Abstract
RNA viruses successfully adapt to various environments by repeatedly producing new mutants, often through generating a number of nucleotide substitutions. To estimate the degree of variation in mutation rates of RNA viruses and to understand the source of such variation, we studied the synonymous substitution rate because synonymous substitution is exempt from functional constraints at the protein level, and its rate reflects the mutation rate to a great extent. We estimated the synonymous substitution rates for a total of 49 different species of RNA viruses, and we found that the rates had tremendous variation by 5 orders of magnitude (from 1.3 × 10−7 to 6.2 × 10−2 /synonymous site/year). Comparing the synonymous substitution rates with the replication frequencies and replication error rates for the RNA viruses, we found that the main source of the rate variation was differences in the replication frequency because the rates of replication error were roughly constant over different RNA viruses. Moreover, we examined a relationship between viral life strategies and synonymous substitution rates to understand which viral life strategies affect replication frequencies. The results show that the variation of synonymous substitution rates has been influenced most by either the difference in the infection modes or the differences in the transmission modes. In conclusion, the variation of mutation rates for RNA viruses is caused by different replication frequencies, which are affected strongly by the infection and transmission modes.
Keywords: RNA virus, evolution, synonymous substitution rate, replication frequency, infection mode, transmission mode
Introduction
A number of emerging viral infection have repeatedly occurred during the last 15 years. They are caused mostly by many kinds of RNA viruses such as severe acute respiratory syndrome (SARS)-related virus, HIV-1, HCV, Ebola virus, Nipah virus, and West Nile virus (Parashar et al. 2000; Lee and Henderson 2001). However, appropriate treatments have usually been delayed because most of the outbreaks have been so sudden that curative medicines could not be found against most RNA viruses in a timely fashion, as has been recently documented by SARS. Therefore, it is of particular importance to study the evolutionary mechanism for producing new variants of RNA viruses.
Mutation by nucleotide substitution is considered to be one of the important evolutionary mechanisms because it is the major source of new mutant productions of RNA viruses. To study the mutation rate, we examined the rate of synonymous substitution in this study because natural selection does not strongly influence the fixation probability of synonymous substitution, at least at the protein level, and therefore the rate of synonymous substitution reflects the mutation rate in a great extent (Miyata and Yasunaga 1980; Bush et al. 1999).
This study examined the degree of variation of synonymous substitution rates among RNA viruses and to identify the main source of the variation. For this purpose, we estimated the synonymous substitution rates for a total of 49 different species of RNA viruses that belong to 39 genera of 15 families.
Materials and Methods
Sequence Data
We focused only on RNA viruses that infect mammals and then selected at least one representative RNA virus species from each genus. Accordingly, we collected the nucleotide sequences for a total of 49 different species of RNA viruses from the National Center for Biotechnology Information Virus Taxonomy and the data originating from two previous papers (Korber et al. 2000; Tanaka et al. 2002).
We also obtained the years of isolation for all strains from the database and the available publications, which were listed for all RNA virus species in appendix A of supplemental materials. We, then, estimated the rate of synonymous substitution for the genes encoding the outer-structural protein. For hepatitis D virus only, however, we used the whole genome sequence because this virus did not have the structural protein. The RNA virus species used in this paper are summarized in tables 1–4. Moreover, these tables show natural hosts, infection modes, and transmission modes, whose references were listed in appendix A of supplemental materials. Examining the infection modes, we always focused upon the infection mode occurring between a given virus species and its natural host. This is because the infection mode from the virus to the natural host is reasonably considered to represent an important feature of the RNA viruses.
Table 1.
Synonymous Substitution Rates for RNA Viruses.
| Virus Species | Synonymous Substitution Rate | Natural Host (Transmission Mode) | Acute Infection | Persistent Infection | Latent Infection | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive Stranded ss RNA Viruses | ||||||||||
| Astroviridae | ||||||||||
| Astrovirus | ||||||||||
| Human astrovirus | 1.03 × 10−2, (0.82–1.20) × 10−2 | Human (fecal-oral route) | ○ | ○ | ||||||
| Caliciviridae | ||||||||||
| Lagovirus | ||||||||||
| Rabbit hemorrhagic disease virus | 8.28 × 10−3, (8.20–8.35) × 10−3 | Rabbit (contagious infection) | ○ | |||||||
| Norovirus | ||||||||||
| Human calicivirus | 3.82 × 10−3, (3.81–3.82) × 10−3 | Human (fecal-oral route) | ○ | |||||||
| Vesivirus | ||||||||||
| Feline calicivirus | 4.64 × 10−3, (4.63–4.65) × 10−3 | Cat (contagious infection) | ○ | ○ | ||||||
| Unclassified genus | ||||||||||
| Hepatitis E virus | 5.23 × 10−3, (4.66–5.96) × 10−3 | Human (fecal-oral route) | ○ | ○ | ||||||
| Flaviviridae | ||||||||||
| Flavivirus | ||||||||||
| Dengue virus | 2.42 × 10−3, (2.40–2.44) × 10−3 | Human, (monkey via vector [mosquito]) | ○ | |||||||
| Yellow fever virus | 1.19 × 10−3, (1.14–1.25) × 10−3 | Monkey (via vector [mosquito]) | ○ | |||||||
| Japanese encephalitis virus | 6.31 × 10−4, (6.24–6.38) × 10−4 | Pig (via vector [mosquito]) | ○ | |||||||
| Tick-borne encephalitis virus | 4.91 × 10−4, (2.93–15.31) × 10−4 | Rodents (via vector [tick]) | ○ | |||||||
| Pestivirus | ||||||||||
| Bovine viral diarrhea virus | 2.34 × 10−3, (2.33–2.34) × 10−3 | Cattle (aerosol infection) | ○ | |||||||
| Hepacivirus | ||||||||||
| Hepatitis C virus | 7.51 × 10−4, (7.37–7.66) × 10−4 | Human (via blood) | ○ | |||||||
| Unclassified genus | ||||||||||
| GB virus C/hepatitis G virus | 1.35 × 10−7, (1.20–1.70) × 10−7 | Human (via blood) | ○ | |||||||
| Picornaviridae | ||||||||||
| Apthovirus | ||||||||||
| Foot-and-mouth disease virus | 8.29 × 10−3, (8.19–8.39) × 10−3 | Ruminant (aerosol infection) | ○ | ○ | ||||||
Estimation of Synonymous Substitution Rate for RNA Viruses
We took two approaches to estimate the rate of synonymous substitution. First, we estimated the rates of synonymous substitution for 46 different species of RNA viruses except Puumala virus, human T-lymphotropic virus 1 (HTLV-1) and GB virus C/hepatitis G virus (HGV), using the time-serial sample data. Multiple alignment was made to match the coding regions by the computer program, CLUSTALW (Thompson, Higgins, and Gibson 1994). For each nucleotide sequence alignment, the phylogenetic tree was constructed by the maximum likelihood method under the premise of the molecular clock (Rambaut 2000). Taking into account the difference in years of isolation among viral sequences, we estimated the divergence time of all nodes in the tree. We then inferred ancestral nucleotide sequences at all nodes of the phylogenetic tree from sequence comparisons by the maximum likelihood method (Yang, Kumar, and Nei 1995). These analyses were conducted using the PAML computer program. The number of synonymous substitutions was estimated for all branches by the Nei-Gojobori method (Nei and Gojobori 1986). The rate of synonymous substitution for each branch was, then, estimated by dividing the number of synonymous substitutions for that branch by the difference in years of the divergence or isolation between both ends of the branch. The error range of the synonymous substitution rates was also estimated, taking into account the standard error of the estimated divergence time at each node.
In the second approach, we estimated the rates of synonymous substitution for the remaining three RNA viruses, Puumala, HTLV-1, and HGV, using the divergence times that have already been reported. These viruses were reported to coevolve with the host species (Horai 1995; Yanagihara et al. 1995; Asikainen et al. 2000; Robertson 2001). Therefore, the divergence time of a virus was considered to correspond to the divergence time of the host. We first constructed each multiple alignment of three RNA viruses to match the coding region by the computer program, CLUSTALW. From the multiple alignment, the phylogenetic tree was constructed by the maximum likelihood method on the basis of the HKY model. The ancestral sequence of the divergence node was estimated by the maximum likelihood approach. The rate of synonymous substitution was estimated by dividing the average number of synonymous substitutions from the ancestral sequence to all tips of the phylogenetic tree by the time period from the known divergence time of the host to the present.
A Test of Substitution Saturation
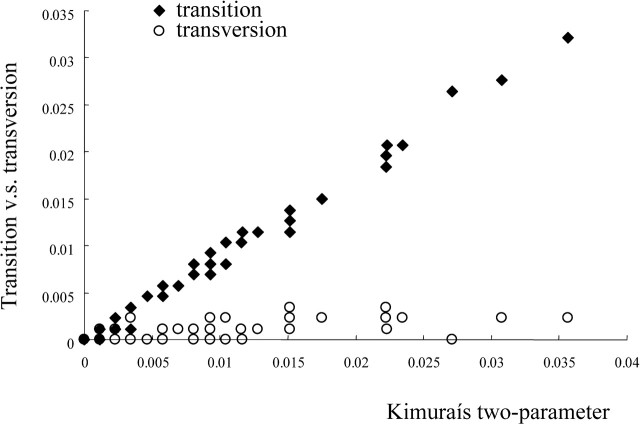
We conducted a statistical test to examine whether the number of substitutions was saturated or not by Xia and Xie's (2001) method. In this method, both transitions and transversions were plotted against evolutionary distances such as the number of nucleotide substitutions. In figure 1, for example, we showed the comparison for transitions and transversions in human enterovirus A. When transitions occur much more frequently than transversions, no saturation of substitution is recognized. On the other hand, when transversions gradually outnumber transitions, substitution saturation is suspected because multiple substitutions may have occurred at each site. Therefore, we conducted the comparison of two regression slopes. If the slope of transitions is significantly steeper than that for transversions against evolutionary distances, the substitution was considered not to be saturated.
Fig. 1.
Transitions-type and transversions-type nucleotide differences plotted against the evolutionary distance of Kimura's two parameter method. No crossing of the different symbols (solid diamonds and open circles), representing transitions and transversions, respectively, suggests rejection of substitution saturation, which is exemplified by the data set of human enterovirus A
Results and Discussion
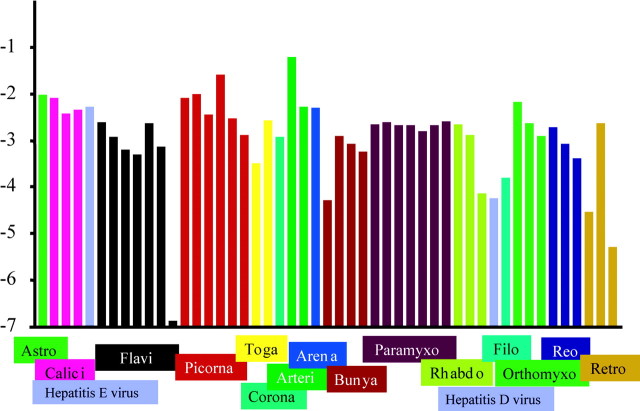
The results showed that the virus species evolving at the highest rate (6.2 × 10−2/synonymous site/year) was porcine reproductive and respiratory syndrome virus, and the virus evolving at the lowest rate (1.3 × 10−7 synonymous site/year) was HGV. The rates for the other RNA viruses estimated here are shown in tables 1–4 and summarized in figure 2. These results indicated that the rates of synonymous substitution varied among RNA viruses by 5 orders of magnitude.
Fig. 2.
Comparison of synonymous substitution rates among RNA viruses. Virus species belonging to the same family were represented by the same color. The end “viridae” of all family names was omitted. For example, Astro indicates Astroviridae. As exceptions, both hepatitis D virus and hepatitis E virus are represented by the same color (gray), since they are not classified into any virus family. The ordinate represents log synonymous substitution rate. Each virus species is ranked by each virus family along the axis of abscissas
Moreover, we conducted a statistical test to examine whether the number of substitutions was saturated or not. This statistical test indicated that, for most data sets, the substitutions were not saturated except hepatitis D virus (appendix B of supplemental materials). For the hepatitis D virus, the substitution could not be rejected with statistical significance of 5%. However, since the P-value of statistical significance (P = 0.19) was not much larger than 0.05, we also listed the rate of synonymous substitution for Hepatitis D virus in Table 3.
Table 3.
Synonymous Substitution Rates for RNA Viruses.
| Virus Species | Synonymous Substitution Rate | Natural Host (Transmission Mode) | Acute Infection | Persistent Infection | Latent Infection | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Orthobunyavirus | ||||||||||
| Cache Valley virus | 8.45 × 10−4, (8.42–8.48) × 10−4 | Sheep, cattle, deer (via vector [mosquito]) | ○ | |||||||
| Phlebovirus | ||||||||||
| Rift Valley fever virus | 5.90 × 10−4, (3.89–12.22) × 10−4 | Cattle (via vector [mosquito]) | ○ | |||||||
| Paramyxoviridae | ||||||||||
| Pneumovirus | ||||||||||
| Bovine respiratory syncytial virus | 2.17 × 10−3, (2.12–2.22) × 10−3 | Cattle (aerosol infection) | ○ | |||||||
| Metapneumovirus | ||||||||||
| Human metapneumovirus | 2.43 × 10−3, (2.24–2.65) × 10−3 | Human (aerosol infection) | ○ | |||||||
| Morbillivirus | ||||||||||
| Canine distemper virus | 2.12 × 10−3, (2.08–2.16) × 10−3 | Dog (aerosol infection) | ○ | ○ | ||||||
| Measles virus | 2.12 × 10−3, (2.11–2.13) × 10−3 | Human (aerosol infection) | ○ | ○ | ||||||
| Respirovirus | ||||||||||
| Humanparainfluenza virus | 1.60 × 10−3, (1.44–1.79) × 10−3 | Human (aerosol infection) | ||||||||
| Rubulavirus | ||||||||||
| Mumps virus | 2.11 × 10−3, (2.11–2.12) × 10−3 | Human (aerosol infection) | ○ | |||||||
| Newcastle disease virus | 2.54 × 10−3, (2.37–2.73) × 10−3 | Bird (aerosol infection) | ○ | |||||||
| Rhabdoviridae | ||||||||||
| Ephemerovirus | ||||||||||
| Bovine ephemeral fever virus | 2.23 × 10−3, (2.22–2.23) × 10−3 | Cattle (via vector [mosquito]) | ○ | |||||||
| Lyssavirus | ||||||||||
| Rabies virus | 1.28 × 10−3, (1.27–1.28) × 10−3 | Mammal (via a bite) | ○ | |||||||
| Vesiculovirus | ||||||||||
| Vesicular stomatitis virus | 7.20 × 10−5, (7.10–7.30) × 10−5 | Ruminant, human (via vector [fly]) | ○ | |||||||
| Unclassified family | ||||||||||
| Deltavirus | ||||||||||
| Hepatitis D virus | 5.80 × 10−5, (3.92–11.51) × 10−5 | Human (via blood) | ○ | |||||||
This tremendous degree of variation in the synonymous substitution rates is much more than the expected one from the previous report. In fact, Jenkins et al.'s (2002) suggested that the variation of synonymous substitution is narrow, only 10−3 to 10−4. This narrow range of variation was obtained because they used only several RNA viruses and, in particular, they did not include any of the slowly and rapidly evolving viruses that were examined in the present study.
Moreover, when the rate variation of RNA viruses was compared with that of nonviral organisms, we found that the former was about 1,000-fold larger than that of the latter. This is because nonviral organisms have been reported to have evolved at varying synonymous substitution rates only by 2 orders of magnitude in the actual range of (0.12–12.4) × 10−9 (Li, Tanimura, and Sharp 1987; Wolfe, Li, and Sharp 1987; Bulmer, Wolfe, and Sharp 1991; Gaut et al. 1996; Pawlowski et al. 1997).
We built three possible hypotheses in the following to understand the reason that such variation existed. The first hypothesis is that the error rate per replication (replication error rate) among the RNA viruses is the main cause of the variation. The second hypothesis is that the number of replications per unit time (replication frequency) among RNA viruses is the main cause of the variation. The third hypothesis is that both of these affect the variation.
To investigate which of these three hypotheses was correct, we compared the rate of replication error with the synonymous substitution rate using eight different RNA viruses, as shown in table 5 (Schrag, Rota, and Bellini 1991; Mansky and Temin 1995; Drake and Holland 1999; Stech et al. 1999; Mansky 2000; Escarmis et al. 2002). The replication error rate of porcine reproductive and respiratory syndrome virus was estimated from the passage number, the number of nucleotide substitutions during the passage, and the time required for viral budding (Dea 1995; Allende 2000). The results showed that the rate of replication error (the order is 10−5) was almost constant among the eight different species of RNA viruses in spite of the wide variation in the synonymous substitution rates among these species. These results indicated that the replication frequency should be the main source of the variation in the synonymous substitution rates because the constancy of the rates of replication error showed that these did not contribute strongly to the variation of the synonymous substitution rates among RNA viruses.
Table 5.
Comparison Between Replication Error Rate and Synonymous Substitution Rate.
| Virus Species | Replication Error Rate (/site/replication) | Synonymous Substitution Rate (/site/year) | ||
|---|---|---|---|---|
| Positive stranded ss RNA viruses | ||||
| Porcine reproductive and respiratory syndrome virus | 3.7 × 10−5 | 6.2 × 10−2 | ||
| Foot-and-mouth disease virus | 3.7 × 10−5 | 8.3 × 10−3 | ||
| Human poliovirus | (2.3–3.05) × 10−5 | 2.6 × 10−2 | ||
| Negative stranded ss RNA viruses | ||||
| Measles virus | (6.0–14) × 10−5 | 2.1 × 10−3 | ||
| Influenza A virus | (17.5–23.5) × 10−5 | 6.8 × 10−3 | ||
| Vesicular stomatitis virus | 10.0 × 10−5 | 7.2 × 10−5 | ||
| Reverse Transcribing viruses | ||||
| Human immunodeficiency virus 1 | 3.4 × 10−5 | 2.4 × 10−3 | ||
| Human T cell lymphotropic virus 1 | 0.7 × 10−5 | 5.2 × 10−6 | ||
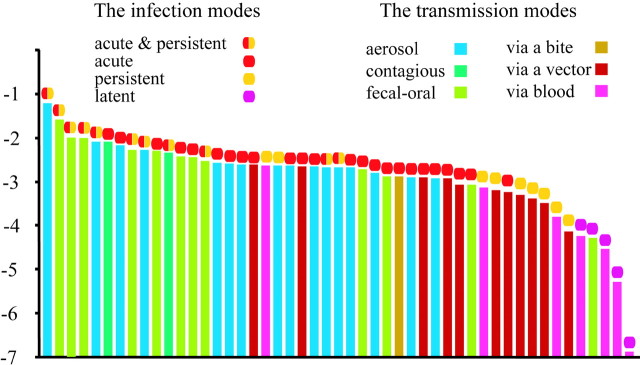
Moreover, we focused on the viral life strategies to the host. These characteristics are considered to affect the replication frequency, because the viral life strategy is strongly related to the infectivity, and the strength of infectivity is related to an increase in the chance for replication. This indicates that the viral life strategy may be related to the replication frequency among RNA viruses. If the relationship between the viral life strategy and the replication frequency is certain, then the viral life strategy should be related to the rates of synonymous substitution among RNA viruses. This is because the main source of the rate variation for RNA viruses was considered to be the replication frequency, as mentioned earlier. Therefore, we compared viral life strategies of RNA virus with the rates of synonymous substitution (fig. 3).
Fig. 3.
Comparison between synonymous substitution rate and the infection and transmission modes. The synonymous substitution rates are ranked in descending order on the abscissa. The viral life strategies are classified into two major categories. The first category is the infection modes such as acute, persistent, and latent infection. The combination of acute and persistent infection is also included in this category. The second category is the transmission modes such as aerosol transmission, contagious transmission, fecal-oral route transmission, transmission via blood (inducing sexual relationship and artificial injection), transmission via a bite and transmission via a vector. The first category, i.e., infection modes, is represented by a circle in the different color above the vertical bars, whereas the second category, i.e., transmission modes, is represented by a vertical bar in a different color. The ordinate represents the log synonymous substitution rate
The viral life strategies examined in the present study were classified into two major categories: infection modes and viral transmission. Infection modes are classified into acute, persistent, and latent infection. In general, the infection modes are not mutually exclusive. In fact, some viruses have both of acute and persistent infection phases. For example, measles viruses induce acute infection in the beginning, and maintain a persistent phase in the same host. When the replication of a virus is strictly limited, by some reasons, in the persistent phases, this infection mode is called latent infection. The viral transmissions were composed of six modes, i.e., aerosol transmission, contagious transmission, fecal-oral route transmission, transmission via blood (sexual relationship and artificial injection), transmission via a bite, and transmission via a vector.
First, we compared the infection modes with the rates of synonymous substitution. The results showed that the rates of synonymous substitution for viruses inducing both acute and persistent infection were higher than those for viruses inducing only acute infection. We also showed that the rates of synonymous substitution for viruses of only acute infection were higher than those for the viruses of only persistent infection and that the rates of synonymous substitution for viruses of only persistent infection were higher than those for the viruses of latent infection. All of those differences were statistically significant (P < 0.05) by the two-tailed Wilcoxon test (fig. 3). Indeed, the replication frequencies for the viruses of only acute infection are considered to be higher than those for the viruses of only persistent infection, because viruses causing an acute symptom are expected to infect the neighboring host cells more frequently than viruses persistently holding a symptom (Overbaugh and Bangham 2001).
The replication frequencies of viruses inducing both acute and persistent infection might be higher than those of viruses inducing only acute infection, to some extent, because viruses inducing both infection could repeatedly replicated themselves in the acute phase. On the other hand, the replication frequencies of latent infection are absolutely lower than those of only persistent infection because the replication of the virus are strictly limited in latent infection. Thus, we reasonably assume that the highest replication frequency is manifested by the viruses inducing both acute and persistent infection, the second highest by viruses inducing only acute infection, the third by viruses inducing only persistent infection, and the fourth by the viruses inducing latent infection. In fact, the rate of synonymous substitution for the viruses inducing both acute and persistent infection is the highest, that for the viruses inducing only acute infection is the second highest, that for the viruses inducing only persistent infection is the third, and that for the viruses inducing latent infection is the fourth. This is because differences in the infection modes are considered to affect the replication frequencies.
Furthermore, we compared the transmission mode with the rate of synonymous substitution among RNA viruses. As mentioned earlier, the transmission modes of RNA viruses were classified into six kinds, i.e., aerosol, contagious, fecal-oral, blood, bite, and vector. In figure 3, the synonymous substitution rates of viruses inducing aerosol, contagious, or fecal-oral route transmission were higher than those of the viruses inducing transmission via blood, bite, or vector, and the differences were significant (P < 0.05) by the two-tailed Wilcoxon test. These results implied that differences in viral transmission modes were also correlated with the rate of synonymous substitution. The correlation can be understood as follows. Viruses that spread rapidly among hosts through aerosol, contagious, or fecal-oral route transmission would quickly replicate because the viruses can infect many individuals surrounding an infected host. On the other hand, viruses that spread slowly among hosts by a transmission via blood, a bite or a vector would replicate slowly compared with viruses inducing a transmission via the aerosol, contagious, or fecal-oral routes. This indicated that the transmission mode affected the replication frequency and that differences in the replication frequencies contributed to the variation of the rate of synonymous substitution for RNA viruses. In fact, there was a good example in which a change of transmission mode seriously affected the evolutionary rate (Salemi et al. 1999). This report is consistent with our results that differences in the transmission mode affect differences in the replication frequency, and differences in the replication frequencies produced the rates of synonymous substitution.
To summarize, the synonymous substitution rates among RNA viruses varied by 5 orders of magnitude. Moreover, in the present study, we proved that the variation in the synonymous substitution rates among RNA viruses was caused by variation of the replication frequency, and that differences in the infection and transmission modes affected the variation of replication frequencies.
Table 2.
Synonymous Substitution Rates for RNA Viruses.
| Virus Species | Synonymous Substitution Rate | Natural Host Transmission Mode | Acute Infection | Persistent Infection | Latent Infection | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterovirus | ||||||||||||||
| Human enterovirus A | 1.00 × 10−2, (0.99–2.01) × 10−2 | Human fecal-oral route | ||||||||||||
| Human enterovirus B | 3.65 × 10−3, (3.60–7.82) × 10−3 | Human fecal-oral route | ||||||||||||
| Human poliovirus | 2.56 × 10−2, (2.56–2.57) × 10−2 | Human fecal-oral route | ||||||||||||
| Swine vesicular disease virus | 2.95 × 10−3, (2.85–8.65) × 10−3 | Pig fecal-oral route | ||||||||||||
| Hepatovirus | ||||||||||||||
| Human hepatitis A virus | 1.30 × 10−3, (1.27–1.33) × 10−3 | Human fecal-oral route | ||||||||||||
| Togaviridae | ||||||||||||||
| Alphavirus | ||||||||||||||
| Eastern equine encephalitis virus | 3.25 × 10−4, (3.03–3.51) × 10−4 | Birds via vector (tick) | ||||||||||||
| Rubivirus | ||||||||||||||
| Rubella virus | 2.64 × 10−3, (2.63–2.65) × 10−3 | Human aerosol infection | ||||||||||||
| Coronaviridae | ||||||||||||||
| Coronavirus | ||||||||||||||
| Bovine coronavirus | 1.20 × 10−3, (1.13–1.43) × 10−3 | Cattle aerosol infection | ||||||||||||
| Arteriviridae | ||||||||||||||
| Arterivirus | ||||||||||||||
| Porcine reproductive and respiratory syndrome virus | 6.21 × 10−2, (6.01–7.81) × 10−2 | Pig aerosol infection | ||||||||||||
| Equine arteritis virus | 5.20 × 10−3, (4.80–7.20) × 10−3 | Horse aerosol infection | ||||||||||||
| Negative stranded ss RNA viruses | ||||||||||||||
| Arenaviridae | ||||||||||||||
| Arenavirus | ||||||||||||||
| Junin virus* | 5.06 × 10−3, (5.02–5.10) × 10−3 | Rodents fecal-oral route | ||||||||||||
| Bunyaviridae | ||||||||||||||
| Hantavirus | ||||||||||||||
| Puumala virus | 5.21 × 10−5, (5.19–7.30) × 10−5 | Rodents fecal-oral route | ||||||||||||
| Nairovirus | ||||||||||||||
| Crimean-Congo hemorrhagic fever virus | 1.23 × 10−3, (1.23–1.24) × 10−3 | Human via vector (tick) | ||||||||||||
* Junin virus induces a latent infection in rodents. However, the virus does not induce such the infection mode in human. In fact, the data used here were the data of the virus strains isolated from human. Therefore, the infectious mode was used for human.
Table 4.
Synonymous Substitution Rates for RNA Viruses.
| Virus Species | Synonymous Substitution Rate | Natural Host (Transmission Mode) | Acute Infection | Persistent Infection | Latent Infection | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Filoviridae | ||||||||||
| Filovirus | ||||||||||
| Ebola-like viruses | 1.54 × 10−4, (1.50–1.58) × 10−4 | Monkey (via blood) | ○ | |||||||
| Orthomyxoviridae | ||||||||||
| Influenza A virus | ||||||||||
| Human Influenza virus A | 6.84 × 10−3, (6.83–6.84) × 10−3 | Human (aerosol infection) | ○ | |||||||
| Influenza B virus | ||||||||||
| Human influenza virus B | 2.30 × 10−3, (2.29–2.31) × 10−3 | Human (aerosol infection) | ○ | |||||||
| Influenza C virus | ||||||||||
| Human influenza virus C | 1.27 × 10−3, (1.27–1.28) × 10−3 | Human (aerosol infection) | ○ | |||||||
| ds RNA viruses | ||||||||||
| Reoviridae | ||||||||||
| Reovirus | ||||||||||
| Human rota virus | 1.93 × 10−3, (1.92–1.94) × 10−3 | Human (fecal-oral route) | ○ | |||||||
| Orthoreo | ||||||||||
| Mamalian orthoreo virus | 8.42 × 10−4, (5.36–19.61) × 10−4 | Human (fecal-oral route) | ○ | |||||||
| Orbivirus | ||||||||||
| Bluetongue virus | 4.22 × 10−4, (3.74–4.96) × 10−4 | Ruminant (via vector [mosquito]) | ○ | |||||||
| Reverse Transcribing viruses | ||||||||||
| Retroviridae | ||||||||||
| Spumavirus | ||||||||||
| Simian foamy virus | 2.9 × 10−5, (2.71–3.21) × 10−5 | Monkey (via blood) | ○ | |||||||
| Lentivirus | ||||||||||
| Human immunodeficiency virus | 2.38 × 10−3, (2.38–2.38) × 10−3 | Human (via blood) | ○ | |||||||
| Deltaretrovirus | ||||||||||
| Human T-lymphotropic virus 1 | 10−6 × (4.60–5.90), 5.2 × 10−6 | Human (via blood) | ○ | |||||||
Supplementary Material
Acknowledgments
We thank Drs. Ken Nishikawa, Kazuho Ikeo, Toshimichi Ikemura, Yoshio Tateno, Toshiyuki Takano, and Masashi Mizokami for their useful comments on our work. This work was suggested in part by a grant given to T.G. from MEXT (Ministry of Education, Sports, Culture and Technology) in Japan.
Footnotes
Dan Graur, Associate Editor
Literature Cited
- Allende, R., W. W. Laegreid, G. F. Kutish, J. A. Galeota, R. W. Wills, and F. A. Osorio. 2000. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J. Virol. 74:10834-10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen, K., T. Hanninen, H. Henttonen, J. Niemimaa, J. Laakkonen, H. K. Andersen, N. Bille, H. Leirs, A. Vaheri, and A. Plyusnin. 2000. Molecular evolution of puumala hantavirus in Fennoscandia: phylogenetic analysis of strains from two recolonization routes, Karelia and Denmark. J. Gen. Virol. 81:2833-2841. [DOI] [PubMed] [Google Scholar]
- Bulmer, M., H. Wolfe, and P. M. Sharp. 1991. Synonymous nucleotide substitution rates in mammalian genes: implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. USA. 88:5974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, R. M., W. M. Fitch, C. A. Bender, and N. J. Cox. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16:1457-1465. [DOI] [PubMed] [Google Scholar]
- Dea, S., N. Sawyer, R. Alain, and R. Athanassious. 1995. Ultrastructural characteristics and morphogenesis of porcine reproductive and respiratory syndrome virus propagated in the highly permissive MARC-145 cell clone. Adv. Exp. Med. Biol. 380:95-98. [DOI] [PubMed] [Google Scholar]
- Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarmis, C., G. Gomez-Mariano, M. Davila, E. Lazaro, and E. Domingo. 2002. Resistance to extinction of low fitness virus subjected to plaque-to-plaque transfers: diversification by mutation clustering. J. Mol. Biol. 315:647-661. [DOI] [PubMed] [Google Scholar]
- Gaut, B. S., B. R. Morton, B. C. McCaig, and M. T. Clegg. 1996. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93:10274-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai, S. 1995. Evolution and the origins of man: clues from complete sequences of hominoid mitochondrial DNA. Southeast Asian J Trop Med Public Health. 26:146-154. [PubMed] [Google Scholar]
- Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- Lee, L. M., and D. K. Henderson. 2001. Emerging viral infections. Curr. Opin. Infect. Dis. 14:467-480. [DOI] [PubMed] [Google Scholar]
- Li, W. H., M. Tanimura, and P. M. Sharp. 1987. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol. 25:330-342. [DOI] [PubMed] [Google Scholar]
- Mansky, L. M. 2000. In vivo analysis of human T-cell leukemia virus type 1 reverse transcription accuracy. J. Virol. 74:9525-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata, T., and T. Yasunaga. 1980. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J. Mol. Evol. 16:23-36. [DOI] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science. 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- Parashar, U. D., L. M. Sunn, and F. Ong, et al. (17 coauthors). 2000. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998–1999 outbreak of severe encephalitis in Malaysia. J. Infect. Dis. 181:1755-1759. [DOI] [PubMed] [Google Scholar]
- Pawlowski, J., I. Bolivar, J. F. Fahrni, C. de Vargas, M. Gouy, and L. Zaninetti. 1997. Extreme differences in rates of molecular evolution of foraminifera revealed by comparison of ribosomal DNA sequences and the fossil record. Mol. Biol. Evol. 14:498-505. [DOI] [PubMed] [Google Scholar]
- Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- Robertson, B. H. 2001. Viral hepatitis and primates: historical and molecular analysis of human and nonhuman primate hepatitis A, B, and the GB-related viruses. J. Viral. Hepat. 8:233-242. [DOI] [PubMed] [Google Scholar]
- Salemi, M., M. Lewis, J. F. Egan, W. W. Hall, J. Desmyter, and A. M. Vandamme. 1999. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc. Natl. Acad. Sci. USA 96:13253-13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag, S. J., P. A. Rota, and W. J. Bellini. 1991. Spontaneous mutation rate of measles virus: direct estimation based on mutations conferring monoclonal antibody resistance. J. Virol. 73:51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stech, J., X. Xiong, C. Scholtissek, and R. G. Webster. 1999. Independence of evolutionary and mutational rates after transmission of avian influenza viruses to swine. J. Virol. 73:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., K. Hanada, M. Mizokami, A. E. Yeo, J. W. Shih, T. Gojobori, and H. J. Alter. 2002. Inaugural Article: A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc. Natl. Acad. Sci. USA 99:15584-15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., W. H. Li, and P. M. Sharp. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84:9054-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001 92:371-373. [DOI] [PubMed] [Google Scholar]
- Yanagihara, R., N. Saitou, V. R. Nerurkar, K. J. Song, I. Bastian, G. Franchini, and D. C. Gajdusek. 1995. Molecular phylogeny and dissemination of human T-cell lymphotropic virus type I viewed within the context of primate evolution and human migration. Cell Mol. Biol. 41:145-161. [PubMed] [Google Scholar]
- Yang, Z., S. Kumar, and M. Nei. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.