Abstract
Background
A number of serious human adenovirus (HAdV) outbreaks have been recently reported: HAdV-B7 (Israel, Singapore, and USA), HAdV-B7d (USA and China), HAdV-D8, -D54, and -C2 (Japan), HAdV-B14p1 (USA, Europe, and China), and HAdV-B55 (China, Singapore, and France).
Methods
To understand the epidemiology of HAdV infections in Singapore, we studied 533 HAdV-positive clinical samples collected from 396 pediatric and 137 adult patients in Singapore from 2012 to 2018. Genome sequencing and phylogenetic analyses were performed to identify HAdV genotypes, clonal clusters, and recombinant or novel HAdVs.
Results
The most prevalent genotypes identified were HAdV-B3 (35.6%), HAdV-B7 (15.4%), and HAdV-E4 (15.2%). We detected 4 new HAdV-C strains and detected incursions with HAdV-B7 (odds ratio [OR], 14.6; 95% confidence interval [CI], 4.1–52.0) and HAdV-E4 (OR, 13.6; 95% CI, 3.9–46.7) among pediatric patients over time. In addition, immunocompromised patients (adjusted OR [aOR], 11.4; 95% CI, 3.8–34.8) and patients infected with HAdV-C2 (aOR, 8.5; 95% CI, 1.5–48.0), HAdV-B7 (aOR, 3.7; 95% CI, 1.2–10.9), or HAdV-E4 (aOR, 3.2; 95% CI, 1.1–8.9) were at increased risk for severe disease.
Conclusions
Singapore would benefit from more frequent studies of clinical HAdV genotypes to identify patients at risk for severe disease and help guide the use of new antiviral therapies, such as brincidofovir, and potential administration of HAdV 4 and 7 vaccine.
Keywords: adenovirus, genotyping, molecular epidemiology, pediatric disease, respiratory disease
Human adenovirus (HAdV) outbreaks are often inexplicable with sparse clinical epidemiological data. We sought to describe clinical HAdV genotypes and identify risk factors associated with severe disease among HAdV-positive patients seen at 2 hospitals in Singapore.
Human adenoviruses (HAdVs) cause diverse illnesses that often vary by HAdV species (A–G) and type (>90 genotypes have been described) [1]. Human adenovirus types 3, 4, 7, 21, 14, and 55 have been associated with severe acute respiratory disease [2–5]. Although a number of HAdV vaccine types have been or are under current study, live enteric-coated vaccines against HAdV-B7 and HAdV-E4 have been used with remarkable effectiveness and safety among US military trainees [6].
Clinical epidemiological data regarding HAdV infections are relatively sparse for Southeast Asia (SEA). However, existing data suggest that HAdV-B3 is ubiquitous and more often associated with milder illness when compared with recent HAdV-B7 infections in Singapore, Malaysia, Taiwan, and China [7–14]. Unlike HAdV-B3 and HAdV-B7, outbreaks of HAdV-E4 have not been as commonly reported globally until recently. Upon its discovery in the 1950s [15], HAdV-E4 was largely considered restricted to and controlled by vaccine in the US military population, with rare detections among civilians, such as school children in the Netherlands in 1958 [16], users of a private swimming pool in the USA in 1977 [17], and conjunctivitis patients in Japan in 1979 [18]. More recently, surveillance for HAdV has improved, and evidence of HAdV-E4 infections among civilian populations in the USA, Italy, West Africa, China, Hong Kong, Taiwan, India, Malaysia, and Singapore have been increasingly reported [8, 19–28]. Due to the historical lack of HAdV genotyping surveillance, it is unknown whether HAdV-E4 is a new or re-emerging pathogen in SEA. In an effort to epidemiologically describe clinical HAdV genotypes in Singapore, we used a hexon and fiber genotyping algorithm [29] to retrospectively and prospectively study HAdV infections among patients from 2 large public hospitals in Singapore.
METHODS
This study was approved by the SingHealth Centralised Institutional Review Board (CIRB Reference 2015/2773) to study archived and prospectively collected HAdV-positive specimens.
Retrospective Patient Sample Collection
Clinical samples previously collected from HAdV-positive patients admitted to Singapore General Hospital (SGH) and KK Women’s and Children’s Hospital (KKH) between 2012 and 2015 were preserved at −80°C and transferred to Duke-NUS Laboratory of One Health Research for study. Clinical records were accessed only to determine clinical sample type.
Prospective Patient Sample Collection
From 2015 to 2018, informed consent was sought from all patients with laboratory-confirmed adenovirus infection at SGH and KKH. A patient with laboratory-confirmed adenovirus infection was defined as a person of any age seeking healthcare and who had evidence of adenovirus infection by polymerase chain reaction (PCR), real-time PCR, commercial direct fluorescent antibody (DFA), culture, specific monoclonal antibody assay, or other clinically approved adenovirus tests. The clinical samples (nasal, pharyngeal, nasopharyngeal, throat, bronchoalveolar, blood, serum, conjunctival, or urine) were first clinically screened with commercial DFA or Seeplex RV15 ACE PCR targeting influenza A and B virus, parainfluenza 1–4 virus, respiratory syncytial virus, metapneumovirus, rhinovirus, enterovirus, human coronavirus OC43 and 229E/NL63, adenovirus, and bocavirus. Residual samples were then preserved at −80°C. Clinical records were reviewed and Specimen Information Forms were completed. Human adenovirus-positive samples were transported on dry ice to the SGH Department of Microbiology where they were divided into two 0.5-mL aliquots (1 kept at SGH and 1 transferred to Duke-NUS’s Laboratory of One Health Research). Human adenovirus-positive patients with an influenza virus codetection were excluded from our study because these samples were required for study at Singapore’s National Public Health Laboratory.
Human Adenovirus Isolation, Deoxyribonucleic Acid Extraction, and Viral Typing
Human adenovirus culture was attempted at Duke-NUS. A 500-μL aliquot of each sample was inoculated into A549 cells with Dulbecco’s modified Eagle’s medium 2% (v/v) fetal bovine serum and incubated at 37°C while controlling for bacterial contamination. Inoculated shell vials were observed for cytopathic effect 72 hours after inoculation and daily afterward for 10 days. Observation of cytopathic effect was used to score HAdV-positive culture.
At Duke-NUS, deoxyribonucleic acid (DNA) was extracted from the culture supernatants of HAdV-positive samples. For culture-negative HAdV samples, DNA was extracted directly from the original clinical sample. Total DNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Inc., Valentica, CA) and typed using conventional PCR [29, 30] targeting the hexon (predicted amplicon size: 764–896 base pairs [bp]) and fiber gene (predicted amplicon size according to species: A 1444–1537 bp; B 670-772 bp; C 1988–2000 bp; D 1205–1221 bp; E 967 bp; F 541–586 bp). Positive PCR bands were purified using a QIAquick PCR Purification kit (QIAGEN, Inc.) before submission to a local sequencing company (AITbiotech Pte Ltd, Singapore) for Sanger sequencing.
At SGH, DNA was extracted from clinical samples using the QIAamp MinElute Virus Spin Kit (QIAGEN, Inc.). The hexon gene typing strategy [29] was independently performed, and in-house sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on the ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). Results from the typing efforts at each institution were then compared and the HAdV genotype was confirmed.
Whole-Genome Sequencing and Phylogenetic Analysis
Deoxyribonucleic acid extracted from culture supernatants from 10 culture-positive HAdV samples were sent for whole-genome sequencing at the Duke-NUS Genome Biology Facility. A Qubit High Sensitivity assay was used to measure DNA concentrations (ng/μL) in each sample before library preparation using the Nextera XT DNA library prep kit. Samples were then studied using the Miseq 250 × 2 bp (500 cycle) Nano kit v2. The quality of short NGS reads were initially checked by FastQC and followed by the removal of the Illumina adaptors using Trimmomatic [31]. The trimmed reads were then mapped against the reference genomes (HAdV-C or HAdV-E) using Geneinous R9.0.3 (Biomatters Ltd., Auckland, New Zealand).
A total of 10 new adenovirus genomes were generated from this study and compared with all available HAdV-C and HAdV-E sequences of fiber, hexon, and penton base and full genomes from GenBank. For each dataset, multiple sequence alignment was initially performed using MAFFT [32] followed by manual alignment. Gene and full genome phylogenies were initially reconstructed using FastTree [33], and the outliers were removed for subsequent analysis. For HAdV-C viruses, 4 final datasets were further subsampled to 84 fiber sequences (1759 bp in length), 72 hexon sequences (2928 bp), 66 penton base sequences (1725 bp), and 46 full genomes (36 445 bp). For HAdV-E viruses, another 4 final datasets were also subsampled to 46 fiber sequences (1278 bp in length), 39 hexon sequences (2811 bp), 39 penton base sequences (1608 bp) and 37 full genomes (36 397 bp). For each dataset, maximum likelihood (ML) phylogenies were reconstructed using RAxML v8.0 [34], and branch support was assessed using 1000 bootstrap (BS) replicates.
Recombination Analysis of Human Adenovirus Species C and E
To detect potential recombinant events, full genome datasets were analyzed separately for HAdV-C and HAdV-E, because these were the species identified from the subset of isolates (n = 10) that were fully sequenced. Human adenovirus-C datasets consisted of 4 novel genomes from Singapore and 22 representative virus isolates from different genotypes (HAdV-C1, HAdV-C2, HAdV-C5, and HAdV-C6). Human adenovirus-E datasets consisted of 6 novel genomes and 40 representative virus isolates of HAdV-E4. The datasets were analyzed using different methods as implemented in Recombination Detection Program (RDP4) [35]. These methods included RDP, GENECONV, Bootscan, MaxChi, Chimaera, SiScan, and 3Seq and their P values estimated. The breakpoints positions and their major and minor parents were identified for putative recombination strains. For each novel genome, a subsampled dataset was subsequently analyzed in RDP to determine their recombination events. To further confirm detected recombination events, independent phylogenetic analyses were performed using ML method as described above.
Statistical Methods
Epidemiological data were imported and categorized for statistical analyses using STATA, version 15.0 (StataCorp, College Station, TX). Logistic regression was performed to examine HAdV genotype frequencies by time period. Pearson’s χ 2 and Fisher’s exact tests were used to examine potential risk factors (gender, age group, immunosuppression, and HAdV genotype) for associations with severe disease (prolonged hospitalization, prolonged fever, or intensive care unit admission). Stepwise, unconditional logistic regression using a saturated model and backward elimination (exclusion P > .05) was used to further examine these risk factors.
RESULTS
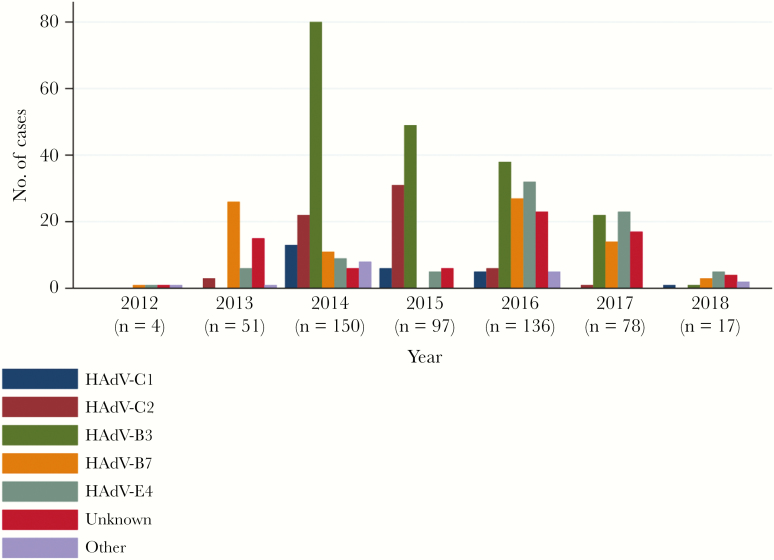
Clinical samples were studied from 533 HAdV-positive patients admitted at either of the 2 hospitals from 2012 to 2018. Partial genome (hexon and fiber) sequencing was attempted on all 533 samples, 458 (86%) of which were successfully genotyped. The most prevalent genotypes identified were HAdV-B3 (35.6%), HAdV-B7 (15.4%), and HAdV-E4 (15.2%) followed by HAdV-C2 (11.8%) and HAdV-C1 (4.9%). See Figure 1 for HAdV genotype frequencies by year.
Figure 1.
Human adenovirus (HAdV) genotype frequencies among HAdV-positive patients hospitalized in Singapore, by year. Years 2012–2013 represent an adult cohort among which 35 cases have been previously described [13]. Year 2014 represents pediatric cases only. Patient recruitment ceased in March of year 2018. “Unknown” represents samples of which genotyping failed. “Other” represents HAdV-C5, -C6, -B11p, -B21, -B35, -D8, or -D53.
Retrospective Patients
Among the 300 retrospective cases (245 pediatric and 55 adult), HAdV-B7 was most prevalent from 2012 to 2013, which was consistent with previous literature for Singapore [36].
Prospective Patients
A total of 233 (151 pediatric and 82 adult) prospective HAdV-positive patients provided informed consent, and their samples (89% respiratory) and clinical data were included in the study (Table 1). Twenty-two immunocompromised patients were included in the study and described separately (Table 2). Human adenovirus-B3 (26.2%) was most prevalent among prospective cases, followed by HAdV-E4 (25.8%) and HAdV-B7 (18.5%) (Table 1). Lastly, immunocompromised patients (adjusted odds ratio [aOR], 11.4; 95% confidence interval [CI], 3.8–34.8) and patients infected with HAdV-C2 (aOR, 8.5; 95% CI, 1.5–48.0), HAdV-B7 (aOR, 3.7; 95% CI, 1.2–10.9), HAdV-E4 (aOR, 3.2; 95% CI, 1.1–8.9), or an unknown HAdV genotype (aOR, 5.7; 95% CI, 2.0–16.2) were found to have an increased risk for severe disease (Table 3).
Table 1.
Medical and Genotyping Information for Prospectively Studied HAdV-Positive Patients Hospitalized in Singapore From 2015 to 2018, by Age Group
| Total | Pediatrica | Adult | |
|---|---|---|---|
| Variables | n = 233 (%) | n = 151 (%) | n = 82 (%) |
| Gender | |||
| Female | 98 (42.1) | 57 (37.8) | 41 (50.0) |
| Male | 135 (57.9) | 94 (62.2) | 41 (50.0) |
| Clinical Presentation | |||
| Respiratory | 204 (87.6) | 128 (84.8) | 76 (92.7) |
| Gastrointestinal | 79 (33.9) | 69 (45.7) | 10 (12.2) |
| Genitourinary | 13 (5.6) | 13 (8.6) | -- |
| Ocular | 59 (25.3) | 58 (38.4) | 1 (1.2) |
| Other | 13 (5.6) | 7 (4.6) | 6 (7.3) |
| Chronic disease (resp/cardiac) | 3 (1.3) | -- | 3 (3.7) |
| Immunocompromisedb | 22 (9.4) | 3 (2.0) | 19 (23.2) |
| Disease Severity | |||
| Prolonged hospitalizationc | 44 (19.0) | 17 (11.3) | 27 (32.9) |
| Prolonged feverd | 23 (9.9) | 13 (8.6) | 10 (12.2) |
| ICU admission | 6 (2.6) | 2 (1.3) | 4 (4.9) |
| HAdV Genotype | |||
| C1 | 6 (2.6) | 4 (2.6) | 2 (2.4) |
| C2 | 8 (3.4) | 6 (4.0) | 2 (2.4) |
| B3 | 61 (26.2) | 57 (37.7) | 4 (4.9) |
| B7 | 44 (18.9) | 32 (21.2) | 12 (14.6) |
| E4 | 60 (25.8) | 38 (25.2) | 22 (26.8) |
| Unknowne | 47 (20.7) | 10 (6.6) | 37 (45.1) |
| Otherf | 7 (3.0) | 4 (2.6) | 3 (3.7) |
Abbreviations: HAdV, human adenovirus; ICU, intensive care unit; resp, respiratory.
aPediatric (≤16 years).
bImmunocompromised within 6 months before clinical sample collection.
cHospitalized for more than 7 consecutive days.
dFever lasting more than 7 days.
eGenotyping failed.
fHAdV-C5, -C6, -B11p, -B21, -B35, -D8, or -D53.
Table 2.
Medical and Genotyping Information for Prospectively Studied HAdV-Positive Immunocompromiseda Patients Hospitalized in Singapore, 2015–2018
| Total | Pediatricb | Adult | |
|---|---|---|---|
| Variables | n = 22 (%) | n = 3 (%) | n = 19 (%) |
| Gender | |||
| Female | 10 (45.5) | 1 (33.3) | 9 (47.4) |
| Male | 12 (54.5) | 2 (66.7) | 10 (52.6) |
| Clinical Presentation | |||
| Respiratory | 13 (59.1) | 1 (33.3) | 12 (63.2) |
| Gastrointestinal | 7 (31.8) | 2 (66.7) | 5 (26.3) |
| Genitourinary | -- | -- | -- |
| Ocular | 1 (4.5) | 1 (33.3) | -- |
| Other | 2 (9.1) | -- | 2 (10.5) |
| Cause of Immunodeficiency | |||
| End-stage renal failure; diabetes mellitus | 1 (4.5) | -- | 1 (5.3) |
| Stem cell transplant | 5 (22.7) | 1 (33.3) | 4 (21.0) |
| Kidney transplant | 1 (4.5) | -- | 1 (5.3) |
| HIV-positive | 2 (9.1%) | -- | 2 (10.5) |
| Nontransplant/other | 13 (59.1) | 2 (66.7) | 11 (57.9) |
| Disease Severity | |||
| Prolonged hospitalizationc | 17 (77.3) | 3 (100) | 14 (73.7) |
| Prolonged feverd | 2 (9.1) | 1 (33.3) | 1 (5.3) |
| ICU admission | 1 (4.5) | -- | 1 (5.3) |
| HAdV Genotype | |||
| C2 | 1 (4.5) | -- | 1 (5.3) |
| C5 | 1 (4.5) | 1 (33.3 | -- |
| B3 | 4 (18.2) | 1 (33.3) | 3 (15.8) |
| B7 | 3 (13.6) | -- | 3 (15.8) |
| E4 | 4 (18.2) | -- | 4 (21.0) |
| Unknowne | 9 (40.9) | 1 (33.3) | 8 (42.1) |
| Otherf | -- | -- | -- |
Abbreviations: HAdV, human adenovirus; HIV, human immunodeficiency virus; ICU, intensive care unit.
aImmunocompromised within 6 months before clinical sample collection.
bPediatric (≤16 years).
cHospitalized for more than 7 consecutive days.
dFever lasting more than 7 days.
eGenotyping failed.
fHAdV-C1, C6, -B11p, -B21, -B35, -D8, or -D53.
Table 3.
Risk Factor Associations for Severe Disease Among Prospectively Studied HAdV-Positive Patients Hospitalized in Singapore, 2015–2018
| Total | Severe Diseasea | Unadjusted | Adjustedb | |
|---|---|---|---|---|
| Risk Factors | n = 233 (%) | n = 65 (%) | OR (95% CI) | OR (95% CI) |
| Gender | ||||
| Female | 98 (42.1) | 22 (33.8) | Reference | -- |
| Male | 135 (57.9) | 43 (66.2) | 1.6 (0.9–2.9) | -- |
| Age Quartiles (Q) | ||||
| ≤2.5 years (Q1) | 70 (30.2) | 18 (27.7) | Reference | -- |
| 3–4.5 years (Q2) | 48 (20.7) | 9 (13.8) | 0.7 (0.3–1.6) | -- |
| 5–43.5 years (Q3) | 56 (24.1) | 13 (20.0) | 0.9 (0.4–2.0) | -- |
| ≥44 years (Q4) | 58 (25.0) | 25 (38.5) | 2.2 (1.0–4.6) | -- |
| Immunocompromisedc | ||||
| Yes | 22 (9.4) | 17 (26.2) | 11.5 (4.0–32.9) | 11.4 (3.8–34.8) |
| No | 211 (90.6) | 48 (73.8) | Reference | Reference |
| HAdV genotype | ||||
| C1 | 6 (2.6) | 1 (1.5) | 1.5 (0.2–15.2) | 2.1 (0.2–21.4) |
| C2 | 8 (3.4) | 5 (7.7) | 12.9 (2.5–65.9) | 14.6 (2.7–80.4) |
| B3 | 61 (26.2) | 7 (10.8) | Reference | Reference |
| B7 | 44 (18.9) | 13 (20.0) | 3.2 (1.2–9.0) | 3.7 (1.2–10.9) |
| E4 | 60 (25.8) | 16 (24.6) | 2.8 (1.1–7.4) | 3.2 (1.1–8.9) |
| Unknownd | 47 (20.7) | 21 (32.3) | 6.2 (2.4–16.5) | 5.7 (2.0–16.2) |
| Othere | 7 (3.0) | 2 (3.1) | 3.1 (0.5–19.0) | 2.8 (0.4–20.4) |
Abbreviations: CI, confidence interval; HAdV, human adenovirus; OR, odds ratio.
aProlonged hospitalization, prolonged fever, or intensive care unit admission.
bAfter stepwise logistic regression using saturated model and backward elimination of covariates with P > .05.
cImmunocompromised within 6 months before clinical sample collection.
dGenotyping failed.
eHAdV-C5, -C6, -B11p, -B21, -B35, -D8, or -D53.
Pediatric Patients
Overall, 396 (74.3%) of the clinical samples were collected from the pediatric population (≤16 years). Among the pediatric cases, the most prevalent genotypes identified were HAdV-B3 (47.0%), HAdV-C2 (14.6%), and HAdV-E4 (13.1%). Among the pediatric samples, 245 (61.9%) were retrospective samples collected from 2014 to 2015, and the remaining 151 (38.1%) were prospectively collected from 2016 to 2018. Pediatric patients hospitalized during 2016–2018 had an increased risk of HAdV-E4 infection (OR, 13.6; 95% CI, 3.9–46.7) and HAdV-B7 infection (OR, 14.6; 95% CI, 4.1–52.0) when compared with pediatric patients hospitalized from 2014 to 2015. Among the pediatric cohort, age <1 year (aOR, 17.9; 95% CI, 1.3–240) and infection with HAdV-C2 (aOR, 23.1; 95% CI, 3.1–173.3) or an unknown HAdV genotype (aOR, 9.8; 95% CI, 1.7–56.3) were risk factors for severe disease (Table 4).
Table 4.
Risk Factor Associations for Severe Disease Among Prospectively Studied HAdV-Positive Pediatric Patients Hospitalized in Singapore, 2016–2018
| Total | Severe Diseasea | Unadjusted | Adjustedb | |
|---|---|---|---|---|
| Risk Factors | n = 151 (%) | n = 29 (%) | OR (95% CI) | OR (95% CI) |
| Gender | ||||
| Female | 57 (37.8) | 9 (31.0) | Reference | -- |
| Male | 94 (62.2) | 20 (69.0) | 1.4 (0.6–3.4) | -- |
| Age Group | ||||
| <1 years | 9 (6.0) | 4 (13.8) | 13.6 (1.2–151) | 17.9 (1.3–240) |
| 1–5 years | 115 (76.2) | 24 (82.8) | 4.5 (0.6–35.4) | 4.9 (0.5–46.4) |
| 6–10 years | 18 (12.0) | 1 (3.4) | Reference | Reference |
| 11–16 years | 8 (5.3) | -- | -- | -- |
| HAdV Genotype | ||||
| C1 | 4 (2.6) | 1 (3.4) | 2.8 (0.3–31.7) | 2.9 (0.3–32.4) |
| C2 | 6 (4.0) | 4 (13.8) | 17 (2.6–113.3) | 23.1 (3.1–173.3) |
| B3 | 57 (37.7) | 6 (20.7) | Reference | Reference |
| B7 | 32 (21.2) | 8 (27.6) | 2.8 (0.9–9.1) | 2.8 (0.9–9.3) |
| E4 | 38 (25.2) | 5 (17.2) | 1.3 (0.4–4.6) | 1.6 (0.4–5.9) |
| Unknownc | 10 (6.6) | 4 (13.8) | 5.7 (1.2–26.0) | 9.8 (1.7–56.3) |
| Otherd | 4 (2.6) | 1 (3.4)e | 2.8 (0.3–31.7) | 4.3 (0.3–55.3) |
Abbreviations: CI, confidence interval; HAdV, human adenovirus; OR, odds ratio.
aProlonged hospitalization, prolonged fever, or intensive care unit admission.
bAfter stepwise logistic regression using saturated model and backward elimination of covariates with P > .05.
cGenotyping failed.
dHAdV-C5, -B11p, or -D8.
eHAdV-C5.
Adult Patients
Overall, 137 (25.7%) of the clinical samples were collected from adults, of which genotypes HAdV-B7 (28.5%), HAdV-E4 (21.2%), and HAdV-C2 (3.6%) were most prevalent. Among the adult samples, 55 (40.1%) were retrospectively collected from 2012 to 2013, whereas the remaining 82 (59.9%) were prospective samples from 2015 to 2018. No significant differences in HAdV genotype frequencies were found between retrospective and prospective adult patients.
Phylogenetic and Genomic Analyses
In 2016, we noticed an increase in HAdV-E4 infections. To learn whether the observed increase might reflect a novel HAdV-E4 strain, 6 HAdV-E4 sample genomes were sequenced (3 from severe and 3 from mildly ill patients). Another 4 HAdV species C samples were chosen for sequencing based on their discrepant hexon and fiber genotyping results between laboratories.
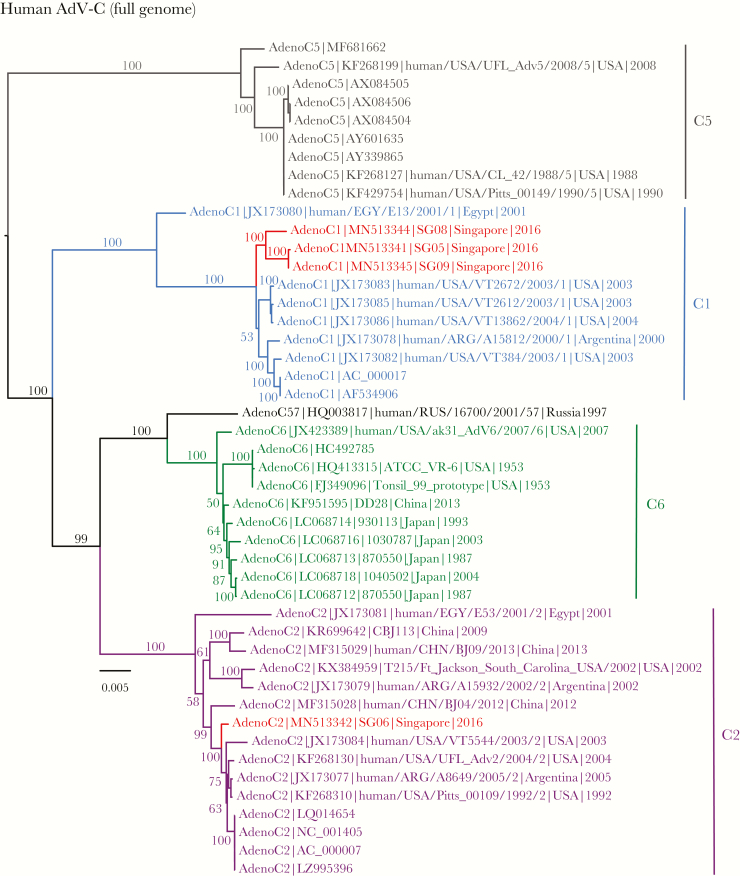
Based on the full genomes of HAdV-C (Figure 2), 3 isolates—SG05, SG08, and SG09—belonged to C1 and were strongly monophyletic, with 100% BS support. These new isolates were also sister to a group of HAdV strains from the United States. Contrarily, isolate SG06 was nested within C2 lineage (ML BS = 100%) and closely related with a Beijing HAdV-C2 strain (human/CHN/BJ04/2012) collected from an infant during a HAdV-C epidemic in Beijing in 2012–2013 [37] as well as earlier C2 strains from the United States isolated from 1992 to 2004. It is notable that SG06 was isolated from a 2-year-old patient requiring oxygen therapy. Hexon, fiber, and penton base topologies (Supplemental Figures 1 and 2) indicated that isolates SG05 and SG09 were most closely related, with strong BS support (87%, 88%, and 99%, respectively). However, penton base phylogeny (Supplemental Figure 2) demonstrated that isolate SG08 was related to C2 lineage, although this lacks statistical support, as opposed to C1 based on full genomes. More notably, the penton base phylogeny demonstrated that isolates SG05 and SG09 were closely related to C2 and C6. Clinical descriptions of the Singapore HAdV-C isolates and their GenBank accession numbers can be found in Supplemental Table 1 and Figure 2.
Figure 2.
Evolutionary relationships of full genomes of human adenovirus (HAdV)-C, inferred by the maximum-likelihood method with the GTR+GAMMA model in RAxML. Colored branches represent viruses isolated from different genotypes. Red isolates denote new isolates collected from patients in Singapore. Bootstrap support values greater than 50% are displayed at major nodes. The scale bar indicates the number of nucleotide substitutions per site.
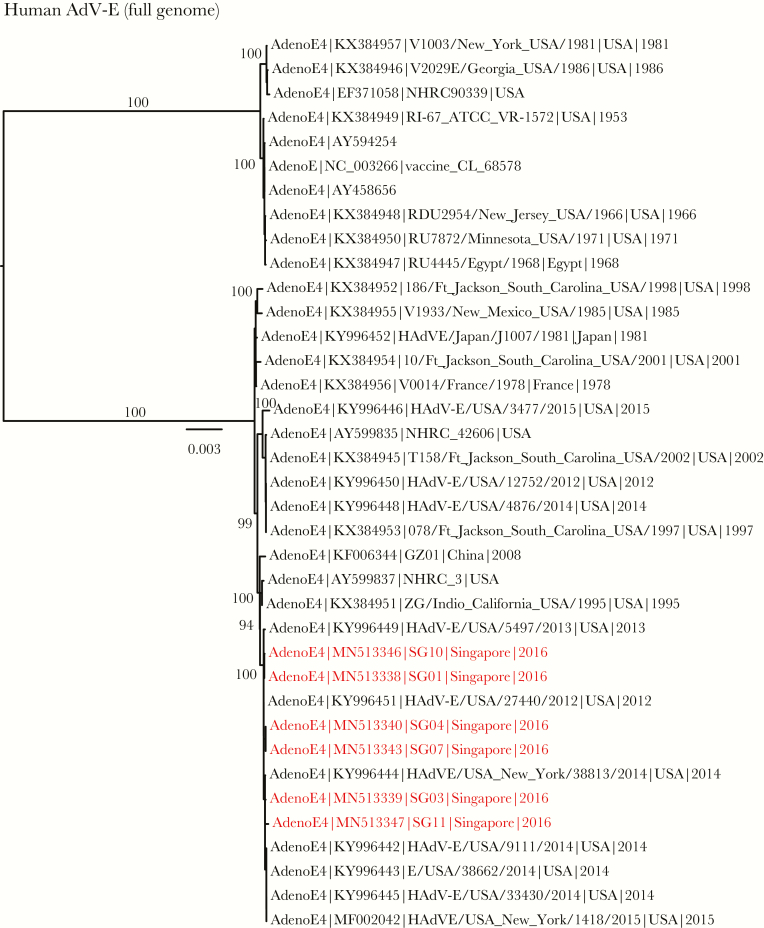
In contrast to HAdV-C, the full genomes of HAdV-E (Figure 3) clearly indicated 6 new isolates (SG01, SG03, SG04, SG07, SG10, and SG11) from Singapore grouped within E4, with strong BS support (ML BS = 100%). These results were concordant with fiber and penton base phylogenies (Supplemental Figures 3 and 4), indicating that these E4 isolates were genetically identical (100% in both genes) and appeared to be clonal. However, the HAdV-E hexon phylogeny (Supplemental Figure 3) indicated that the 6 E4 isolates were relatively segregated, despite being largely monophyletic. These 6 isolates also exhibited 99.8%–100% nucleotide similarities, suggesting that some genetic variations existed among them. Clinical descriptions of the HAdV-E isolates and their GenBank accession numbers can be found in Supplemental Table 1 and Figure 3.
Figure 3.
Evolutionary relationships of full genomes of human adenovirus (HAdV)-E, inferred by the maximum-likelihood method with the GTR+GAMMA model in RAxML. Red isolates denote new isolates collected from patients in Singapore. Bootstrap support values greater than 50% are displayed at major nodes. The scale bar indicates the number of nucleotide substitutions per site.
Global mutational analysis using Genome Mutation Mapper ([GMM]; unpublished beta software) was then used to investigate nucleotide-level variations (single-nucleotide polymorphisms and insertions/deletions) between the 6 HAdV-E4 isolates (SG01, SG03, SG04, SG07, SG10, and SG11) and a 2002 HAdV-E4 reference field strain [38]. Based on the GMM results, SG01 and SG10 are clones, “lineal” to SG04 and SG07, which are clonal. Clones SG01 and SG10 are also lineal to SG03, which is lineal to SG11. It is notable that SG11 has both an insertion and a deletion near the 30.7-kDa gene.
In addition, to provide nexus with the earlier reports of HAdV-E4 in the literature and to compare genome types [5, 38], in silico restriction enzyme fragment analyses were performed using SnapGene (version 4.3), revealing that SG03 and SG11 are novel separate genome type variants, with differing DraI and BamHI patterns and given designations “4a1 DraI/BamHI v1” and “4a1 DraI/BamHI v2”. The others are genome typed as HAdV-E4a1. These patterns support the lineages described by the GMM results. For reference, the 2002 HAdV-E4 field strain is genome type HAdV-E4a1[5], and the patterns for the prototype strains differ from the HAdV-E4a strains across all restriction enzymes applied, with one exception, a match of HAdV-E4p MIL and the HAdV-E4a strains using EcoRI.
Recombination Analyses
Our RDP analyses detected recombination events in 4 new HAdV-C isolates from Singapore. Recombination analysis of SG05 isolate clearly identified 2 parental strains (BJ04 and C1), and the estimated breakpoint was located at the position 17874 (Supplemental Figure 5A, Supplemental Table 2). To confirm this result, phylogenetic analysis was independently performed for the gene regions before and after the breakpoint. The ML topologies (Supplemental Figure 5B) indicate that SG05 recombinant was descended from C1, but it also comprised genetic components from the BJ04 recombinant strain. The BJ04 strain was crucially important [37] because it has been shown to bear gene sequences from 3 different genotypes: HAdV-C1 at (position 1–18076, comprising E1A, E1B, penton base), HAdV-C2 (18077–33397, comprising hexon and fiber), and HAdV-C6 (33398–end, comprising the E4 gene region). The E4 region of SG05 recombinant also contained C6 because this region was recombined with the BJ04 strain. Another recombinant, SG09, was found to be almost identical (99.9% nucleotide identity) to SG05, therefore they both displayed the same breakpoint location (Supplemental Table 2) and RDP graph (data not shown) as well as the phylogenetic position as expected (Supplemental Figure 1). These findings indicate that SG05 and SG09 isolates were recombinants of C1 and BJ04 parental strains.
Recombination analysis of isolate SG06 (Supplemental Figure 6A) suggested that this strain was recombinant of C2 and BJ04 parental strains, although the SG06 backbone was largely derived from C2. The ML phylogenies (Supplemental Figure 6B) confirmed, with strong BS support (ML BS = 100%), that sequence position 12502–35973 of SG06 isolate was nested within C2, whereas sequence position 7536–12501 of SG06 isolate was closely related with the BJ04 and BJ09 strains. In contrast, the RDP analysis of isolate SG08 (Supplemental Figure 7A) displayed the parental C1 subtype as the main backbone, although recombination with C6 was found towards the end of the full genome. The RDP results were also concordant with the 2 ML phylogenies that are incongruent with each other (Supplemental Figure 7B). All recombination results were statistically significant (P < .05) (Supplemental Table 2). In marked contrast, no recombination events were detected in the 6 HAdV-E isolates.
DISCUSSION
This is the first large-scale study to systematically describe clinical HAdV genotypes in Singapore. Our results highlight the prevalence of HAdV-B3 and an increase in morbidity due to HAdV-E4 and HAdV-B7 pediatric infections in Singapore. More important, patients with HAdV-C2, HAdV-E4, and HAdV-B7 were more likely to have severe disease. Hence, it seems prudent for public health officials and clinicians to consider using antiviral therapies and HAdV vaccines in Singapore. For instance, brincidofovir, an experimental antiviral therapy, is being used in the USA against severe HAdV infections with good outcomes [39–41]. In addition, a very effective “Adenovirus Type 4 and Type 7 Vaccine, Live, Oral” has been used among US military personnel since 1971. The new version of this vaccine, used since 2011, has resulted in a dramatic decline of respiratory infections among US military recruits [42]. Although this nonattenuated vaccine is not indicated to prevent infections for other populations (concerns for safety among the immunocompromised), other vaccines [6] are being developed that might prove useful in Singapore.
Given the lack of HAdV surveillance in SEA, it is unknown whether HAdV-E4 is a new or re-emerging pathogen in the region. The high prevalence of HAdV-E4 infections identified in our study is intriguing. The HAdV-E4 genes and genome have been shown to be almost identical to several chimpanzee adenoviruses, including the SAdV-E25 genome at 91% and the SAdV-E26 at 92%, with the 1952 prototype as reference [38, 43]. These values are consistent with HAdV-E4 being the only HAdV amongst several chimpanzee adenoviruses phylogenetically grouped within HAdV species E, supporting a zoonotic origin, consistent with current hypotheses that cross-species infections may lead to the emergence of novel human pathogens [44]. Furthermore, sequence analysis of 2 recent circulating HAdV-E4 strains revealed a recombination event that provides a conserved DNA replication motif (NF-I), likely from a respiratory pathogen of HAdV species B, that is present in almost all HAdVs but absent in SAdVs as well as the HAdV-E4 prototype and contemporaneously circulating isolates, including the “vaccine” strain [38, 45]. This HAdV-like motif, as well as the conserved core origin and NF-III motif, were present in all 6 of the fully sequenced Singapore HAdV-E4 genomes. The NF-I human transcription factor and its binding have been extensively characterized molecularly and biochemically as required for optimal HAdV DNA replication [46, 47]. This putative genome-based host adaptation may be the “tipping point” that has allowed HAdV-E4 entry into a global general population, which is presumably immunologically naive to its epsilon (hexon) antigen, because HAdV-E4 had been known to circulate sporadically outside the US military basic trainee populations [20, 48–50]. The recombination event is likely a recent one, with the earliest appearance in 1978 (KX384956 V0014/France/1978) as noted in a resurvey [23] of 2 sets of recently released genomes that include archived and currently circulating military and civilian HAdV-E4 isolates [20, 49], and it may be important as an example of a molecular evolution pathway by which zoonosis may be a source of emergent human pathogens [44].
Our RDP analyses detected separate recombination events in each of our 4 HAdV-C isolates, which suggests the frequent exchange of genetic components among different genotypes. SG05, SG09, and SG06 were shown to be recombinants of a clinically relevant parental strain, BJ04, a HAdV-C2 strain isolated from an infant during a 2012–2013 epidemic in Beijing [37]. It is notable that SG06 was the only HAdV-C2 strain fully sequenced in our study and was isolated in 2016 from a severely ill 2-year-old Singapore patient requiring oxygen therapy. Given the observed increased risk for severe disease among the 6 prospective pediatric patients infected with HAdV-C2, 4 infections of which occurred within a 1-month period, we speculate that these patients might represent a cluster.
Although our study revealed important findings regarding the epidemiology and clinical severity of HAdVs in Singapore, we were limited in our approach because we did not rule out all possible coinfections (other than influenza virus). It is also possible that some of the HAdV infections were not associated with the acute clinical presentations. Our study could have been strengthened by testing blood (or another sterile compartment such as CSF) from all study patients or by testing matched patient controls to demonstrate a stronger probability of disease association. However, the prevalent genotypes identified (ie, HAdV-B3, HAdV-B7, and HAdV-E4) were genotypes that are commonly known to cause acute respiratory infections. Finally, mixed HAdV infections could not be ruled out as the cause of the untypable patient samples (14%). This theory is supported by the observed increased risk for severe disease among patients infected with an “unknown” HAdV genotype, as well as the high prevalence (40.9%) of unknown HAdV genotype infections among immunocompromised patients in our study. Another reason for an untypable sample is inadequate HAdV DNA for amplification, resulting in an unknown HAdV genotype classification. The majority of these untypable samples were adult samples, which is consistent with the theory that infected adults shed less virus when compared with children.
CONCLUSIONS
Our study demonstrates how HAdV genotyping can be performed in a clinical setting and can be useful in detecting novel HAdV incursions. For Singapore and other countries considering new HAdV treatment and control measures, we strongly recommend periodic but routine HAdV genotype surveillance with a goal of collecting the HAdV prevalence data necessary to make informed decisions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Dr. Chan Kian Seng from the Singapore General Hospital (SGH) Molecular Laboratory for storing SGH residual samples; Dr. Nancy W. Tee, Dr. Loo Liat Hui, Soong Han Yang, and Jiang Zheng Hao at the KK Clinical Microbiology Laboratory for storing KK Women’s and Children’s Hospital residual samples; Emmerie Wong and Fatima Bautista at the KK Research Center for patient recruitment and data collection; Yelen Veronica and Iris Lim Bee Theng at KK hospital for locating patient charts; Dr. Adriana Kajon from the Lovelace Respiratory Institute for advice on the restriction enzyme fragment analysis; and PLinfa Wang from Duke-NUS Medical School for funding the whole-genome sequencing service costs.
Financial support. This work was funded by SingHealth and Duke-NUS Medical School (SingHealth/Duke-NUS/RCG/2015/0007; to J. G. L. and G. C. G.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Ismail AM, Lee JS, Lee JY, et al. Adenoviromics: mining the human adenovirus species D genome. Front Microbiol 2018; 9:2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lynch JP 3rd, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med 2016; 37:586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gray GC, McCarthy T, Lebeck MG, et al. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004–2006. Clin Infect Dis 2007; 45:1120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S, Tian X. Vaccine development for human mastadenovirus. J Thorac Dis 2018; 10:2280–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kajon AE, Lu X, Erdman DD, et al. Molecular epidemiology and brief history of emerging adenovirus 14-associated respiratory disease in the United States. J Infect Dis 2010; 202:93–103. [DOI] [PubMed] [Google Scholar]
- 6. Gray GC. Adenovirus 4 and 7 vaccine: new body armor for U.S. Marine Corps Officer Trainees. J Infect Dis 2019. doi:10.1093/infdis/jiz061 [DOI] [PubMed] [Google Scholar]
- 7. Ng OT, Thoon KC, Chua HY, et al. Severe pediatric adenovirus 7 disease in singapore linked to recent outbreaks across Asia. Emerg Infect Dis 2015; 21:1192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li L, Woo YY, de Bruyne JA, et al. Epidemiology, clinical presentation and respiratory sequelae of adenovirus pneumonia in children in Kuala Lumpur, Malaysia. PLoS One 2018; 13:e0205795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foong Ng K, Kee Tan K, Hong Ng B, Nair P, Ying Gan W. Epidemiology of adenovirus respiratory infections among hospitalized children in Seremban, Malaysia. Trans R Soc Trop Med Hyg 2015; 109:433–9. [DOI] [PubMed] [Google Scholar]
- 10. Yusof MA, Rashid TR, Thayan R, et al. Human adenovirus type 7 outbreak in Police Training Center, Malaysia, 2011. Emerg Infect Dis 2012; 18:852–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsou TP, Tan BF, Chang HY, et al. Community outbreak of adenovirus, Taiwan, 2011. Emerg Infect Dis 2012; 18:1825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu P, Ma C, Nawaz M, et al. Outbreak of acute respiratory disease caused by human adenovirus type 7 in a military training camp in Shaanxi, China. Microbiol Immunol 2013; 57:553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalimuddin S, Chan YFZ, Wu IQ, et al. A report of adult human adenovirus infections in a tertiary hospital. Open Forum Infect Dis 2017; 4:ofx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao S, Wan C, Ke C, et al. Re-emergent human adenovirus genome type 7d caused an acute respiratory disease outbreak in Southern China after a twenty-one year absence. Sci Rep 2014; 4:7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hilleman MR, Werner JH. Recovery of new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med 1954; 85:183–8. [DOI] [PubMed] [Google Scholar]
- 16. Van Der Veen J, Van Der Ploeg G. An outbreak of pharyngoconjunctival fever caused by types 3 and 4 adenovirus at Waalwijk, The Netherlands. Am J Hyg 1958; 68:95–105. [DOI] [PubMed] [Google Scholar]
- 17. D’Angelo LJ, Hierholzer JC, Keenlyside RA, Anderson LJ, Martone WJ. Pharyngoconjunctival fever caused by adenovirus type 4: report of a swimming pool-related outbreak with recovery of virus from pool water. J Infect Dis 1979; 140:42–7. [DOI] [PubMed] [Google Scholar]
- 18. Aoki K, Kato M, Ohtsuka H, Ishii K, Nakazono N, Sawada H. Clinical and aetiological study of adenoviral conjunctivitis, with special reference to adenovirus types 4 and 19 infections. Br J Ophthalmol 1982; 66:776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seah VX, Chew Y, Thoon KC, et al. 2341. Trends in adenovirus infections in singapore children and outcomes of cidofovir treatment in the severely ill. Open Forum Infect Dis 5:Suppl 1S695–6. [Google Scholar]
- 20. Kajon AE, Lamson DM, Bair CR, et al. Adenovirus type 4 respiratory infections among civilian adults, Northeastern United States, 2011–2015. Emerg Infect Dis 2018; 24:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. La Rosa G, Muscillo M, Iaconelli M, et al. Molecular characterization of human adenoviruses isolated in Italy. New Microbiol 2006; 29:177–84. [PubMed] [Google Scholar]
- 22. Wang B, Li J, Wu S, et al. Seroepidemiological investigation of HAdV-4 infection among healthy adults in China and in Sierra Leone, West Africa. Emerg Microbes Infect 2018; 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Kang J, Dehghan S, et al. A survey of recent adenoviral respiratory pathogens in Hong Kong reveals emergent and recombinant human adenovirus type 4 (HAdV-E4) circulating in civilian populations. Viruses 2019; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen HL, Chiou SS, Hsiao HP, et al. Respiratory adenoviral infections in children: a study of hospitalized cases in southern Taiwan in 2001--2002. J Trop Pediatr 2004; 50:279–84. [DOI] [PubMed] [Google Scholar]
- 25. Thounaojam AD, Balakrishnan A, Mun AB. Detection and molecular typing of human adenovirus associated with respiratory illness in Kerala. Jpn J Infect Dis 2016; 69:500–4. [DOI] [PubMed] [Google Scholar]
- 26. Wang YF, Shen FC, Wang SL, et al. Molecular epidemiology and clinical manifestations of adenovirus respiratory infections in Taiwanese children. Medicine (Baltimore) 2016; 95:e3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Lu X, Sun Y, et al. A swimming pool-associated outbreak of pharyngoconjunctival fever caused by human adenovirus type 4 in Beijing, China. Int J Infect Dis 2018; 75:89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J, Lu X, Jiang B, et al. Adenovirus-associated acute conjunctivitis in Beijing, China, 2011–2013. BMC Infect Dis 2018; 18:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCarthy T, Lebeck MG, Capuano AW, Schnurr DP, Gray GC. Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J Clin Virol 2009; 46:80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu W, McDonough MC, Erdman DD. Species-specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol 2000; 38:4114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009; 26:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 2015; 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajkumar V, Chiang CS, Low JM, et al. Risk factors for severe adenovirus infection in children during an outbreak in Singapore. Ann Acad Med Singapore 2015; 44:50–9. [PubMed] [Google Scholar]
- 37. Mao N, Zhu Z, Rivailler P, et al. Whole genomic analysis of two potential recombinant strains within human mastadenovirus species C previously found in Beijing, China. Sci Rep 2017; 7:15380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dehghan S, Seto J, Liu EB, et al. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 2013; 443:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hiwarkar P, Amrolia P, Sivaprakasam P, et al. Brincidofovir is highly efficacious in controlling adenoviremia in pediatric recipients of hematopoietic cell transplant. Blood 2017; 129:2033–7. [DOI] [PubMed] [Google Scholar]
- 40. Florescu DF, Pergam SA, Neely MN, et al. Safety and efficacy of CMX001 as salvage therapy for severe adenovirus infections in immunocompromised patients. Biol Blood Marrow Transplant 2012; 18:731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grimley MS, Chemaly RF, Englund JA, et al. Brincidofovir for asymptomatic adenovirus viremia in pediatric and adult allogeneic hematopoietic cell transplant recipients: a randomized placebo-controlled phase II trial. Biol Blood Marrow Transplant 2017; 23:512–21. [DOI] [PubMed] [Google Scholar]
- 42. Radin JM, Hawksworth AW, Blair PJ, et al. Dramatic decline of respiratory illness among US military recruits after the renewed use of adenovirus vaccines. Clin Infect Dis 2014; 59:962–8. [DOI] [PubMed] [Google Scholar]
- 43. Purkayastha A, Ditty SE, Su J, et al. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J Virol 2005; 79:2559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bailey ES, Fieldhouse JK, Choi JY, Gray GC. A mini review of the zoonotic threat potential of influenza viruses, coronaviruses, adenoviruses, and enteroviruses. Front Public Health 2018; 6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Purkayastha A, Su J, McGraw J, et al. Genomic and bioinformatics analyses of HAdV-4vac and HAdV-7vac, two human adenovirus (HAdV) strains that constituted original prophylaxis against HAdV-related acute respiratory disease, a reemerging epidemic disease. J Clin Microbiol 2005; 43:3083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stillman BW, Topp WC, Engler JA. Conserved sequences at the origin of adenovirus DNA replication. J Virol 1982; 44:530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rawlins DR, Rosenfeld PJ, Wides RJ, Challberg MD, Kelly TJ Jr. Structure and function of the adenovirus origin of replication. Cell 1984; 37:309–19. [DOI] [PubMed] [Google Scholar]
- 48. Potter RN, Cantrell JA, Mallak CT, Gaydos JC. Adenovirus-associated deaths in US military during postvaccination period, 1999–2010. Emerg Infect Dis 2012; 18:507–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hang J, Vento TJ, Norby EA, et al. Adenovirus type 4 respiratory infections with a concurrent outbreak of coxsackievirus A21 among United States Army Basic Trainees, a retrospective viral etiology study using next-generation sequencing. J Med Virol 2017; 89:1387–94. [DOI] [PubMed] [Google Scholar]
- 50. Barraza EM, Ludwig SL, Gaydos JC, Brundage JF. Reemergence of adenovirus type 4 acute respiratory disease in military trainees: report of an outbreak during a lapse in vaccination. J Infect Dis 1999; 179:1531–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.