Abstract
Parasites and pathogens are a fundamental driving force in the ecology and evolution of mammalian populations, and understanding disease processes in natural populations is an urgent priority in the face of increased rates of infectious disease emergence. In this review, we argue that mammalogists are uniquely placed to contribute to addressing these challenges because in-depth knowledge of mammal species is fundamental to the development of wild model systems that could accelerate discovery in disease ecology. The use of animal models—species for which a broad range of diagnostic, molecular, and genetic tools have been developed—in tightly controlled laboratory environments has been instrumental in driving progress in the biomedical sciences. However, in natural populations, disease processes operate in the context of enormous genetic, phenotypic, and environmental variability. Understanding diseases in animal populations (including humans) thus requires investment in “wild animal models” that explicitly include individual variation and relevant environmental gradients. Wild mammal groups such as primates and rodents have already been identified as potentially useful models of infectious diseases in the wild. Here, we discuss the enormous potential that ungulates hold as candidates for wild model systems. The diversity, broad geographic distribution, and often high abundance of species in this group make them a highly accessible target for disease research. Moreover, a depth of background knowledge, close relationships to domesticated animals, and ongoing management of many wild ungulate species provide context, tools, and opportunity for cutting-edge research at the interface of ecological and biomedical sciences. Studies of wild ungulates are already helping to unravel some key challenges in infectious disease research, including the role of parasites in trophic cascades, the consequences of climate change for disease dynamics, and the systems biology of host–parasite interactions. Other areas where ungulate studies may provide new insight include research on the sources and drivers of emerging infectious diseases.
Keywords: climate change, disease ecology, host–parasite interactions, mammals, systems biology, trophic cascade
Parasites are the most abundant and diverse trophic group on the planet. They are ubiquitous in all natural populations and act as a fundamental driving force in mammalian ecology and evolution (Barber and Dingemanse 2010; Tompkins et al. 2011; Dunn et al. 2012). Human activities are drastically altering biotic communities via biodiversity loss and species invasions, including pathogens. Anticipating and managing the resulting changes in disease dynamics is one of the most daunting challenges of our time. Meeting this challenge will require continued progress in understanding disease processes in complex ecological communities. Motivated by these urgent basic and applied questions, infectious disease ecology and evolution are experiencing rapid growth as areas of scientific enquiry. The biomedical sciences investigate host–pathogen interactions at suborganismal (cellular and molecular) scales with the aim of discovering targets for intervention that can modify these interactions to prevent pathogen invasion (e.g., vaccines) or promote their expulsion (e.g., antimicrobial drugs). Ultimately, the goal is to help individual patients prevent or manage disease. By contrast, the role of disease ecological and evolutionary studies is to increase our understanding of the role that host–parasite interactions play in driving population and evolutionary dynamics and to apply these insights to disease management at the population scale, for biological conservation, veterinary health, and public health.
Disease ecologists ask fundamental questions about disease processes in host individuals, populations, and communities such as under what conditions can a given parasite invade a host population and how fast will it spread? How will a disease affect host population dynamics? How do host behavior, population dynamics, and physiology affect disease dynamics in natural populations? What roles do different host species play in spreading or attenuating infection? And how do host–parasite and parasite–parasite interactions at the molecular level scale up to affect population-level disease dynamics? Consequently, variation among individuals and environments is of fundamental interest in disease ecology, whereas experimental approaches in biomedicine typically go to great lengths to limit such variability, in an effort to discern molecular-level interactions more clearly. Biomedicine has established a number of successful laboratory model systems, developing a sophisticated array of molecular and genetic tools that have transformed the study of disease processes at suborganismal scales. In this review, we argue that disease ecology, too, can benefit greatly from model study systems. Model systems in disease ecology and evolution, however, must expressly incorporate individual variability in genotype, behavior, and physiology and be observable across meaningful environmental and temporal gradients and scales.
What makes a good model system?
Animal models have been an important component of infectious disease research for decades. Common models in disease research include small laboratory animals such as mice (Mus musculus) and rats (Rattus rattus—Niewiesk and Prince 2002; Buer and Balling 2003), nonhuman primates such as macaques (Macaca) and marmosets (Callithrix—Nath et al. 2000), and other vertebrates such as pigs (Sus scrofa domesticus) and zebra fish (Danio rerio—Lieschke and Currie 2007; Meurens et al. 2012). These organisms all possess traits that make them well suited to studying host–pathogen interactions in captive settings. For example, small laboratory animals are often readily accessible, easily manipulated, and relatively simple and inexpensive to maintain under laboratory conditions. Larger organisms such as macaques are more expensive to maintain in captivity, but share a high degree of genetic and physiological similarity to humans, making them an invaluable model for a number of human diseases (Gardner and Luciw 2008). Importantly, almost all of the common animal models are associated with a large repository of resources and tools that facilitate studies of infection and immunity at the molecular, genetic, and cellular levels. In the laboratory mouse for instance, decades of basic research on all aspects of biology, a sequenced genome, and an abundance of mutant strains form the backdrop of ever-expanding research into links between genes and disease (Chinwalla et al. 2002; White et al. 2013b).
Despite extensive insights derived from the use of animal models in biomedicine, the applicability of these models, particularly mouse models, to complex conditions in humans has come under recent scrutiny (Coers et al. 2009; Seok et al. 2013). From an ecological and evolutionary perspective, this ongoing debate prompts even broader reflection about how studies of infectious diseases in laboratory and captive settings apply to natural populations in the real world. Unlike the laboratory, the real world is complex and most animals, including humans, interact with parasites and pathogens in this highly variable context. This reality has fueled ideas that “wild” animal models may be as important as laboratory models in the study of infectious disease processes (Wolfe et al. 1998; Pedersen and Babayan 2011). Whereas laboratory animal models have paved the way toward advancing our understanding of disease processes occurring at the genetic, molecular, and cellular levels, the promise of wild models lies in their potential to help define how mechanisms revealed in the laboratory operate within the genetic, phenotypic, and environmental contexts that characterize natural populations. The use of wild models may also facilitate the inclusion of a more diverse set of organisms in infectious disease research, allowing for increasingly powerful comparative studies. However, the challenge is to find wild study systems that will balance realistic complexity with tractability and can thus serve as effective models for discovering how infectious disease processes function in nature.
If wild models can contribute to innovation in infectious disease research, a question that arises is, what types of organisms would make good wild animal models? If by definition model species in the laboratory setting are organisms that are simple, readily accessible, and easily manipulated, by extension, good wild models might be highly abundant, easy to find, and amenable to observational, experimental, or longitudinal studies. In addition, good wild models might be characterized by close genetic, physiological, or other similarity to humans or their domestic animals. Moreover, if wild models are to act as bridges between the laboratory and the real world, the ideal wild models should be associated with resources and tools that allow for molecular-level approaches to be applied in the field. Several groups of mammals squarely fit these criteria (e.g., wild rodents, wild primates). Mammalogists are thus uniquely positioned to spearhead efforts to develop new wild models for disease ecological research. Here, we focus on the ungulates as a mammalian group that has much to contribute to our understanding of infectious disease processes in the wild.
How do ungulates measure up as wild models?
Terrestrial hoofed mammals (ungulates) from the orders Cetartiodactyla (even-toed ungulates) and Perissodactyla (odd-toed ungulates) are a diverse and widely distributed group. With between 250 and 450 species recognized (Groves and Grubb 2011), ungulates occur in almost every terrestrial ecoregion, ranging from tropical forest to tundra. The wide distribution and abundance of many ungulates make them highly accessible for disease ecological research. The fact that rhesus macaques (Macaca mulatta) are widely distributed across Asia contributes to their use as the major nonhuman primate model of infectious disease (Mitruka et al. 1976; e.g., Middle East respiratory syndrome—Falzarano et al. 2013; de Wit et al. 2014; tuberculosis—White et al. 2013a; Min et al. 2014, coinfections—schistosomes and simian-human immunodeficiency virus, and malaria—Ayash-Rashkovsky et al. 2007; Chenine et al. 2008; Semenya et al. 2012). Likewise, the widespread occurrence of baboons (Papio) in Africa and the Middle East, coupled with typically large population sizes, may explain the increasing number of field-based infectious disease studies focusing on this group (Weyher et al. 2006; Tung et al. 2009; Drewe et al. 2012; Akinyi et al. 2013). Importantly, numerous ungulate species share these characteristics—broad geographic range and large population sizes—making them easy to locate and observe. Moreover, for some ungulate species like white-tailed deer (Odocoileus virginianus), red deer (Cervus elaphus), and wild boar (S. scrofa), a high level of accessibility coupled with an extensive knowledge base on basic behavior, ecology, life history, physiology, population dynamics, and evolution provide an attractive menu of candidate wild model systems that can be used to investigate questions about infectious diseases processes in a range of environments.
Another important criterion for a good animal model is tractability. In the context of a wild model, the organism must be amenable to studies that include repeated observation, direct manipulation, or both. The power of laboratory models has always been that they allow for direct manipulation and experimentation in controlled settings. Given the inherent variability of natural environments, achieving fundamental insights from wild “model” organisms requires the ability to experimentally manipulate individuals in the field or to track individuals through time, both of which are techniques that maximize power for drawing inferences about disease processes in the wild. Both approaches are challenging in natural populations, but recent studies of wild rodent systems in Europe illustrate that key insights can be gained from applying these study designs in natural populations. For instance, Telfer et al. (2010) used longitudinal studies of wild voles (Microtus agrestis) to demonstrate that interactions among parasites occurring within hosts are as important as, if not more than, host and environmental factors in determining infection risk.
Ungulates are typically larger and more long-lived than rodents and some primates. This makes studying fitness consequences of infectious diseases a more substantial time commitment and attaining significant sample sizes more challenging. Heritability and genetic selection pressure of pathogens are likewise more difficult to study in species with a long generation time, and capturing large animals can be difficult and time consuming, making longitudinal studies more costly. Nonetheless, important insights into infectious disease processes have emerged from experimental and longitudinal studies of a growing number of free-ranging ungulates (Table 1). These case studies show that wild ungulate populations can be used to address topics ranging from the fitness consequence of infection in wild populations to the drivers of immunological variability in nature (Table 1). However, this type of work could expand to many more species. Even though ungulates are much larger in size than rodents and longer lived, traits that make field studies more logistically challenging and expensive, these perceived disadvantages offer some important benefits. For instance, in studies where multiple physiological, immunological, and other variables must be tracked through time, the large body size of ungulates allows for the collection of sufficient sample volumes (e.g., blood, feces) from live animals, an objective that can be difficult to achieve using smaller study organisms. Also, studying long-lived model species like ungulates makes it possible to examine links between life span and infectious disease processes, questions that are particularly relevant for understanding chronic diseases including many observed in human populations. Finally, from a practical standpoint, the logistical difficulties of working with ungulates are tempered by the fact that this is the only mammalian group for which management of wild species is a worldwide phenomenon (Apollonio et al. 2010). Ungulate management varies by species and geographic region but is typically managed by governmental wildlife organizations and may include controlled hunting, disease management, radiotracking, population censuses, and animal translocations. Often, these management activities can go hand in hand with disease research, reducing the cost of accessing wild ungulates and enhancing study feasibility. Indeed, collaboration between researchers and managers is routine in studies of ungulate ecology. Recently, such collaborations have given rise to a number of important insights into the ecology and evolution of infectious disease in wild populations (Jolles et al. 2005; Robinson et al. 2012; Schaschl et al. 2012). Thus, ungulate systems where ongoing management activities are taking place provide particularly attractive targets for disease ecological research.
Table 1.
Examples of longitudinal and/or experimental studies of infectious disease in free-ranging ungulates.
| Species | Geographic location | Type of study | Major finding | Implication | Reference |
|---|---|---|---|---|---|
| Reindeer (Rangifer tarandus platyrhynchus) | Svalbard, Norway | Experimental and longitudinal | Anthelminthic treatment of the parasite Ostertagia gruehneri increased female fecundity but not overwinter survival. Parasite abundance was positively associated with the magnitude of the fecundity effect | Demonstrates that density- dependent parasite- mediated reduction in host fecundity is a plausible mechanism of host population regulation | Albon et al. (2002) |
| Soay sheep (Ovis aries) | St. Kilda, Scotland, United Kingdom | Longitudinal | Individuals with the higher antibody levels were more likely to survive harsh winters, but higher antibody levels were associated with reduced fecundity | Highlights the role of environmental variability as a mechanism maintaining immunoheterogeneity | Graham et al. (2010) |
| African buffalo (Syncerus caffer) | Hluhluwe- iMfolozi Park, South Africa | Experimental | Anthelminthic treatment of gastrointestinal nematode parasites increased circulating levels of interferon gamma, a molecule involved in the immune defense against microparasites | Suggests that immunological trade-offs described in humans occur in wild mammals and have the potential to influence disease dynamics | Ezenwa et al. (2010) |
| Bighorn sheep (Ovis canadensis) | Idaho, United States | Longitudinal | Among adult females, previous exposure to pneumonia was associated with a decrease in the risk of future disease-related mortality; however, a lamb’s risk of dying was positively associated with its mother’s previous exposures | Uses individual-based data to reconstruct how hosts respond immunologically to an unknown causative agent | Plowright et al. (2013) |
Central to the tractability of using a wild species as a model of infectious disease is the availability of resources necessary for probing the mechanistic basis of host–pathogen interactions. These resources include, but are not limited to, diagnostic assays for identifying pathogen exposure and infection, molecular and immunological reagents for quantifying physiological, immunological, and biochemical responses of the host to infection, and drugs and vaccines that can be used for manipulating host susceptibility or infection status. Many livestock species, including cattle (Bos taurus), sheep (Ovis aries), goats (Capra hircus), pig (S. scrofa domesticus), and horses (Equus caballus), serve as models of both human and animal diseases, and a plethora of resources have been developed for use in these species. For example, the genomes of the cow, sheep, pig, and horse have all been fully or partially sequenced (National Animal Genome Research Program 1998–2014), and the development of immunological reagents for veterinary species is a major focus of many biotechnology companies. Importantly, the close phylogenetic relationships between wild and domesticated ungulates mean that many of the resources developed for livestock can be applied to studies of their free-ranging relatives with modest additional investment. Indeed, there are an increasing number of examples where molecular, genetic, and immunological tools from livestock have been successfully adopted for use in their wild relatives (Grobler et al. 2002; Poissant et al. 2009; Olsen and Johnson 2012). Transferring molecular tools from livestock to related wild species does involve optimizing protocols to take account of species-specific concentrations of target substrates (e.g., Beechler et al. 2012) or differences in reagent affinity for species-specific proteins (e.g., antibody affinity in enzyme-linked immunosorbent assays). The validity of diagnostic tests must be demonstrated in wildlife species (e.g., Michel et al. 2011; Gorsich et al. 2014), and vaccine and drug efficacy carefully evaluated. While genetic resources from related domestic species are potentially valuable in analyzing genomic and transcriptomic data from wild ungulates, interspecific variation at genetic loci of interest can thwart attempts to identify target genes for important traits—such as disease resistance—in wildlife (Brown et al. 2013). These caveats notwithstanding, modifying existing assays is typically a lesser hurdle than designing them from scratch: the transferability of molecular and genetic tools from domestic species makes wild ungulates one of only a few mammalian groups where cutting-edge research at the interface of ecological and biomedical sciences can take place.
How are wild ungulates already contributing to infectious disease research?
Studies conducted on wild ungulates are addressing some of the major current challenges in infectious disease ecology and evolution (Table 1). Below we showcase 3 examples that illustrate how this group is contributing to fundamental insights in the field: longitudinal and experimental studies in Soay sheep (O. aries) are uncovering linkages across organizational levels, from host genetics and immunity to population-level disease outcomes; research in arctic ungulates is revealing striking sensitivity of host–parasite interactions to climate change, and the central position of ungulates in terrestrial food webs is enabling insights into the role of ungulate infections as drivers and outcomes of trophic cascades.
From genes to populations: toward a systems biology of infectious disease.
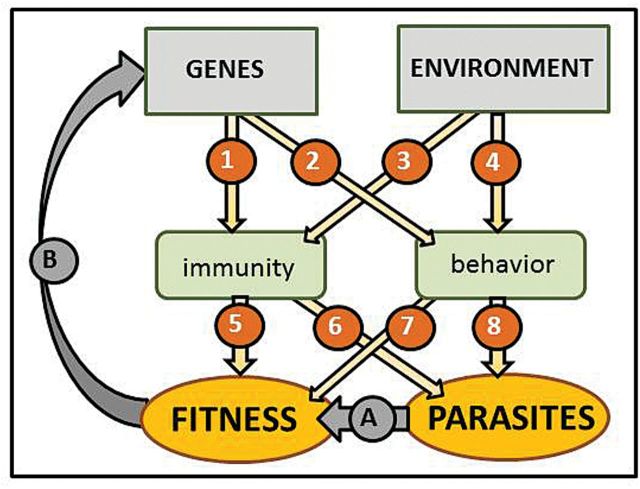
“Systems biology” seeks to combine detailed knowledge of the components involved in a biological system and their interactions, with mathematical models to enable us to understand the emergent properties of the system as a whole (Ideker et al. 2001; Kitano 2002). The approach has been applied primarily to suborganismal biological systems, such as cells or tissues (e.g., cancer biology—Wang et al. 2013; cancer and personalized medicine—Benes 2013; viral pathogenesis—Law et al. 2013; human microbiome—Weinstock 2012), but it is just as relevant in disease ecology (Fig. 1). In the face of rapid environmental change, a mechanistic understanding of disease dynamics as emerging from the interplay of genetic, environmental, immunologic, and behavioral factors that define host–parasite interactions is urgently needed to improve our ability to anticipate and manage shifts in infectious diseases. This is a formidable challenge that cannot be addressed adequately in laboratory-based systems because host genetics and environmental variation are likely to interact in shaping immunity and host behavior (Fig. 1, arrows 1–4). Host behavior defines the opportunities for parasite transmission through contacts among hosts, with vectors, or with environmental reservoirs of infectious material. Immunity then determines which contacts between hosts and parasites actually lead to infection and how infections progress within the host. Ultimately, the host–parasite interaction results in fitness effects on hosts and parasites, which scale up to shape population dynamics for both (Fig. 1, arrows 5–8). When these interaction mechanisms are poorly understood, our predictions of disease dynamics under changing biotic and abiotic environmental conditions rely on statistical extrapolation from current and past conditions. However, given potentially new parameter combinations and nonlinear functional relationships between environmental drivers and host immunologic or behavioral responses, such predictions must be viewed with caution. A mechanistic framework linking host genetics and environmental variation with immunologic and behavioral responses that determine disease dynamics could complement statistical approaches, improving our ability to predict how infectious disease patterns are likely to change when host population or environmental parameters are pushed outside previously observed conditions by anthropogenic forcing.
Fig. 1.
A “systems biology” approach in disease ecology. Host genetics and the environment affect immunity and host behavior (1–4)—which determine susceptibility and transmission opportunities for parasites (6, 8). Parasites cause heterogeneities in host fitness (A), driving population genetic structure (B). Immunity and behavior may affect fitness directly (5, 7), e.g., through allocation trade-offs against other fitness components such as growth or reproduction. In combination, variation in host traits and the environment thus drive host population and disease dynamics. (Our focus is on host and environmental traits as drivers of disease dynamics. For simplicity, direct effects of genes on fitness and of environment on fitness and parasites are not depicted as arrows.)
Disease studies in feral Soay sheep have been at the forefront of progress toward addressing this formidable challenge (Fig. 2). The Soay sheep study population is located in the island of Hirta (636 ha) in the St. Kilda archipelago in the United Kingdom. The population was founded in 1932 when 107 sheep were introduced from the neighboring island of Soay. The population is unmanaged and fluctuates between 600 and 2,000 individuals. Sheep are parasitized by gastrointestinal helminths (primarily Teladorsagia circumcincta, Trichostrongylus axei, and Trichostrongylus vitrinus—Craig et al. 2006), several species of parasitic protozoa (Craig et al. 2007), and ectoparasitic sheep keds (the dipteran Melophagus ovinus—Wilson et al. 2004). Long-term population monitoring of Soay sheep, quantifying individual fitness, parasite burdens, and immunity (Clutton-Brock and Pemberton 2004), has been combined with parasite removal experiments and detailed genetic analyses. These studies have established many of the mechanistic details underpinning the population dynamics of sheep and their parasites.
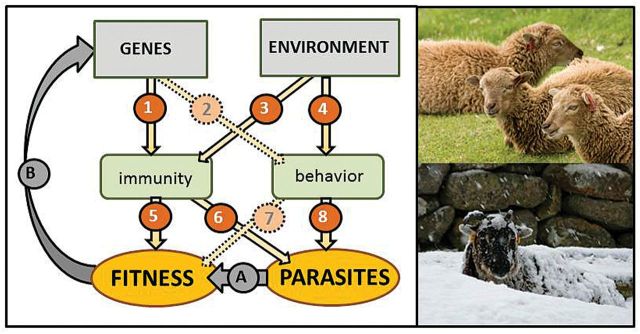
Fig. 2.
Soay sheep (Ovis aries) and their parasites—a model system in disease ecology. Many of the mechanistic links illustrated in this schematic have been explored in Soay sheep. Gaps remain, shown as dotted arrows, in linking host genetics with behaviors relevant to parasite transmission and host fitness. References (by arrow number): 1) Coltman et al. (1999, 2001a), Paterson et al. (1998), Beraldi et al. (2007), and Brown et al. (2013); 3) Hayward et al. (2009) and Hazlerigg et al. (2013); 4) Hutchings et al. (2002a, 2002b); 5) Coltman et al. (2001b) and Graham et al. (2010); 6) References for 1) and Hayward et al. (2009); and 8) Hutchings et al. (2002a); A) References for 1) and B) Coltman et al. (1999).
Strong environmental variation has allowed detection of links between genes, environment, and immunity in Soay sheep (arrows 1 and 3 in Fig. 2). Soay sheep population dynamics are driven strongly by nutritional restriction during harsh winters. Mortality risk during these events varies with parasite burdens, which depend on genetic background (Paterson et al. 1998; Coltman et al. 1999; Coltman et al. 2001a). Variation in parasite resistance in Soay lambs is moderately heritable. However, in adult sheep, the evidence for heritable variation in parasite burdens is weak (Beraldi et al. 2007), suggesting that environmental factors and infection history may outweigh host genetic background in determining parasite burdens throughout the lifetime of the sheep. Consistent with this, cumulative environmental stress (Hayward et al. 2009) and season (Hazlerigg et al. 2013) have both been shown to affect parasite resistance in Soays. A recent study (Brown et al. 2013) attempted to identify genes associated with variation in parasite burdens and immune traits in Soay sheep, using a candidate gene approach. Genes known to be involved in parasite resistance in domestic sheep and cattle were evaluated. While several genes did show significant associations with parasite burden, these associations were no more common in candidate genes than in randomly selected “control” genes. The genetic basis for parasite resistance may thus not match for domestic and natural populations.
Studies of parasite resistance in Soay sheep have revealed context-dependent fitness consequences of immunogenetic variation (arrow 5 in Fig. 2). Population-level variation in genes associated with parasite resistance may be maintained by trade-offs, if parasite immunity is costly to the animal and if environmental variation or animal characteristics such as age and sex modify the fitness effects of different alleles. Investigations into the genetic basis for parasite resistance in Soay sheep have demonstrated such context-dependent trade-offs between parasite resistance and fitness: high antibody responsiveness was shown to be heritable in Soays and associated with improved survival of female sheep during harsh winters (Graham et al. 2010). However, balancing this apparent benefit, heritable high antibody responsiveness also correlated with reduced reproduction. Thus, for female Soays, the “optimal” level of antibody responsiveness depends on winter conditions, which vary enormously from year to year. Interestingly, rams with highly reactive antibodies reap no survival benefit, and an earlier study found that rams with high parasite resistance do not appear to pay a price in terms of growth or adult body size (Coltman et al. 2001b). Trade-offs maintaining immunoheterogeneity in the Soay sheep population may thus be driven disproportionately by female rather than male sheep.
In addition to immunogenetic variation, age and sex-dependent avoidance of pasture contaminated with parasite infectious stages contributes to variability in parasite burdens (arrows 4 and 8 in Fig. 2; Hutchings et al. 2002a, 2002b). However, the extent to which variation in behavioral avoidance of parasites is heritable (arrow 2 in Fig. 2) and the fitness costs of pasture “selectivity” are as yet unclear (arrow 7 in Fig. 2), and more studies are needed.
Two decades of intensive, individual-based research on the Soay sheep population of Hirta have elucidated many of the mechanisms connecting environmental variation with host genetics, immunity and behavior, parasitic infections, and their fitness consequences (Fig. 2). Soay sheep are thus a wild model system that is advancing toward understanding emergent properties of the whole system, such as the ecological and evolutionary dynamics of host and parasite populations. The suitability of Soay sheep as a wild model for research on the systems biology of infectious disease is due to several population traits: The sheep are highly observable because they habituate readily and their environment is sparsely vegetated. Population size is relatively easy to assess, because the sheep occur on an island, which limits population size and prevents migration. The biotic context for disease studies is comparatively simple, with no other ungulate species, no predators, and a limited number of parasite species in the community. Parasite effects on hosts are thus not obscured by predation or interspecific competition. Strong environmental variability sets the stage for fluctuating selective pressures, and genetic resources, diagnostic methods, and antiparasitic drugs developed for domestic sheep can be applied to feral Soays with minimal modification. Individual-based longitudinal and experimental studies in other natural ungulate populations, comprising a range of environmental conditions, will be needed to test the generality of findings from this unusually tractable model system.
Effects of climate change on infectious disease dynamics.
There can be little doubt that the current rapid change in global climates will affect infectious disease dynamics, since climatic conditions underpin the geographic ranges, behavior, physiology, and reproductive rates of organisms—including hosts, vectors, and parasites. However, in contrast to the earlier view that climate warming should generally result in a sicker world (Harvell et al. 2002; Patz et al. 2005; McMichael et al. 2006), current evidence points to complex relationships between climate and disease that may be driven by both abiotic and biotic components of the focal systems and that can result in positive or negative associations between climate change and disease, depending on the thermal tolerances of hosts, vectors, and pathogens (Altizer et al. 2013). Host–parasite interactions involving ectothermic hosts and vector-borne or environmentally transmitted parasites are likely to be intrinsically more sensitive to temperature fluctuations than directly transmitted infections of endothermic hosts (Rohr et al. 2011). Variation among species in temperature sensitivity of immunity may introduce significant noise into these simple contrasts, and disease emergence at the leading edge of vector and parasite range shifts may be balanced by reductions in cases at the receding edge (Harvell et al. 2009).
While general predictions concerning the response of infectious disease dynamics to climate change are thus elusive and vigorously debated, significant progress is emerging from studies of Arctic ungulates and their parasites, paired with innovative theoretical approaches. Arctic ecosystems present tractable study systems for climate–disease research because temperature warming in the arctic is proceeding most rapidly (Klein et al. 2008; Post et al. 2009), host and parasite communities are comparatively simple, and organismal range shifts occur overwhelmingly from south to north rather than in both directions. Ungulates are prominent players in arctic regions, both in terms of their ecological function, and as sources of food and animal products for human populations (Kutz et al. 2012). As such, shifts in parasite dynamics in wild ungulates due to climate change may jeopardize regional food security and increase zoonotic disease risk as well as affect the health and viability of the wildlife populations themselves (Davidson et al. 2011; Jenkins et al. 2011).
Evidence for changes in disease dynamics due to climate change is accumulating for multiple arctic ungulate species. For example, musk oxen (Ovibos moschatus) are experiencing unprecedented outbreaks of pneumonia, both verminous (caused by lungworms) and bacterial (caused by Pasteurella—Ytrehus et al. 2008). Several mechanisms driven by climate warming appear to be important in causing these outbreaks. Musk oxen die-offs due to pasteurellosis have been associated with warm weather events, prompting the hypothesis that increased respiration in an effort to achieve cooling may have allowed more bacteria to enter the lung. Stress-induced immunosuppression may then have allowed these opportunistic pathogens to proliferate to the point of clinical disease (Ytrehus et al. 2008). In the case of lungworms, longer warmer summers appear to have allowed larval development time to accelerate from a 2-year to a 1-year cycle, greatly exacerbating parasite exposure risk to the musk oxen (Kutz et al. 2004, 2005). Mathematical modeling approaches combining conventional compartment models for macroparasite–host dynamics (Anderson and May 1978, 1991; Dobson and Hudson 1992) with temperature responses of larval survival and development according to the metabolic theory of ecology (Gillooly et al. 2001; Brown et al. 2004; McCoy and Gillooly 2008; Munch and Salinas 2009) can reproduce these shifts in helminth life cycles (Molnar et al. 2013), providing theoretical validation for the idea that temperature dependency of parasite life cycles is sufficient to cause the observed shifts in infection patterns.
Similarly, northward range expansions of protostrongylid nematodes (muscleworms and lungworms) and filarioid nematodes (legworms) are exposing populations of caribou (Rangifer tarandus), Dall’s sheep (Ovis dalli), and moose (Alces americanus) to new or increased infectious disease burdens (Jenkins et al. 2006; Laaksonen et al. 2010; Halvorsen 2012; Verocai et al. 2012). In some cases, effects of the parasites themselves on host health may be exacerbated by reduced foraging time due to harassment by insect vectors, which increase during warmer weather (Witter et al. 2012a, 2012b). Emergence and reemergence of infectious diseases in the wake of climate change are by no means limited to the Arctic (e.g., global changes in distribution of orbiviruses—MacLachlan and Guthrie 2010; Falconi et al. 2011), but owing to more pronounced temperature increases at high latitudes, arctic wildlife serves as a sensitive sentinel for impacts of climate change on wildlife disease dynamics, zoonoses, and human health (Hoberg et al. 2008; Kutz et al. 2012). Empirical and theoretical studies focusing on Arctic ungulates are providing some of the most compelling documentation of changing host–parasite relationships in response to climate change. Cutting-edge work connecting physiological mechanisms with outcomes for disease dynamics at the population level is feasible in Arctic ungulate study systems due to highly observable host animals and relatively simple parasite communities coupled with a strong regional climate signal.
Disease, trophic cascades, and ecosystem outcomes.
The ubiquity and sheer abundance of parasites in ecological communities begs the question of what role infectious agents play in structuring communities and mediating ecosystem-level fluxes of energy and nutrients. In light of this, parasites are a surprisingly recent addition to trophic web analyses (Sukhdeo 2012), and the majority of such studies have focused on a few well-resolved aquatic and marine food webs (Dunne et al. 2013). In terrestrial ecosystems dominated by large mammals, ungulates occupy a central position as important consumers of primary production and as prey base for a broad range of medium- to large-sized predators (Drent and Olff 1999). As such, both bottom-up and top-down mechanisms may play a role in regulating ungulate populations (Sinclair et al. 2003; Fritz et al. 2011; Hopcraft et al. 2012). Dynamics and ecosystem-level impacts of ungulate infectious diseases, then, should vary with the host species’ roles in the food web. Where ungulate populations exert intense pressure on their food, infectious diseases that reduce animal numbers may release vegetation from curbs on recruitment imposed by ungulate consumption. Such trophic cascades should be especially pronounced where diseases take a large net toll, as may be expected under low predation pressure (due to predator extirpation or large ungulate body size) where disease acts as an independent mortality factor.
Infectious disease studies of ungulates based on 2 ecosystems boasting diverse ungulate and predator assemblages—the Greater Yellowstone Ecosystem in the western United States and East Africa’s Serengeti-Mara ecosystem—are providing tests of 2 central ideas: enhancing predator populations may offer innovative options for disease control through competitive interactions between predators and pathogens and diseases may drive ecosystem dynamics by mediating recruitment limitation of woody plants.
A recent increase in brucellosis prevalence in elk (C. elaphus) herds in the Greater Yellowstone Ecosystem is serious cause for concern (Cross et al. 2010a). Brucellosis (pathogen: Brucella abortus) is a bacterial infection that causes morbidity and abortions in a broad range of host species including cervid and bovid ungulates as well as humans. Preventing spillover of the infection from elk to cattle is likely to prove even more challenging than limiting disease spread from bison because elk are far more abundant than bison and range freely over large swathes of the landscape. The observed increase in brucellosis has been linked to increases in elk population density over the past 20 years and massive expansion of elk winter aggregations coinciding with the seasonal transmission peak for B. abortus (Cross et al. 2010a, 2010b). These increases in elk density and B. abortus seroprevalence have occurred in elk populations outside Yellowstone National Park. At the same time, elk populations inside the park (White et al. 2012) and migratory elk that spend part of the year in the park (Middleton et al. 2013a) have experienced low recruitment and declines in numbers and group size. Direct and indirect effects of predation by wolves and grizzly bears in Yellowstone National Park, climate-driven changes in plant phenology, and competition with elk feeding on irrigated pasture on private land have been implicated as mechanisms driving poor performance of elk in the park (Creel et al. 2011; Griffin et al. 2011; Middleton et al. 2013a, 2013b; Wilmers and Levi 2013). Brucellosis in elk in the park remained stable at low prevalence (Barber-Meyer et al. 2007), and elk declines in the park have been accompanied by increased recruitment of woody vegetation (Ripple and Beschta 2012). While more work is needed to understand the causes underlying elk population dynamics in Yellowstone National Park (Massey et al. 2013), these correlations suggest that predator protection may be effective as part of a strategy both for limiting the spread of brucellosis in elk and for restoring western deciduous woodlands in the United States. Ongoing efforts to restore predators to western United States wild lands and elsewhere may provide opportunities for studying the effects of predators on ungulate disease dynamics through a series of natural experiments.
Ungulate–pathogen–vegetation dynamics in the Serengeti-Mara ecosystem present a contrasting picture. The Serengeti-Mara ecosystem comprises a dynamic patchwork of grassland and wooded habitats. Biotic (herbivores, including elephants) and abiotic (fire, rainfall) drivers influence grassland–woodland dynamics in African savannas (Sankaran et al. 2005; Bucini and Hanan 2007; Bond 2008). In the Serengeti-Mara ecosystem, it is indirect effects of rinderpest, an infectious disease of artiodactyls (even-toed ungulates), that act as a dominant driver of woody plant dynamics (Holdo et al. 2009). Rinderpest is caused by a Morbillivirus that was introduced into the Serengeti-Mara ecosystem with European cattle in the late 19th century. The disease decimated wild bovid populations until the early 1960s, when it disappeared from wildlife, following vaccination of cattle in surrounding areas (McNaughton 1992). The irruption of wildebeest (Connochaetes taurinus) in this ecosystem following rinderpest eradication caused a sharp reduction in fuel load due to increased grazing, which in turn reduced fire frequency and intensity. This promoted woody plant recruitment, shifting more area from grassland to woodland and altering ecosystem-wide nutrient dynamics (Holdo et al. 2009). Infectious diseases can thus play contrasting roles as causes or consequences of trophic cascades in different ecosystems, depending on the interplay between disease and predation, the trophic niche of the host (grazer or browser), and the abiotic context that underlies ecosystem dynamics. Ungulates have proven to be ideal models for exploring these types of dynamics.
Conclusion and future directions
Both past and current works highlight the point that ungulate model systems are poised to help pave the way forward in disease ecology and evolution. There are also clear opportunities for the future where ungulates may provide fundamental insight. For example, emerging infectious diseases are one of the most pressing public health concerns of our time (Jones et al. 2008), and in a now classic assessment of the characteristics of emerging pathogens of humans, Taylor et al. (2001) estimated that over 60% of human emerging infectious diseases are transmissible between humans and animals, and 57% of these emerging zoonoses were linked to wild ungulate hosts (Cleaveland et al. 2001). These statistics suggest that wild ungulates are an important conduit of pathogens to humans making them a relevant focus group for pathogen discovery studies, similar to current work being done in wild primates (Calvignac-Spencer et al. 2012). Future studies investigating the diversity of infectious agents in wild ungulate populations may shed light on likely taxonomic and geographic sources of new pathogens. Because one explanation for why wild ungulates are such an important source of human pathogens is their relatedness to livestock species (Wolfe et al. 2007), studies of wild ungulates are also well poised to provide insights into the drivers of emergence. For instance, are contact rates between wild and domestic ungulates important predictors of novel pathogen spillover into human populations?
In a translational context, wild model systems have much to contribute to the discovery of new approaches to reduce the global impact of disease in human populations, impacts that might come about via direct effects of disease on human health or indirect effects mediated by economic losses (e.g., via livestock production). Wild animals contend with a diverse suite of parasites and pathogens under a range of environmental conditions, and studying model organisms in their natural environment may unveil new mechanisms by which hosts resist and tolerate infection. Studying ungulates in particular could inform disease control and prevention in livestock species. Overall, insights from wild mammal systems are fueling new discoveries and driving new paradigms in many fields in the biological sciences (Manceau et al. 2011; Seifert et al. 2012). For scientists focused on the study of infectious diseases, the utility of using natural systems to probe complex interactions between hosts and pathogens is increasingly recognized (Jackson et al. 2011). The skill sets and knowledge of disease ecologists and mammalogists are highly complementary and present strong opportunities for collaborative research aimed at understanding disease processes in natural populations: disease ecologists study parasite population dynamics and offer experimental and quantitative skills to develop and parameterize mathematical models that represent disease dynamics. Mammalogists bring to the table in-depth knowledge about the biology of mammalian host populations as well as essential skills for working with free-ranging mammals and analyzing the resulting complex data sets. The time is ripe, therefore, for mammalogists and disease ecologists to join forces and develop a range of wild ungulate systems as new wild models of infectious disease.
Supplementary Material
Acknowledgments
We thank R. Ostfeld for organizing the symposium that sparked this contribution. Picture credits (Fig. 2): A. Ozgul and G. Prior. This work was supported by the National Science Foundation Ecology of Infectious Diseases Program (EF-1102493, EF-0723928). VOE also received support from a National Science Foundation CAREER Award (IOS-1101836).
Literature Cited
- Akinyi M. Y., Tung J., Jeneby M., Patel N. B., Altmann J., Alberts S. C. 2013. Role of grooming in reducing tick load in wild baboons (Papio cynocephalus). Animal Behaviour 85:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albon S. D., Stein A., Irvine R. J., Langvatn R., Ropstad E., Halvorsen O. 2002. The role of parasites in the dynamics of reindeer population. Proceedings of the Royal Society of London, B. Biological Sciences 269:1625–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altizer S., Ostfeld R. S., Johnson P. T. J., Kutz S., Harvell C. D. 2013. Climate change and infectious diseases: from evidence to a predictive framework. Science 341:514–519. [DOI] [PubMed] [Google Scholar]
- Anderson R. M., May R. M. 1978. Regulation and stability of host-parasite population interactions: I. Regulatory processes. Journal of Animal Ecology 47:219–247. [Google Scholar]
- Anderson R. M., May R. M. 1991. Infectious diseases of humans: dynamics and control. Oxford University Press, New York. [Google Scholar]
- Apollonio M., Andersen R., Putman R. 2010. European ungulates and their management in the 21st century. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Ayash-Rashkovsky M., et al. 2007. Coinfection with Schistosoma mansoni reactivates viremia in rhesus macaques with chronic simian-human immunodeficiency virus clade C infection. Infection and Immunity 75:1751–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber I., Dingemanse N. J. 2010. Parasitism and the evolutionary ecology of animal personality. Philosophical Transactions of the Royal Society B 365:4077–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber-Meyer S. M., White P. J., Mech D. 2007. Survey of selected pathogens and blood parameters of northern Yellowstone elk: wolf sanitation effect implications. American Midland Naturalist 158:369–381. [Google Scholar]
- Beechler B. R., Broughton H., Bell A., Ezenwa V. O., Jolles A. E. 2012. Innate immunity in free-ranging African buffalo (Syncerus caffer): associations with parasite infection and white blood cell counts. Physiological and Biochemical Zoology 85:255–264. [DOI] [PubMed] [Google Scholar]
- Benes C. H. 2013. Functionalizing genomic data for personalization of medicine. Clinical Pharmacology & Therapeutics 93:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraldi D., et al. 2007. Quantitative trait loci (QTL) mapping of resistance to strongyles and coccidia in the free-living Soay sheep (Ovis aries). International Journal for Parasitology 37:121–129. [DOI] [PubMed] [Google Scholar]
- Bond W. J. 2008. What limits trees in C4 grasslands and savannas? Annual Review of Ecology and Evolution, and Systematics 39:641–659. [Google Scholar]
- Brown E. A., et al. 2013. Detecting genes for variation in parasite burden and immunological traits in a wild population: testing the candidate gene approach. Molecular Ecology 22:757–773. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B. 2004. Toward a metabolic theory of ecology. Ecology 85:1771–1789. [Google Scholar]
- Bucini G., Hanan N. P. 2007. A continental-scale analysis of tree cover in African savannas. Global Ecology and Biogeography 16:593–605. [Google Scholar]
- Buer J., Balling R. 2003. Mice, microbes and models of infection. Nature Reviews Genetics 4:195–205. [DOI] [PubMed] [Google Scholar]
- Calvignac-Spencer S., Leendertz S., Gillespie T., Leendertz F. 2012. Wild great apes as sentinels and sources of infectious disease. Clinical Microbiology and Infection 18:521–527. [DOI] [PubMed] [Google Scholar]
- Chenine A.-L., et al. 2008. Acute Schistosoma mansoni infection increases susceptibility to systemic SHIV clade C infection in rhesus macaques after mucosal virus exposure. PLoS Neglected Tropical Diseases 2: e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinwalla A. T., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. [DOI] [PubMed] [Google Scholar]
- Cleaveland S., Laurenson M. K., Taylor L. H. 2001. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Proceedings of the Royal Society of London, B. Biological Sciences 356:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock T. H., Pemberton J. M. 2004. Soay sheep: dynamics and selection in an island population. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Coers J., Starnbach M. N., Howard J. C. 2009. Modeling infectious disease in mice: co-adaptation and the role of host-specific IFNγ responses. PLoS Pathogens 5:e1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman D. W., Pilkington J. G., Kruuk L. E. B., Wilson K., Pemberton J. M. 2001b. Positive genetic correlation between parasite resistance and body size in a free-living ungulate population. Evolution 55:2116–2125. [DOI] [PubMed] [Google Scholar]
- Coltman D. W., Pilkington J. G., Smith J. A., Pemberton J. M. 1999. Parasite-mediated selection against inbred Soay sheep in a free-living island population. Evolution 53:1259–1267. [DOI] [PubMed] [Google Scholar]
- Coltman D. W., Wilson K., Pilkington J. G., Stear M. J., Pemberton J. M. 2001a. A microsatellite polymorphism in the gamma interferon gene is associated with resistance to gastrointestinal nematodes in a naturally-parasitized population of Soay sheep. Parasitology 122:571–582. [DOI] [PubMed] [Google Scholar]
- Craig B. H., Pilkington J. G., Kruuk L. E. B., Pemberton J. M. 2007. Epidemiology of parasitic protozoan infections in Soay sheep (Ovis aries L.) on St Kilda. Parasitology 134:9–21. [DOI] [PubMed] [Google Scholar]
- Craig B. H., Pilkington J. G., Pemberton J. M. 2006. Gastrointestinal nematode species burdens and host mortality in a feral sheep population. Parasitology 133:485–496. [DOI] [PubMed] [Google Scholar]
- Creel S., Cunningham D. A., Winnie J. A. 2011. A survey of the effects of wolf predation risk on pregnancy rates and calf recruitment in elk. Ecological Applications 2:2847–2853. [Google Scholar]
- Cross P. C., et al. 2010a. Probable causes of increasing brucellosis in free-ranging elk of the Greater Yellowstone Ecosystem. Ecological Applications 20:278–288. [DOI] [PubMed] [Google Scholar]
- Cross P. C., Heisey D. M., Scurlock B. M., Edwards W. H., Ebinger M. R., Brennan A. 2010b. Mapping brucellosis increases relative to elk density using hierarchical Bayesian models. PLoS One 5:e10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R., Simard M., Kutz S. J., Kapel C. M. O., Hamnes I. S., Robertson L. J. 2011. Arctic parasitology: why should we care? Trends in Parasitology 27:238–244. [DOI] [PubMed] [Google Scholar]
- de Wit E., et al. 2014. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proceedings of the National Academy of Sciences 110:16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. P., Hudson P. J. 1992. Regulation and stability of a free-living host-parasite system: Trichostrongylus tenuis in red grouse. II. Population models. Journal of Animal Ecology 61:487–498. [Google Scholar]
- Drent R. H., Olff H. 1999. Herbivores: between plants and predators. Symposia of the British Ecological Society. Blackwell Science, Oxford, United Kingdom. [Google Scholar]
- Drewe J. A., O’Riain M. J., Beamish E., Currie H., Parsons S. 2012. Survey of infections transmissible between baboons and humans, Cape Town, South Africa. Emerging Infectious Diseases 18:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn A. M., et al. 2012. Invasions and infections: indirect effects of parasites in invasions. Functional Ecology 26:1262–1274. [Google Scholar]
- Dunne J. A., et al. 2013. Parasites affect food web structure primarily through increased diversity and complexity. PLoS Biology 11:e1001579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezenwa V. O., Etienne R. S., Luikart G., Beja-Pereira A., Jolles A. E. 2010. Hidden consequences of living in a wormy world: nematode-induced immune suppression facilitates tuberculosis invasion in African buffalo. American Naturalist 176:613–624. [DOI] [PubMed] [Google Scholar]
- Falconi C., López-Olvera J. R., Gortázar C. 2011. BTV infection in wild ruminants, with emphasis on red deer: a review. Veterinary Microbiology 151:209–219. [DOI] [PubMed] [Google Scholar]
- Falzarano D., et al. 2013. Treatment with interferon-alpha 2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nature Medicine 19:1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz H., Loreau M., Chamaille-Jammes S., Valeix M., Clobert J. 2011. A food web perspective on large herbivore community limitation. Ecography 34:196–202. [Google Scholar]
- Gardner M. B., Luciw P. A. 2008. Macaque models of human infectious disease. ILAR Journal 49:220–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly J. F., Brown J. H., West G. B., Savage V. M., Charnov E. L. 2001. Effects of size and temperature on metabolic rate. Science 293:2248–2251. [DOI] [PubMed] [Google Scholar]
- Gorsich E. E., Bengis R., Ezenwa V. O., Jolles A. E. 2014. Evaluation of sensitivity and specificity of an ELISA for diagnosis of brucellosis in African buffalo using Bayesian latent class modeling. Journal of Wildlife Disease. [DOI] [PubMed] [Google Scholar]
- Graham A. L., Hayward A. D., Watt K. A., Pilkington J. G., Pemberton J. M., Nussey D. H. 2010. Fitness correlates of heritable variation in antibody responsiveness in a wild mammal. Science 330:662–665. [DOI] [PubMed] [Google Scholar]
- Griffin K. A., et al. 2011. Neonatal mortality of elk driven by climate, predator phenology and predator community composition. Journal of Animal Ecology 80:1246–1257. [DOI] [PubMed] [Google Scholar]
- Grobler D. G., Michel A. L., De Klerk L. M., Bengis R. G. 2002. The gamma-interferon test: its usefulness in a bovine tuberculosis survey in African buffaloes (Syncerus caffer) in the Kruger National Park. Onderstepoort Journal of Veterinary Research 69:221–227. [PubMed] [Google Scholar]
- Groves C., Grubb P. 2011. Ungulate taxonomy. Johns Hopkins University Press, Baltimore, Maryland. [Google Scholar]
- Halvorsen O. 2012. Reindeer parasites, weather and warming of the Arctic. Polar Biology 35:1749–1752. [Google Scholar]
- Harvell C. D., et al. 2002. Climate warming and disease risks for terrestrial and marine biota. Science 296:2158–2162. [DOI] [PubMed] [Google Scholar]
- Harvell C. D., Altizer S., Cattadori I. M., Harrington L., Weil E. 2009. Climate change and wildlife diseases: when does the host matter the most? Ecology 90:912–920. [DOI] [PubMed] [Google Scholar]
- Hayward A. D., Wilson A. J., Pilkington J. G., Kruuk L. E. B. 2009. Ageing in a variable habitat: environmental stress affects senescence in parasite resistance in St Kilda Soay sheep. Proceedings of the Royal Society of London, B. Biological Sciences 276:3477–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlerigg D. G., Wyse C. A., Dardente H., Hanon E. A., Lincoln G. A. 2013. Photoperiodic variation in CD45-positive cells and cell proliferation in the mediobasal hypothalamus of the Soay sheep. Chronobiology International 30:548–558. [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., Polley L., Jenkins E. J., Kutz S. J., Veitch A. M., Elkin B. T. 2008. Integrated approaches and empirical models for investigation of parasitic diseases in northern wildlife. Emerging Infectious Diseases 14:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdo R. M., et al. 2009. A disease-mediated trophic cascade in the Serengeti and its implications for ecosystem C. PLoS Biology 7:e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopcraft J. G. C., Anderson T. M., Perez-Vila S., Mayemba E., Olff H. 2012. Body size and the division of niche space: food and predation differentially shape the distribution of Serengeti grazers. Journal of Animal Ecology 81:201–213. [DOI] [PubMed] [Google Scholar]
- Hutchings M. R., Gordon I. J., Kyriazakis I., Robertson E., Jackson F. 2002a. Grazing in heterogeneous environments: infra- and supra-parasite distributions determine herbivore grazing decisions. Oecologia 132:453–460. [DOI] [PubMed] [Google Scholar]
- Hutchings M. R., Milner J. M., Gordon I. J., Kyriazakis I., Jackson F. 2002b. Grazing decisions of Soay sheep, Ovis aries, on St Kilda: a consequence of parasite distribution? Oikos 96:235–244. [Google Scholar]
- Ideker T., Galitski T., Hood L. 2001. A new approach to decoding life: systems biology. Annual Review of Genomics and Human Genetics 2:343–372. [DOI] [PubMed] [Google Scholar]
- Jackson J. A., et al. 2011. The analysis of immunological profiles in wild animals: a case study on immunodynamics in the field vole, Microtus agrestis. Molecular Ecology 20:893–909. [DOI] [PubMed] [Google Scholar]
- Jenkins E. J., Schurer J. M., Gesy K. M. 2011. Old problems on a new playing field: helminth zoonoses transmitted among dogs, wildlife, and people in a changing northern climate. Veterinary Parasitology 182:24–69. [DOI] [PubMed] [Google Scholar]
- Jenkins E. J., Veitch A. M., Kutz S. J., Hoberg E. P., Polley L. 2006. Climate change and the epidemiology of protostrongylid nematodes in northern ecosystems: Parelaphostrongylus odocoilei and Protostrongylus stilesi in Dall’s sheep (Ovis d. dalli). Parasitology 132:387–401. [DOI] [PubMed] [Google Scholar]
- Jolles A. E., Cooper D. V., Levin S. A. 2005. Hidden effects of chronic tuberculosis in African buffalo. Ecology 86:2358–2364. [Google Scholar]
- Jones K. E., et al. 2008. Global trends in emerging infectious diseases. Nature 451:990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. 2002. Systems biology: a brief overview. Science 295:1662–1664. [DOI] [PubMed] [Google Scholar]
- Klein D. R., Bruun H. H., Lundgren R., Philipp M. 2008. Climate change influences on species interrelationships and distributions in high-Arctic Greenland. Advances in Ecological Research 40:81–100. [Google Scholar]
- Kutz S. J., et al. 2012. Parasites in ungulates of Arctic North America and Greenland: a view of contemporary diversity, ecology, and impact in a world under change. Advances in Parasitology 79:99–252. [DOI] [PubMed] [Google Scholar]
- Kutz S. J., Hoberg E. P., Nagy J., Polley L., Elgin B. 2004. “Emerging” parasitic infections in arctic ungulates. Integrative and Comparative Biology 44:109–118. [DOI] [PubMed] [Google Scholar]
- Kutz S. J., Hoberg E. P., Polley L., Jenkins E. J. 2005. Global warming is changing the dynamics of Arctic host-parasite systems. Proceedings of the Royal Society of London, B. Biological Sciences 272:2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laaksonen S., et al. 2010. Climate change promotes the emergence of serious disease outbreaks of filarioid nematodes. EcoHealth 7:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law G. L., Korth M. J., Benecke A. G., Katze M. G. 2013. Systems virology: host-directed approaches to viral pathogenesis and drug targeting. Nature Reviews Microbiology 11:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke G. J., Currie P. D. 2007. Animal models of human disease: zebrafish swim into view. Nature Reviews Genetics 8:353–367. [DOI] [PubMed] [Google Scholar]
- MacLachlan N. J., Guthrie A. J. 2010. Re-emergence of bluetongue, African horse sickness, and other orbivirus diseases. Veterinary Research 41:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M., Domingues V. S., Mallarino R., Hoekstra H. E. 2011. The developmental role of agouti in color pattern evolution. Science 331:1062–1065. [DOI] [PubMed] [Google Scholar]
- Massey J., Cubaynes S., Coulson T. 2013. Will central Wyoming elk stop migrating to Yellowstone, and should we care? Ecology 94:1271–1274. [DOI] [PubMed] [Google Scholar]
- McCoy M. W., Gillooly J. F. 2008. Predicting natural mortality rates of plants and animals. Ecology Letters 11:710–716 (Corrigendum. Ecology Letters 12:731–733). [DOI] [PubMed] [Google Scholar]
- McMichael A. J., Woodruff R. E., Hales S. 2006. Climate change and human health: present and future risks. Lancet 367:859–869. [DOI] [PubMed] [Google Scholar]
- McNaughton S. J. 1992. The propagation of disturbance in savannas through food webs. Journal of Vegetation Science 3:301–314. [Google Scholar]
- Meurens F., Summerfield A., Nauwynck H., Saif L., Gerdts V. 2012. The pig: a model for human infectious diseases. Trends in Microbiology 20:50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A. L., Cooper D., Jooste J., de Klerk L.-M., Jolles A. E. 2011. Approaches towards optimising the gamma interferon assay for diagnosing Mycobacterium bovis infection in African buffalo (Syncerus caffer). Preventive Veterinary Medicine 98:142–151. [DOI] [PubMed] [Google Scholar]
- Middleton A. D., et al. 2013a. Animal migration amid shifting patterns of phenology and predation: lessons from a Yellowstone elk herd. Ecology 94:1245–1256. [DOI] [PubMed] [Google Scholar]
- Middleton A. D., et al. 2013b. Grizzly bear predation links the loss of native trout to the demography of migratory elk in Yellowstone. Proceedings of the Royal Society of London, B. Biological Sciences 280:20130870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min F., Zhang Y., Pan J., Wang J., Yuan W. 2014. Mycobacterium tuberculosis infection in rhesus monkeys (Macaca mulatta) and evaluation of ESAT-6 and CFP10 as immunodiagnostic antigens. Experimental Animals 62:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitruka B. M., Rawnsley H. M., Vadehra D. V. 1976. Animals for medical research: models for the study of human disease. John Wiley, Hoboken, New Jersey. [Google Scholar]
- Molnar P. K., Kutz S. J., Hoar B. M., Dobson A. P. 2013. Metabolic approaches to understanding climate change impacts on seasonal host-macroparasite dynamics. Ecology Letters 16:9–21. [DOI] [PubMed] [Google Scholar]
- Munch S. B., Salinas S. 2009. Latitudinal variation in lifespan within species is explained by the metabolic theory of ecology. Proceedings of the National Academy of Sciences 106:13860–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath B. M., Schumann K. E., Boyer J. D. 2000. The chimpanzee and other non-human-primate models in HIV-1 vaccine research. Trends in Microbiology 8:426–431. [DOI] [PubMed] [Google Scholar]
- National Animal Genome Research Program. 1998–2014. NRSP-8 Bioinformatics Coordination Program http://www.animalgenome.org/ Accessed 1 November 2013.
- Niewiesk S., Prince G. 2002. Diversifying animal models: the use of hispid cotton rats (Sigmodon hispidus) in infectious diseases. Laboratory Animals 36:357–372. [DOI] [PubMed] [Google Scholar]
- Olsen S., Johnson C. 2012. Efficacy of dart or booster vaccination with strain RB51 in protecting bison against experimental Brucella abortus challenge. Clinical and Vaccine Immunology 19:886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S., Wilson K., Pemberton J. M. 1998. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.). Proceedings of the National Academy of Sciences 95:3714–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz J. A., Campbell-Lendrum D., Holloway T., Foley J. A. 2005. Impact of regional climate change on human health. Nature 438:310–317. [DOI] [PubMed] [Google Scholar]
- Pedersen A. B., Babayan S. A. 2011. Wild immunology. Molecular Ecology 20:872–880. [DOI] [PubMed] [Google Scholar]
- Plowright R. K., Manlove K., Cassirer E. F., Cross P. C., Besser T. E., Hudson P. J. 2013. Use of exposure history to identify patterns of immunity to pneumonia in bighorn sheep (Ovis canadensis). PLoS One 8:e61919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J., et al. 2009. Genome-wide cross-amplification of domestic sheep microsatellites in bighorn sheep and mountain goats. Molecular Ecology Resources 9:1121–1126. [DOI] [PubMed] [Google Scholar]
- Post E., et al. 2009. Ecological dynamics across the arctic associated with recent climate change. Science 325:1355–1358. [DOI] [PubMed] [Google Scholar]
- Ripple W. J., Beschta R. L. 2012. Trophic cascades in Yellowstone: the first 15 years after wolf reintroduction. Biological Conservation 145:205–213. [Google Scholar]
- Robinson S. J., Samuel M. D., Johnson C. J., Adams M., McKenzie D. I. 2012. Emerging prion disease drives host selection in a wildlife population. Ecological Applications 22:1050–1059. [DOI] [PubMed] [Google Scholar]
- Rohr J. R., et al. 2011. Frontiers in climate change—disease research. Trends in Ecology and Evolution 26:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran M., et al. 2005. Determinants of woody cover in African savannas. Nature 438:846–849. [DOI] [PubMed] [Google Scholar]
- Schaschl H., Suchentrunk F., Morris D. L., Slimen H. B., Smith S., Arnold W. 2012. Sex-specific selection for MHC variability in Alpine chamois. BMC Evolutionary Biology 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A. W., Kiama S. G., Seifert M. G., Goheen J. R., Palmer T. M., Maden M. 2012. Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489:561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenya A. A., et al. 2012. Schistosoma mansoni infection impairs antimalaria treatment and immune responses of rhesus macaques infected with mosquito-borne Plasmodium coatneyi. Infection and Immunity 80:3821–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J., et al. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proceedings of the National Academy of Sciences 110:3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair A. R. E., Mduma S., Brashares J. S. 2003. Patterns of predation in a diverse predator-prey system. Nature 425:288–290. [DOI] [PubMed] [Google Scholar]
- Sukhdeo M. V. K. 2012. Where are the parasites in food webs? Parasites & Vectors 5:239–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L. H., Latham S. M., Woolhouse M. E. 2001. Risk factors for human disease emergence. Philosophical Transactions of the Royal Society of London, B. Biological Sciences 356:983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer S., et al. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330:243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins D. M., Dunn A. M., Smith M. J., Telfer S. 2011. Review: wildlife diseases: from individuals to ecosystems. Journal of Animal Ecology 80:19–38. [DOI] [PubMed] [Google Scholar]
- Tung J., Primus A., Bouley A. J., Severson T. F., Alberts S. C., Wray G. A. 2009. Evolution of a malaria resistance gene in wild primates. Nature 460:388–391. [DOI] [PubMed] [Google Scholar]
- Verocai G. G., et al. 2012. Defining parasite biodiversity at high latitudes of North America: new host and geographic records for Onchocerca cervipedis (Nematoda: Onchocercidae) in moose and caribou. Parasites and Vectors 5:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Zhou J., Zaman N., Beitel L. K., Trifiro M., Paliouras M. 2013. Cancer systems biology in the genome sequencing era. Part 1, dissecting and modeling of tumor clones and their networks. Seminars in Cancer Biology 23:279–285. [DOI] [PubMed] [Google Scholar]
- Weinstock G. M. 2012. Genomic approaches to studying the human microbiota. Nature 489:250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyher A. H., Ross C., Semple S. 2006. Gastrointestinal parasites in crop raiding and wild foraging Papio anubis in Nigeria. International Journal of Primatology 27:1519–1534. [Google Scholar]
- White A. D., et al. 2013a. Evaluation of the safety and immunogenicity of a candidate tuberculosis vaccine, MVA85A, delivered by aerosol to the lungs of macaques. Clinical and Vaccine Immunology 20:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. K., et al. 2013b. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. J., Proffitt K. M., Lemke T. O. 2012. Changes in elk distribution and group sizes after wolf restoration. American Midland Naturalist 167:174–187. [Google Scholar]
- Wilmers C. C., Levi T. 2013. Do irrigation and predator control reduce the productivity of migratory ungulate herds? Ecology 94:1264–1270. [DOI] [PubMed] [Google Scholar]
- Wilson K., Grenfell B. T., Pilkington J. G., Boyd H. E. G., Gulland F. M. D. 2004. Parasites and their impact. Pp. 113–165 in Soay sheep: dynamics and selection in an island population (Clutton-Brock T. H., Pemberton J. M., eds.). Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Witter L. A., Johnson C. J., Croft B., Gunn A., Gillingham M. P. 2012a. Behavioural trade-offs in response to external stimuli: time allocation of an Arctic ungulate during varying intensities of harassment by parasitic flies. Journal of Animal Ecology 81:284–295. [DOI] [PubMed] [Google Scholar]
- Witter L. A., Johnson C. J., Croft B., Gunn A., Poirier L. M. 2012b. Gauging climate change effects at local scales: weather-based indices to monitor insect harassment in caribou. Ecological Applications 22:1836–1851. [DOI] [PubMed] [Google Scholar]
- Wolfe N. D., Dunavan C. P., Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe N. D., Escalante A. A., Karesh W. B., Kilbourn A., Spielman A., Lal A. A. 1998. Wild primate populations in emerging infectious disease research: the missing link? Emerging Infectious Diseases 4:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytrehus B., Bretten T., Bersjo B., Isaksen K. 2008. Fatal pneumonia epizootic in musk ox (Ovibos moschatus) in a period of extraordinary weather conditions. EcoHealth 5:213–223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.