C. 1999. Poultry integrated pest management: status and future. Integr. Pest Manage. Rev. 4: 53–73. [Google Scholar]
- Barin A., Arabkhazaeli F., Rahbari S., Madani S. A. 2010. The housefly, Musca domestica, as a possible mechanical vector of Newcastle disease virus in the laboratory and field. Med. Vet. Entomol. 24: 88–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boase C. 2007. Houseflies: a review of dispersion behaviour. Int. Pest Control 49: 210–213. [Google Scholar]
- Calibeo-Hayes D., Denning S. S., Stringham S. M., Guy J. S., Smith L. G., Watson D. W. 2003. Mechanical transmission of turkey coronavirus by domestic houseflies (Musca domestica Linnaeus). Avian Dis. 47: 149–153. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., King D. J., Afonso C., Swayne D., Cardona C. J., Kuney D. R., Gerry A. C. 2007. Detection and isolation of exotic Newcastle disease virus from field-collected flies. J. Med. Entomol. 44: 840–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., King D. J., Cardona C. J., Gerry A. C. 2008. Persistence of exotic Newcastle disease virus (ENDV) in laboratory infected Musca domestica and Fannia canicularis. Avian Dis. 52: 375–379. [DOI] [PubMed] [Google Scholar]
- Davidson I., Braverman Y. 2005. Insect contribution to horizontal transmission of Reticuloendotheliosis virus. J. Med. Entomol. 42: 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Fuentes F. P., Terra W. R. 1987. Physiological adaptations for digesting bacteria. Water fluxes and distribution of digestive enzymes in Musca domestica larval midgut. Insect Biochem. 17: 809–817. [Google Scholar]
- Greenberg B. 1973. Flies and disease, vol. II. Princeton University Press, Princeton, NJ. [Google Scholar]
- Hald B., Olsen A., Madsen M. 1998. Typhaea stercorea (Coleoptera: Mycetophagidae), a carrier of Salmonella enterica serovar Infantis in a Danish broiler house. J. Econ. Entomol. 91: 660–664. [DOI] [PubMed] [Google Scholar]
- Hald B., Skovgard H., Pedersen K., Bunkenborg H. 2008. Influxed insects as vectors for Campylobacter jejuni and Campylobacter coli in Danish broiler houses. Poult. Sci. 87: 1428–1434. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Sasaki T., Saito N., Tamura K., Suzuki K., Watanabe H., Agui N. 1999. Houseflies: not simple mechanical vectors of enterohemorrhagic Escherichia coli O157:H7. Am. J. Trop. Med. Hyg. 61: 625–629. [DOI] [PubMed] [Google Scholar]
- Malik A., Singh N., Satya S. 2007. House fly (Musca domestica): a review of control strategies for a challenging pest. J. Environ. Sci. Health B 42: 453–469. [DOI] [PubMed] [Google Scholar]
- Morales A. C., Hilt D. A., Williams S. M., Pantin-Jackwood M. J., Suarez D. L., Spackman E., Stallknecht D. E., Jackwood M. W. 2009. Biologic characterization of H4, H6, and H9 type low pathogenicity avian influenza viruses from wild birds in chickens and turkeys. Avian Dis. 53: 552–562. [DOI] [PubMed] [Google Scholar]
- Nazni W. A., Luke H., Wan Rozita W. M., Abdullah A. G., Sa'diyah I., Azahari A. H., Zamree I., Tan S. B., Lee H. L., Sofian M. A. 2005a. Determination of the flight range and dispersal of the house fly, Musca domestica (L.) using mark release recapture technique. Trop. Biomed. 22: 53–61. [PubMed] [Google Scholar]
- Nazni W. A., Seleena B., Lee H. L., Jeffery J., Rogayah T.A.T., Sofian M. A. 2005b. Bacteria fauna from the house fly, Musca domestica (L.). Trop. Biomed. 22: 225–231. [PubMed] [Google Scholar]
- Pantin-Jackwood M. J., Swayne D. E. 2009. Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev. Sci. Technol. 28: 113–136. [PubMed] [Google Scholar]
- Sawabe K., Hoshino K., Isawa H., Sasaki T., Hayashi T., Tsuda Y., Kurahashi H., Tanabayashi K., Hotta A., Saito T., et al. 2006. Detection and isolation of highly pathogenic H5N1 avian influenza A viruses from blow flies collected in the vicinity of an infected poultry farm in Kyoto, Japan, 2004. Am. J. Trop. Med. Hyg. 75: 327–332. [PubMed] [Google Scholar]
- Sawabe K., Tanabayashi K., Hotta A., Hoshino K., Isawa H., Sasaki T., Yamada A., Kurahashi H., Shudo C., Kobayashi M. 2009. Survival of avian H5N1 influenza A viruses in Calliphora nigribarbis (Diptera: Calliphoridae). J. Med. Entomol. 46: 852–855. [DOI] [PubMed] [Google Scholar]
- Sinha M. 1976. Digestive enzymes in the gut and salivary glands of Sarcophaga ruficornis FAB. and Musca domestica L. (Diptera: Insecta). Appl. Entomol. Zool. 11: 260–262. [Google Scholar]
- Slomka M. J., Coward V. J., Banks J., Londt B. Z., Brown I. H., Voermans J., Koch G., Handberg K. J., Jorgensen P. H., Cherbonnel-Pansart M., et al. 2007. Identification of sensitive and specific avian influenza polymerase chain reaction methods through blind ring trials organized in the European Union. Avian Dis. 51: 227–234. [DOI] [PubMed] [Google Scholar]
- Spackman E., Swayne D. E., Suarez D. L., Senne D. A., Pedersen J. C., Killian M. L., Pasick J., Handel K., Pillai S. P., Lee C. W., et al. 2007. Characterization of low-pathogenicity H5N1 avian influenza viruses from North America. J. Virol. 81: 11612–11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler A. R., Trampel D. W., Roth J. A. 2008. The onset of virus shedding and clinical signs in chickens infected with high-pathogenicity and low-pathogenicity avian influenza viruses. Avian Pathol. 37: 555–577. [DOI] [PubMed] [Google Scholar]
- Stafford K. C. 2008. Fly management handbook. A guide to biological, dispersal, and management of the house fly and related flies for farmers, municipalities, and public health officials. Connecticut Agricultural Experiment Station, New Haven, CT. [Google Scholar]
- Stallknecht D. E. 2003. Ecology and epidemiology of avian influenza viruses in wild bird populations: waterfowl, shorebirds, pelicans, cormorants, etc. Avian Dis. 47: 61–69. [Google Scholar]
- Swayne D. E. 2007. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 51: 242–249. [DOI] [PubMed] [Google Scholar]
- Swayne D. E., Beck J. R. 2005. Experimental study to determine if low-pathogenicity and high-pathogenicity avian influenza viruses can be present in chicken breast and thigh meat following intranasal virus inoculation. Avian Dis. 49: 81–85. [DOI] [PubMed] [Google Scholar]
- Swayne D. E., King D. J. 2003. Avian influenza and Newcastle disease. J. Am. Vet. Med. Assoc. 222: 1534–1540. [DOI] [PubMed] [Google Scholar]
- Terra W. R., Espinoza-Fuentes F. P., Ferreira C. 1988. Midgut amylase, lysozyme, aminopeptidase, and trehalase from larvae and adults of Musca domestica. Arch. Insect Biochem. Physiol. 9: 283–297. [Google Scholar]
- Wanaratana S., Panyim S., Pakpinyo S. 2011. The potential of house flies to act as a vector of avian influenza subtype H5N1 under experimental conditions. Med. Vet. Entomol. 25: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D. W., Nino E. L., Rochon K., Denning S., Smith L., Guy J. S. 2007. Experimental evaluation of Musca domestica (Diptera: Muscidae) as a vector of Newcastle disease virus. J. Med. Entomol. 44: 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. D., Schmidtmann E. T., Richard R. D., Lehman R. D. 1986. Isolation of avian influenza from insects, pp. 221–223. InArbovirus research in Australia: Proceedings 4th Symposium, 6–9 May 1986, Brisbane, Australia. [Google Scholar]