Abstract
Cancer cells are known to adopt aerobic glycolysis in order to fuel tumor growth, but the molecular basis of this metabolic shift remains largely undefined. O-GlcNAcase (OGA) is an enzyme harboring O-linked β-N-acetylglucosamine (O-GlcNAc) hydrolase and cryptic lysine acetyltransferase activities. Here, we report that OGA is upregulated in a wide range of human cancers and drives aerobic glycolysis and tumor growth by inhibiting pyruvate kinase M2 (PKM2). PKM2 is dynamically O-GlcNAcylated in response to changes in glucose availability. Under high glucose conditions, PKM2 is a target of OGA-associated acetyltransferase activity, which facilitates O-GlcNAcylation of PKM2 by O-GlcNAc transferase (OGT). O-GlcNAcylation inhibits PKM2 catalytic activity and thereby promotes aerobic glycolysis and tumor growth. These studies define a causative role for OGA in tumor progression and reveal PKM2 O-GlcNAcylation as a metabolic rheostat that mediates exquisite control of aerobic glycolysis.
Keywords: OGA, OGT, PKM2, acetylation, O-GlcNAc, aerobic glycolysis, cancer metabolism
Introduction
Metabolic reprogramming is an emerging hallmark of cancer10, 33. In order to generate the biomass required for rapid proliferation, cancer cells consume an exceptionally large amount of glucose and process it primarily through glycolysis, even in the presence of oxygen. Although aerobic glycolysis produces ATP inefficiently, it allows for increased diversion of glycolytic intermediates into biosynthetic pathways that produce the amino acid, lipid, and nucleotide building blocks required for the construction of new cells. This dramatic shift from catabolic oxidative phosphorylation characteristic of normal cells to anabolic aerobic glycolysis favored by cancer cells is a phenomenon known as the Warburg effect31.
Cells also metabolize a small percentage of glucose through the hexosamine biosynthetic pathway (HBP), which enables the dynamic post-translational modification of cytoplasmic, nuclear, and mitochondrial proteins by O-linked β-N-acetylglucosamine (O-GlcNAc)11, 38. O-GlcNAc transferase (OGT) catalyzes the addition of GlcNAc moieties to the hydroxyl groups of serine and threonine residues, while O-GlcNAcase (OGA) catalyzes the removal of these sugar modifications from target proteins. Cellular levels of UDP-GlcNAc, the end product of the HBP and donor substrate for O-GlcNAcylation, fluctuate in response to changes in the availability of nutrients such as glucose, glutamine, free fatty acids, and uridine. Thus, O-GlcNAc has been proposed to function as a cellular nutrient sensor3, 20, 25. As a result, numerous studies have investigated the role of O-GlcNAc in regulating cellular metabolism, several of which implicate aberrant O-GlcNAcylation in the pathogenesis of metabolic disorders such as diabetes and obesity16, 24, 26, 37, 41.
Since elevated cellular O-GlcNAcylation levels have been observed in various cancers5, 22, 27, 28, 39, one question of interest has been whether O-GlcNAc plays a role in regulating cancer cell metabolism. While previous studies have demonstrated that OGT can drive metabolic reprogramming in cancer cells and promote tumor growth through phosphofructokinase 1 (PFK1) and hypoxia-inducible factor 1 alpha (HIF-1α)8, 40, the fuion of the classically opposing enzyme OGA in cancer has not yet been established. Here, we report the surprising finding that OGA, an enzyme harboring O-GlcNAc hydrolase and cryptic lysine acetyltransferase activities12, 30, can promote aerobic glycolysis and drive tumor growth through its associated acetyltransferase activity.
Post-translational modifications have been shown to regulate several key control points in the metabolic network of cancer cells. One such control point is pyruvate kinase muscle isoform 2 (PKM2), one of two splice variants of the PKM gene (PKM1/2)36. While PKM1 exists only as highly active tetramers, PKM2 can function either as highly active tetramers or as less active dimers and monomers1, 2. In cancer cells, the less active oligomers control the conversion of phosphoenolpyruvate (PEP) to pyruvate, thereby impeding carbon flow into the tricarboxylic acid (TCA) cycle. Previous studies have implicated tyrosine phosphorylation- and lysine acetylation-mediated suppression of PKM2 catalytic activity in aerobic glycolysis and tumor progression6, 7, 14, 18, 19. Here, we reveal that, at high glucose, OGA can facilitate PKM2 acetylation, leading to enhanced O-GlcNAcylation of PKM2 by OGT and reduced PKM2 tetramerization and catalytic activity. Additionally, we show that O-GlcNAcylation of PKM2 drives metabolic reprogramming in cancer cells and promotes tumor growth. These studies elucidate the causative role of OGA in tumor progression, further establishing the importance of O-GlcNAc in regulating cancer cell metabolism.
Results
Expression of O-GlcNAc cycling and HBP enzymes is upregulated in human tumors
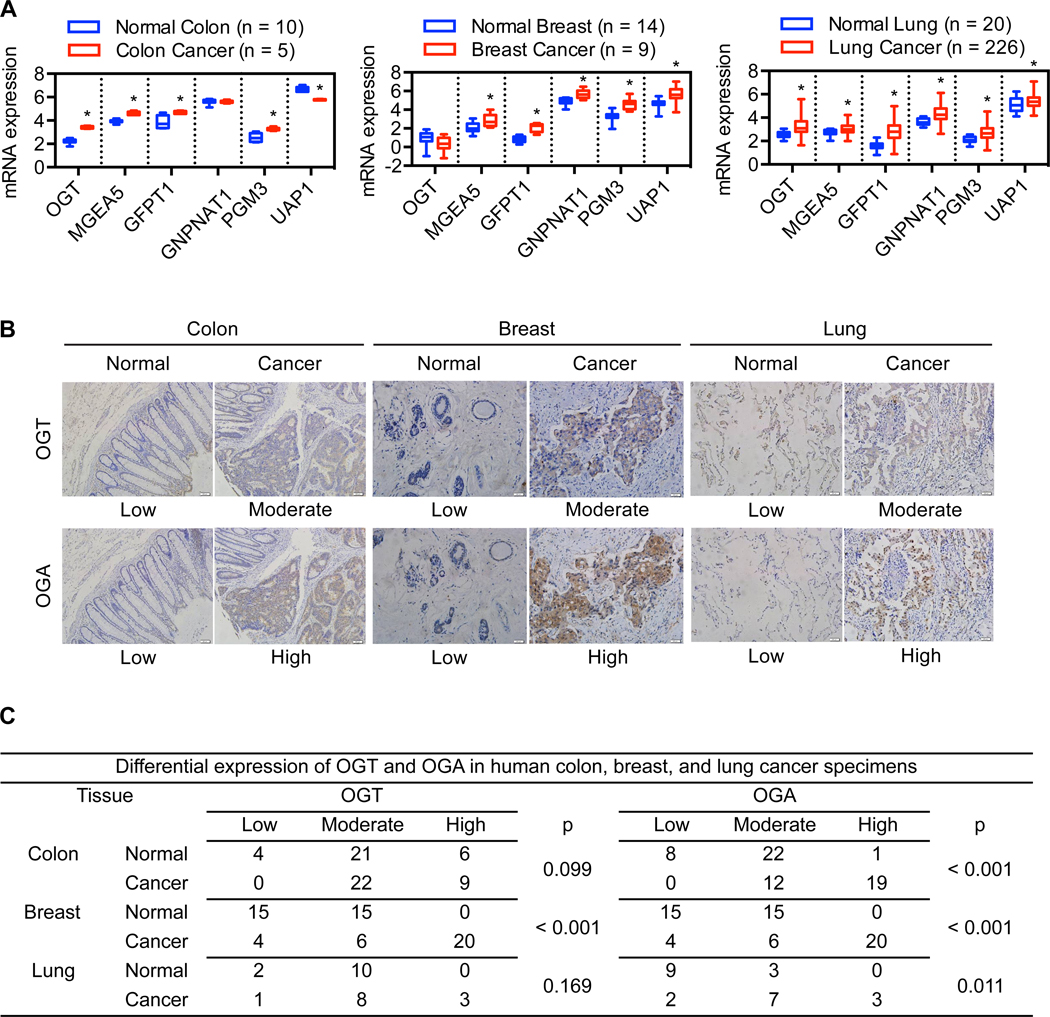
Protein O-GlcNAcylation is controlled by the interplay between the two O-GlcNAc cycling enzymes OGT and OGA. Availability of UDP-GlcNAc, the donor substrate for O-GlcNAcylation and end product of the HBP, also plays a critical role in determining cellular O-GlcNAcylation levels25. To gain insight into the role of O-GlcNAc signaling in human cancers, we analyzed mRNA expression of the O-GlcNAc cycling and HBP enzymes in human tumors using publicly available microarray datasets. Our analysis revealed that mRNA expression of OGT, OGA, and major HBP enzymes (GFPT1, GNPNAT1, PGM3, and UAP1) is significantly increased in colon, breast, and lung cancers as compared to their respective normal tissues (Figure 1A). Overall, mRNA expression of OGT, OGA, and GFPT1 is elevated in a wide range of tumor types (Table S1–S3). Consistent with these observations, immunohistochemical analysis of human tumor specimens demonstrated that OGA is more frequently expressed at a moderate to high level in colon, breast, and lung cancers as compared to adjacent normal tissues and that OGT expression is significantly increased in breast cancers (Figure 1B–C). Similarly, most lymphomas analyzed exhibited moderate to high expression of OGT and/or OGA (Figure S1A–B). These results indicate that the expression levels of O-GlcNAc cycling and HBP enzymes are simultaneously upregulated in many human tumors.
Figure 1. Expression of O-GlcNAc cycling and HBP enzymes is upregulated in human tumors.
(A) Box-and-whisker plots comparing the mRNA expression levels of the indicated genes in human tumors and their respective normal tissues. (B) Representative images of immunohistochemical analysis of OGT and OGA expression levels in human tumors and their respective normal tissues (magnification: 200X). (C) Stratification of human tumors and their respective normal tissues into low, moderate, and high expression of OGT or OGA. For A, multiple t-tests plus Holm-Sidak correction for multiple comparisons was used to compare cancer with normal for each tumor type (*p < 0.05). For C, chi-square test was used to compare cancer with normal; p-values are shown.
OGA-associated acetyltransferase activity promotes tumor growth and aerobic glycolysis
Both gain- and loss-of-function studies have shown that OGT can promote cell proliferation and tumor growth5, 8, 40. Since OGA expression is simultaneously upregulated in human tumors, we next sought to determine whether OGA also plays a role in driving tumor progression. To test this idea, we generated a Tet-Off stable cell line overexpressing OGA for use in xenograft studies. HeLa cells were used to construct this cell line because of their low basal tumorigenic potential. In vitro, Tet-Off HeLa cells proliferated at a faster rate in the absence of doxycycline than in the presence of doxycycline (Figure S2A). In vivo, nude mice injected subcutaneously with Tet-Off HeLa cells without being fed doxycycline developed tumors faster than those injected with vector control cells (Figure S2B–F). This was corroborated by a faster rate of cell proliferation in OGA-overexpressing tumors as evaluated by 5-bromodeoxyuridine (BrdU) incorporation (Figure S2F). In contrast, Tet-Off HeLa cells injected into doxycycline-fed mice showed no expression of exogenous OGA protein and no growth advantage over vector control cells (Figure S2B–F). In humans, high OGA expression levels are strongly associated with high pathological grades in colon adenocarcinoma and breast ductal carcinoma and advanced clinical stages of lung adenocarcinoma (Table S4–S6). Collectively, these data suggest that OGA may support tumor progression.
OGA is an enzyme that possesses both O-GlcNAc hydrolase and cryptic lysine acetyltransferase activities12, 30. To assess the impact of these two enzymatic activities on cell proliferation, we generated MDA-MB-231 breast cancer and H1299 lung cancer cells stably expressing wild-type OGA (OGAwt) or an OGA mutant defective in either O-GlcNAcase or cryptic acetyltransferase activity (OGAcd and OGAhd, respectively). For both cancer cell types, OGAhd-expressing cells proliferated at a slower rate than OGAwt- and OGAcd-expressing cells, suggesting that OGA-associated acetyltransferase activity is important for cell proliferation (Figure S2G). To further evaluate the contributions of these two enzymatic activities to the tumor-promoting capacity of OGA, we constructed Tet-Off HeLa cells expressing OGAwt, OGAcd, or OGAhd and performed xenograft studies. As expected, expression of OGAwt decreased global O-GlcNAcylation and increased global acetylation, while these effects were blunted in OGAcd- and OGAhd-expressing cells, respectively (Figure S2H). Over a 30-day period, OGAwt and OGAcd cells formed tumors much faster than vector control cells, whereas OGAhd and control cells formed tumors at a similar rate (Figure 2A). The volume and weight of OGAwt and OGAcd tumors were greater than those of OGAhd and control tumors at the end of the 30-day period (Figure 2B–C and S2I). These data suggest that OGA promotes tumor growth through its associated acetyltransferase activity.
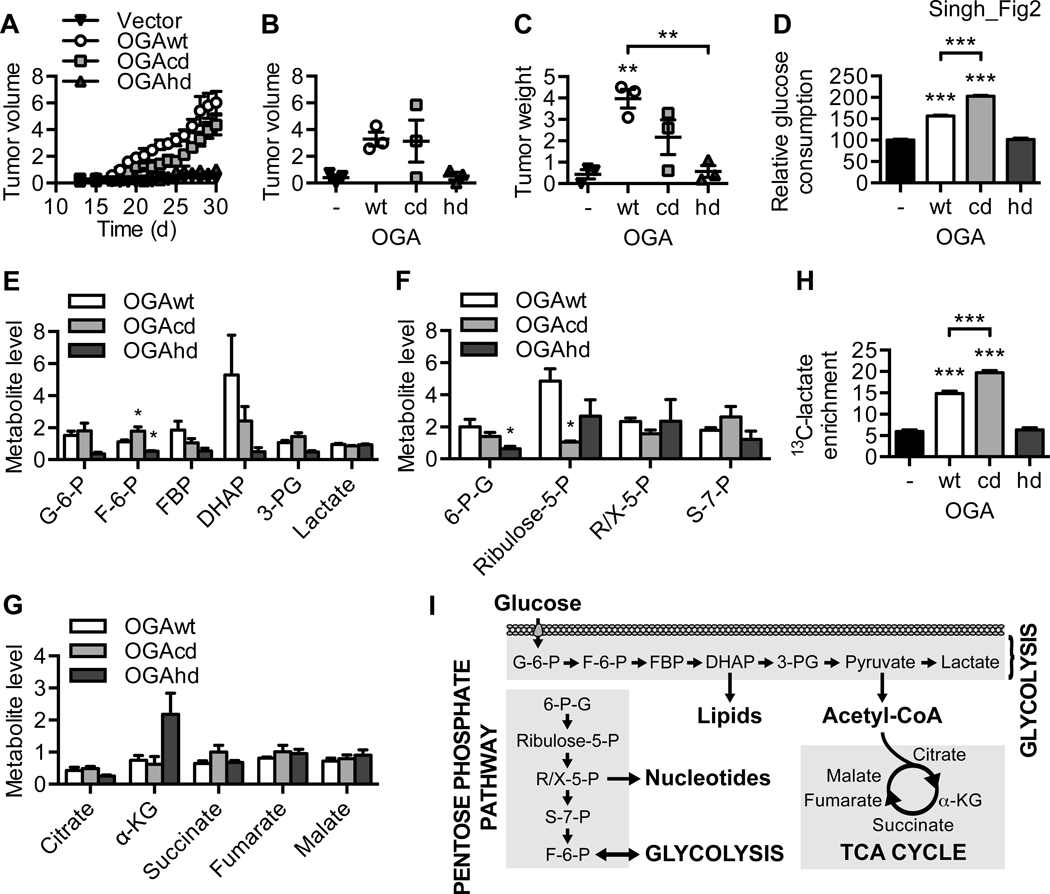
Figure 2. OGA-associated acetyltransferase activity promotes tumor growth and aerobic glycolysis.
(A) Tet-Off HeLa cells expressing vector, OGAwt, OGAcd, or OGAhd were injected subcutaneously into the flanks of nude mice. Daily progression of the xenograft tumors is shown (n = 3). (B) Volume and (C) weight of the xenograft tumors at the end of the 30-day period (n = 3). (D) Relative glucose consumption (n = 3), levels of (E) glycolysis, (F) pentose phosphate pathway, and (G) TCA cycle metabolites (n = 4), and (H) steady-state fractional enrichment of 13C-lactate (n = 5) in OGAwt, OGAcd, and OGAhd cells. (I) Schematic representation of glucose metabolism via glycolysis, the pentose phosphate pathway, and the TCA cycle. All values represent mean ± SEM. For B-D and H, ordinary one-way ANOVA plus Tukey’s multiple comparisons test was used to compare each column with every other column (**p < 0.01, ***p < 0.001 as compared to vector and OGAhd unless otherwise indicated). For E-G, ordinary one-way ANOVA plus Dunnett’s multiple comparisons test was used to compare each column with OGAwt for each metabolite (*p < 0.05).
Proliferating cancer cells are known to consume an exceptionally large amount of glucose, leading to increased glycolytic and pentose phosphate pathway (PPP) flux and elevated lactate production31. To determine whether OGA can drive metabolic reprogramming in cancer cells, we first performed an in vitro glucose consumption assay, which demonstrated that HeLa cells expressing either OGAwt or OGAcd consume significantly more glucose than OGAhd and control cells (Figure 2D). We then performed quantification of major glycolytic and PPP metabolites in OGAwt, OGAcd, and OGAhd cells grown in excess high glucose medium for 24 hours. We found that the relative levels of major glycolytic metabolites, including glucose-6-phosphate (G-6-P), fructose-6-phosphate (F-6-P), fructose-1,6-bisphosphate (FBP), dihydroxyacetone phosphate (DHAP), and 3-phosphoglycerate (3-PG), were reduced in OGAhd cells as compared to OGAwt cells (Figure 2E). While there were no significant differences in the relative levels of most major PPP metabolites, the relative amount of 6-phosphogluconate (6-P-G) was also found to be significantly reduced in OGAhd cells, suggesting that less glucose may be entering the PPP in these cells (Figure 2F). A panel of TCA cycle metabolites showed no significant differences among OGAwt, OGAcd, and OGAhd cells (Figure 2G). Consistent with the results of our xenograft studies, these metabolic analyses indicate that OGA-associated acetyltransferase activity promotes aerobic glycolysis and may help redirect glucose into anabolic pathways such as the PPP to support cell proliferation and tumor growth.
Unexpectedly, we did not observe any significant differences in the relative levels of lactate in our metabolomics analysis (Figure 2E). Since proliferating cancer cells are known to use lactate as a carbon source and cell culture medium does not contain lactate29, it is possible that cancer cells will consume some of the lactate generated by glycolysis if they are allowed to grow for more than 24 hours without medium change. Therefore, to measure lactate production more accurately, we quantified fractional 13C-lactate enrichment using 13C-nuclear magnetic resonance (13C-NMR) spectroscopy after treating cells with uniformly labeled 13C-glucose for only 4 hours. We found that 13C-lactate enrichment was significantly higher in OGAwt and OGAcd cells than in OGAhd and control cells (Figure 2H and S2J). Overall, these results demonstrate that OGA-associated acetyltransferase activity promotes tumor growth and drives metabolic reprogramming in cancer cells.
OGA enhances PKM2 acetylation and O-GlcNAcylation
To identify potential targets of OGA-associated acetyltransferase activity that may be responsible for its tumor-promoting capacity, we transiently expressed either OGAwt or OGAhd in HeLa cells and performed a differential proteomic analysis of OGA-binding proteins (Figure S3A–B). A majority of the proteins that co-purified with OGAwt did not co-purify with OGAhd, implicating these proteins as potential targets of OGA-associated acetyltransferase activity (Figure 3A and Table S7–S8). One protein that interacted exclusively with OGAwt was PKM2. Since previous studies have linked PKM2 to aerobic glycolysis and tumor progression36, we reasoned that one possible explanation for the tumor-promoting capacity of OGA-associated acetyltransferase activity could be its targeting of PKM2. To determine whether the ability of OGA to promote cell proliferation is dependent on PKM2, we knocked down PKM2 in HeLa cells expressing OGAwt using shPKM2. Quantification of in vitro colony formation revealed that depletion of PKM2 abrogates OGA-induced cell proliferation, suggesting that PKM2 may be a critical target of OGA in cancer cells (Figure 3B and S3C).
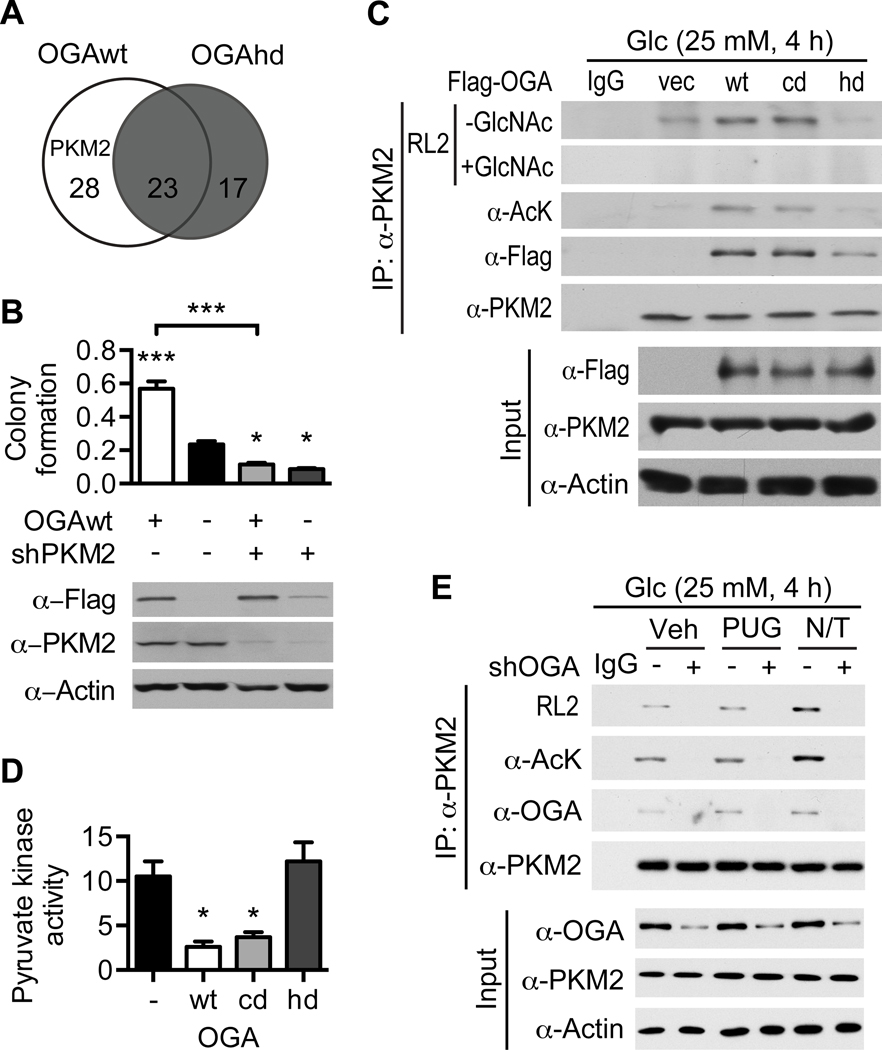
Figure 3. OGA enhances PKM2 acetylation and O-GlcNAcylation.
(A) Venn diagram comparing HeLa cell proteins that co-purified with OGAwt to those that co-purified with OGAhd. (B) Clonogenic assay of HeLa cells expressing OGAwt and infected with lentivirus encoding shPKM2 (n = 3). (Top) Quantification of colony formation by measurement of crystal violet dye incorporation. (Bottom) Western blot confirming expression of OGAwt and knockdown of PKM2. (C) HeLa cells were transiently transfected with the indicated plasmids and treated with 25 mM glucose for 4 hours. PKM2 was immunoprecipitated and analyzed by Western blotting with the indicated antibodies. The specificity of the RL2 antibody was confirmed by competition with 0.1 M free GlcNAc. (D) Pyruvate kinase activity in vector, OGAwt, OGAcd, and OGAhd xenograft tumor homogenate (n = 3). (E) MCF7 cells were infected with adenovirus encoding shOGA and treated with 25 mM glucose plus the indicated inhibitors for 4 hours. PKM2 was immunoprecipitated and analyzed by Western blotting with the indicated antibodies. All values represent mean ± SEM. For B, ordinary one-way ANOVA plus Tukey’s multiple comparisons test was used to compare each column with every other column (*p < 0.05, ***p < 0.001 as compared to -OGAwt/-shPKM2 unless otherwise indicated). For D, ordinary one-way ANOVA plus Tukey’s multiple comparisons test was used to compare each column with every other column (*p < 0.05 as compared to vector and OGAhd).
To determine whether OGA can affect PKM2 acetylation in a cellular context, we transiently expressed OGAwt, OGAcd, or OGAhd in HeLa cells. We found that expression of either OGAwt or OGAcd markedly increased PKM2 acetylation, whereas expression of OGAhd had no effect and was associated with an impaired physical interaction between OGA and PKM2 (Figure 3C). Comparison of PKM2 acetylation levels among control, OGAwt, OGAcd, and OGAhd xenograft tumors revealed that expression of OGAwt or OGAcd but not OGAhd also increases PKM2 acetylation in vivo (Figure S3D). Increased PKM2 acetylation in OGAwt and OGAcd tumors was associated with decreased pyruvate kinase activity in these tumors, suggesting that OGA-induced acetylation of PKM2 may suppress its catalytic activity (Figure 3D). Since previous studies have linked reduced pyruvate kinase activity to aerobic glycolysis and tumor progression6, 7, 14, 18, 19, we reasoned that suppression of PKM2 catalytic activity by OGA-induced acetylation could plausibly contribute to the tumorigenic and metabolic phenotypes we observed.
To test whether OGA can directly acetylate PKM2 in vitro, we performed a cell-free acetyltransferase activity assay using purified OGA and recombinant PKM2. We found that addition of OGAwt to PKM2 accelerated the enzymatic reaction, indicating that OGA is indeed associated with acetyltransferase activity toward PKM2 (Figure S3E). However, in this cell-free system, the hd mutation did not affect the ability of OGA to induce PKM2 acetylation, suggesting that OGA does not possess intrinsic acetyltransferase activity. These results argue that, in a cellular context, other factors associated with OGA confer acetyltransferase activity toward PKM2.
Unexpectedly, we also observed an increase in PKM2 O-GlcNAcylation in cells and xenograft tumors expressing OGAwt or OGAcd but not in those expressing OGAhd (Figure 3C and S3D). This finding raised the possibility that OGA-induced acetylation of PKM2 also modulates PKM2 O-GlcNAcylation. To test this idea further, MCF7 breast cancer cells were depleted of OGA using shOGA and treated with the O-GlcNAcase inhibitor PUGNAc or the lysine deacetylase inhibitors nicotinamide and trichostatin A (NAM/TSA) (Figure S3F). MCF7 cells were used for OGA knockdown because their basal OGA expression level was the highest among the seven cell lines tested (Figure S3G). In MCF7 cells, NAM/TSA treatment increased both acetylation and O-GlcNAcylation of PKM2 and depletion of OGA reduced PKM2 acetylation and O-GlcNAcylation in all three treatment groups, indicating that OGA-induced acetylation of PKM2 enhances PKM2 O-GlcNAcylation (Figure 3E). In MCF10A non-tumorigenic breast epithelial cells, depletion of OGA had no effect on PKM2 acetylation in any treatment group and increased PKM2 O-GlcNAcylation in the vehicle- and PUGNAc-treated groups, suggesting that the response observed in MCF7 cells may be specific to cancer cells (Figure S3H). However, in HeLa cells, OGA knockdown only reduced PKM2 acetylation and O-GlcNAcylation in the NAM/TSA-treated group (Figure S3I). We reasoned that this was indicative of a low basal level of OGA-associated acetyltransferase activity in HeLa cells, which is consistent with our finding that OGA overexpression dramatically enhances the low basal tumorigenic potential of HeLa cells and that HeLa cells have the lowest basal OGA expression level among all cell lines tested (Figure S2B–F and S3G).
We also found that, in all three cell lines, PUGNAc treatment had no appreciable effect on PKM2 O-GlcNAcylation (Figure 3E). This observation suggests that, under the high glucose conditions used in these experiments, PKM2 may not be a major substrate of OGA O-GlcNAcase activity. Overall, these results demonstrate that PKM2 is a critical target of OGA-associated acetyltransferase activity in cancer cells and that acetylation of PKM2 enhances its O-GlcNAcylation.
Glucose availability modulates the effect of OGA on PKM2
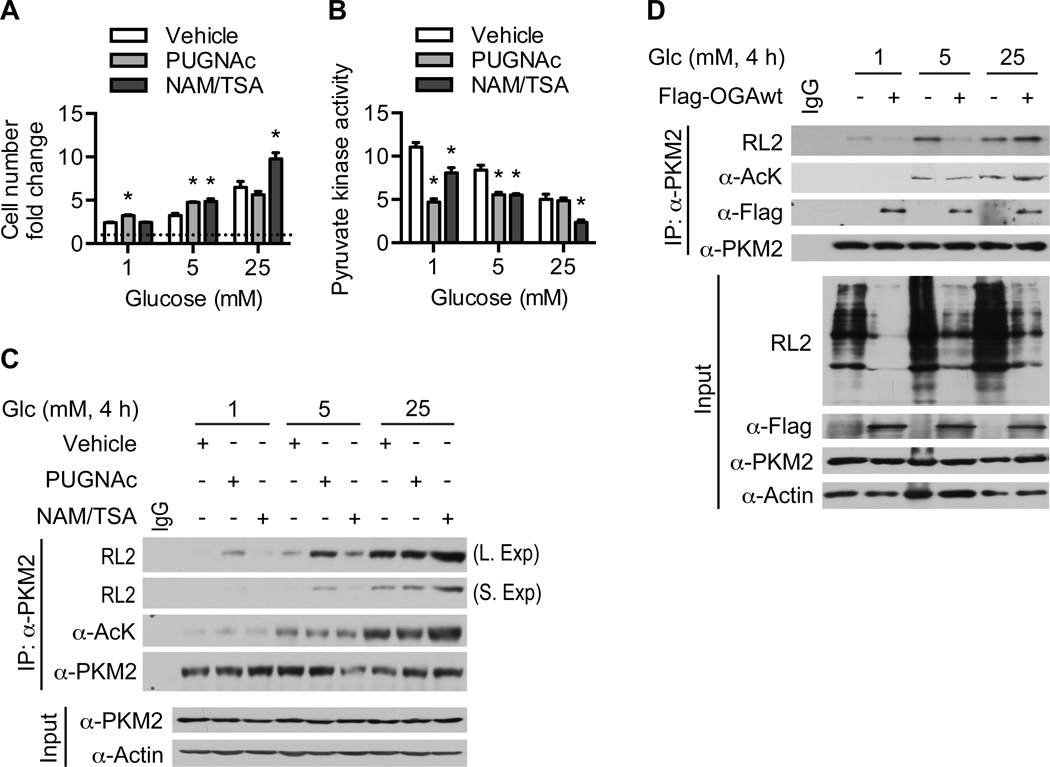
Since PKM2 appears to be a major substrate of OGA-associated acetyltransferase activity but not of OGA O-GlcNAcase activity under high glucose conditions, we next examined whether the effect of OGA on PKM2 is dependent on glucose availability. To test this idea, we treated HeLa cells with PUGNAc or NAM/TSA at different glucose concentrations. PUGNAc treatment enhanced cell proliferation slightly at low (1 mM) and medium (5 mM) glucose but not at high (25 mM) glucose, whereas NAM/TSA treatment increased cell proliferation slightly at medium glucose and dramatically at high glucose (Figure 4A). These increases in cell proliferation were paralleled by decreases in pyruvate kinase activity, consistent with our previous observation that pyruvate kinase activity is suppressed in fast-growing OGAwt and OGAcd xenograft tumors (Figure 4B and 3D). Although PUGNAc treatment raised global O-GlcNAcylation levels at all three glucose concentrations, it only increased PKM2 O-GlcNAcylation at low and medium glucose (Figure S4 and 4C). Conversely, NAM/TSA treatment raised global lysine acetylation levels at all three glucose concentrations, but only increased PKM2 acetylation at high glucose (Figure S4 and 4C). These data suggest that PKM2 is primarily a target of OGA O-GlcNAcase activity at low and medium glucose and OGA-associated acetyltransferase activity at high glucose. Transient expression of OGAwt in HeLa cells decreased PKM2 O-GlcNAcylation at low and medium glucose but increased PKM2 acetylation and O-GlcNAcylation at high glucose, further supporting this notion (Figure 4D). Taken together, these results demonstrate that the effect of OGA on cell proliferation and PKM2 acetylation and O-GlcNAcylation is dependent on glucose availability.
Figure 4. Glucose availability modulates the effect of OGA on PKM2.
(A) Cell number fold change of HeLa cells cultured in the indicated glucose concentrations plus the indicated inhibitors for 72 hours (n = 3). (B) Pyruvate kinase activity (n = 3) and (C) Western blot analysis of immunoprecipitated PKM2 in HeLa cells treated with the indicated glucose concentrations plus the indicated inhibitors for 4 hours. (D) HeLa cells were transiently transfected with Flag-OGAwt and treated with the indicated glucose concentrations for 4 hours. PKM2 was immunoprecipitated and analyzed by Western blotting with the indicated antibodies. All values represent mean ± SEM. For A-B, ordinary one-way ANOVA plus Dunnett’s multiple comparisons test was used to compare each column with vehicle for each glucose concentration (*p < 0.05).
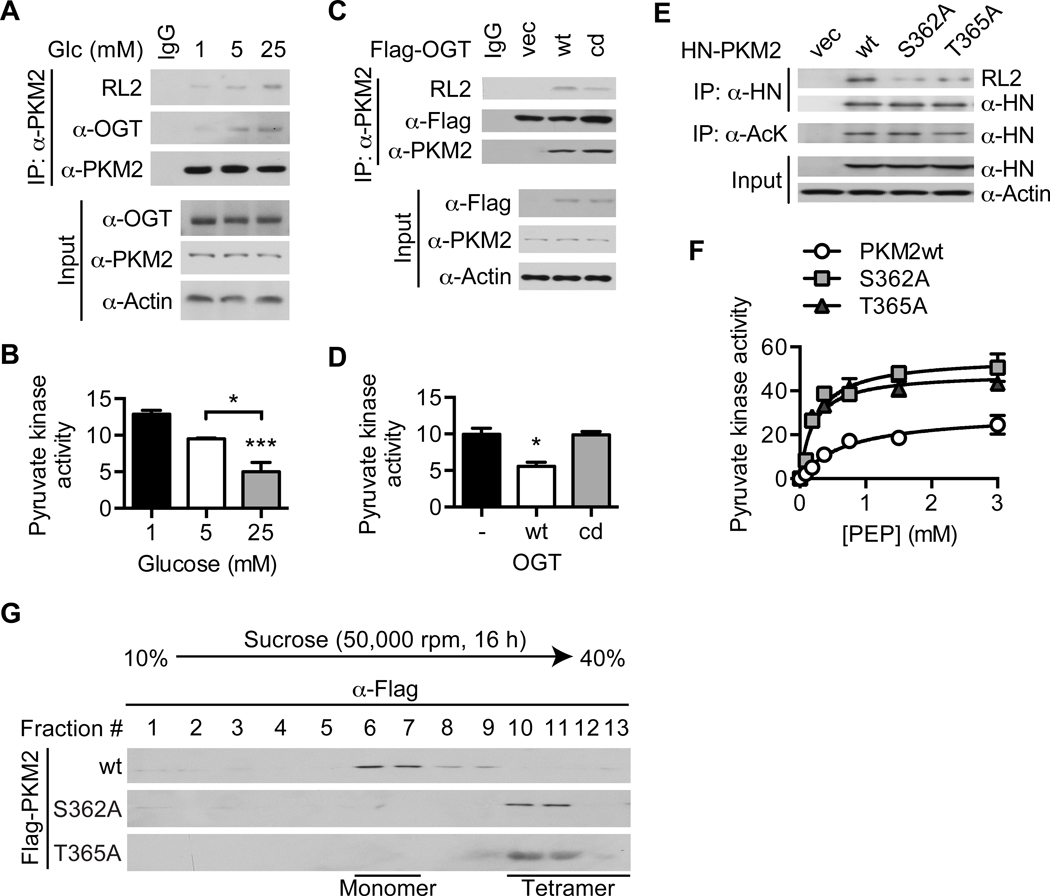
O-GlcNAcylation of PKM2 inhibits its catalytic activity and tetramerization
Since OGA-induced acetylation of PKM2 enhances PKM2 O-GlcNAcylation and increased PKM2 O-GlcNAcylation correlates with decreased pyruvate kinase activity, we next sought to determine whether O-GlcNAcylation of PKM2 directly regulates its catalytic activity (Figure 3D, S3D, and 4B–C). In HeLa cells, PKM2 O-GlcNAcylation increased and pyruvate kinase activity decreased as glucose concentration was elevated (Figure 5A–B and S5A). Transient expression of wild-type OGT (OGTwt) in HeLa cells enhanced PKM2 O-GlcNAcylation and reduced pyruvate kinase activity, whereas expression of a catalytic-dead mutant of OGT (OGTcd) had no effect (Figure 5C–D and S5B). Collectively, these data suggest that O-GlcNAcylation is a glucose-sensitive regulator of PKM2 catalytic activity.
Figure 5. O-GlcNAcylation of PKM2 inhibits its catalytic activity and tetramerization.
(A) Western blot analysis of immunoprecipitated PKM2 and (B) pyruvate kinase activity (n = 3) in HeLa cells treated with the indicated glucose concentrations for 4 hours. (C) Western blot analysis of immunoprecipitated PKM2 and (D) pyruvate kinase activity (n = 2) in HeLa cells transiently transfected with the indicated plasmids. (E) Western blot analysis of immunoprecipitated HN-PKM2 and (F) effect of PEP concentration on pyruvate kinase activity (n = 2) in HeLa cells transiently transfected with the indicated plasmids. (G) HeLa cells were transiently transfected with the indicated plasmids. Comparative analysis of PKM2 oligomeric forms using sucrose density gradient ultracentrifugation of whole cell lysates followed by Western blotting with an anti-Flag antibody is shown. All values represent mean ± SEM. For B and D, ordinary one-way ANOVA plus Tukey’s multiple comparisons test was used to compare each column with every other column (*p < 0.05, ***p < 0.001 as compared to 1 mM for B and vector and OGTcd for D unless otherwise indicated).
To identify O-GlcNAcylated PKM2 amino acid residues, we purified HN-tagged PKM2 transiently expressed in human embryonic kidney 293T (HEK293T) cells and analyzed it using collision-induced dissociation mass spectrometry (CID-MS) (Figure S5C). Two conserved O-GlcNAc sites, S362 and T365, were identified (Figure S5D–E). Compared to wild-type PKM2 (PKM2wt), the S362A and T365A mutants transiently expressed in HeLa cells showed decreased O-GlcNAcylation and increased catalytic activity (Figure 5E–F and S5F). Since PKM2 is known to be most active in its tetrameric form1, 2, we then performed sucrose density gradient ultracentrifugation on whole-cell lysates of HeLa cells transiently expressing wild-type or mutant PKM2 to determine whether O-GlcNAcylation modulates the activity of PKM2 by altering its oligomerization. We found that PKM2wt exists primarily in the form of less active monomers, while the S362A and T365A mutants are predominantly fully active tetramers (Figure 5G and S5G). Taken together, these results demonstrate that O-GlcNAcylation of PKM2 inhibits its catalytic activity by blocking its tetramerization.
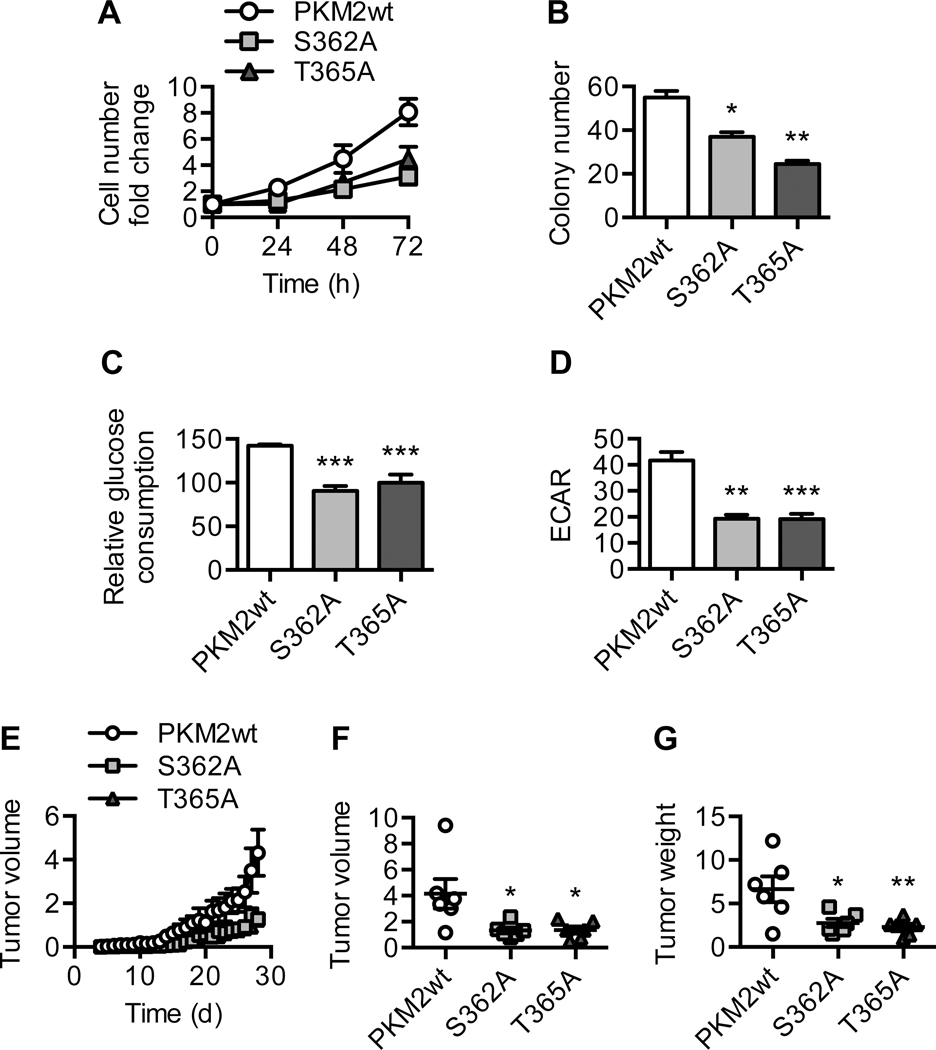
PKM2 O-GlcNAcylation promotes aerobic glycolysis and tumor growth
Since enhanced PKM2 O-GlcNAcylation and reduced pyruvate kinase activity correlate with increased cell proliferation, we next examined the functional significance of PKM2 O-GlcNAcylation in terms of cell proliferation and tumor growth (Figure 4A–C). To do so, we generated Tet-Off HeLa cell lines stably expressing PKM2wt, S362A, or T365A. Compared to PKM2wt cells, S362A and T365A cells showed marked reduction in their in vitro proliferation rates and colony formation potential (Figure 6A–B and S6A). This was associated with reduced glucose consumption and glycolytic flux in these cells (Figure 6C–D). To determine whether PKM2 O-GlcNAcylation is critical for tumor growth in vivo, we injected nude mice subcutaneously with PKM2wt, S362A, or T365A cells. We found that, over a 4-week period, tumors derived from S362A or T365A cells had reduced growth rates compared to those derived from PKM2wt cells (Figure 6E). The volume and weight of S362A and T365A tumors were also significantly lower than those of PKM2wt tumors at the end of the 4-week period, even though the expression levels of exogenous PKM2 protein in these tumors were comparable (Figure 6F–G and S6B). These results demonstrate that O-GlcNAcylation of PKM2 at S362 and T365 drives metabolic reprogramming in cancer cells and promotes tumor growth.
Figure 6. PKM2 O-GlcNAcylation promotes aerobic glycolysis and tumor growth.
(A) Cell number fold change of Tet-Off HeLa cells expressing PKM2wt, S362A, or T365A over 72 hours (n = 2). (B) Clonogenic assay of PKM2wt, S362A, and T365A cells (n = 2). Quantification of colony number after crystal violet staining is shown. (C) Relative glucose consumption (n = 6) and (D) basal extracellular acidification rate (ECAR) (n = 3) of PKM2wt, S362A, and T365A cells. (E) PKM2wt, S362A, or T365A cells were injected subcutaneously into the flanks of nude mice. Daily progression of the xenograft tumors is shown (n = 6). (F) Volume and (G) weight of the xenograft tumors at the end of the 4-week period (n = 6). All values represent mean ± SEM. For B-D and F-G, ordinary one-way ANOVA plus Dunnett’s multiple comparisons test was used to compare each column with PKM2wt (*p < 0.05, **p < 0.01, ***p < 0.001).
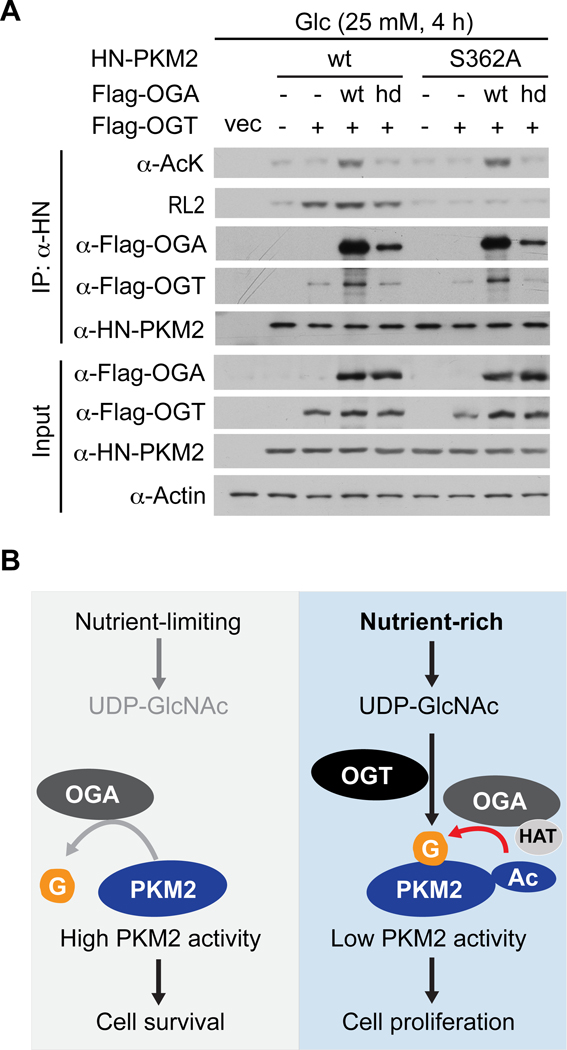
OGA cooperates with OGT to regulate PKM2 O-GlcNAcylation
Thus far, we have demonstrated that the tumor-promoting capacity of OGA depends on its associated acetyltransferase activity and requires PKM2. We have also shown that OGA-induced acetylation of PKM2 at high glucose enhances PKM2 O-GlcNAcylation and that O-GlcNAcylation of PKM2 promotes tumor growth by suppressing PKM2 catalytic activity. However, the mechanism through which OGA-induced acetylation of PKM2 leads to enhanced PKM2 O-GlcNAcylation has not yet been examined.
Our previous work demonstrated that OGA can physically associate with OGT34. Thus, we hypothesized that OGA-induced acetylation of PKM2 at high glucose recruits OGT and thereby enhances PKM2 O-GlcNAcylation. Indeed, transient expression of OGAwt in HeLa cells at high glucose increased PKM2wt acetylation, association with OGTwt, and O-GlcNAcylation, and these effects were lost with OGAhd (Figure 7A). Mutation of S362 abolished OGA-induced O-GlcNAcylation but not acetylation of PKM2, indicating that O-GlcNAcylation of PKM2 does not affect its acetylation (Figure 7A). Taken together, these results suggest that, under sufficient nutrient availability, acetylation of PKM2 induced by OGA-associated acetyltransferase activity facilitates OGT recruitment and enhances PKM2 O-GlcNAcylation, leading to suppression of PKM2 catalytic activity (Figure 7B).
Figure 7. OGA cooperates with OGT to regulate PKM2 O-GlcNAcylation.
(A) HeLa cells were transiently transfected with the indicated plasmids and treated with 25 mM glucose for 4 hours. HN-PKM2 was immunoprecipitated and analyzed by Western blotting with the indicated antibodies. (B) Working model. Under nutrient-limiting conditions, the O-GlcNAcase activity of OGA targets and activates PKM2, thereby increasing carbon flow into the TCA cycle and promoting cell survival. Under nutrient-rich conditions, the acetyltransferase activity associated with OGA targets and inactivates PKM2 through enhanced OGT-mediated O-GlcNAcylation, thereby increasing aerobic glycolysis and promoting cell proliferation.
Discussion
Altered metabolism is a fundamental characteristic of cancer10, 33. It is largely achieved by reprogramming gene expression in the metabolic network. However, a major unanswered question in the field is how various post-translational modifications of metabolic enzymes contribute to metabolic alterations in cancer. The present study illuminates a key role for the O-GlcNAc modification in cancer metabolism. Specifically, using cell-based studies, mouse xenograft models, and extensive human cancer surveys, we 1) elucidate the causative role of OGA in tumor progression, 2) reveal the unexpected importance of OGA-associated acetyltransferase activity to aerobic glycolysis and tumor growth, 3) demonstrate that PKM2 is a target of OGT and OGA, and 4) show that glucose availability can regulate cancer cell metabolism through the interplay between acetylation and O-GlcNAcylation of PKM2.
There is currently no consensus on the role of OGA in cancer. Yang et al. demonstrated that OGA protein levels are elevated in human colorectal cancers and that low OGA gene expression is associated with improved patient survival39, suggesting that OGA supports tumor progression. On the other hand, several groups have reported decreased OGA gene expression in specific human cancers and a correlation between low OGA transcript levels and poor patient outcome8, 15, implicating OGA in tumor suppression. However, these studies have generally been limited to small-scale measurements of either gene or protein expression in a single cancer type, which may be responsible for the discrepancies observed. Our study provides, to the best of our knowledge, the largest and most comprehensive profile of OGA expression in human cancers: it spans numerous cancer types and includes both gene expression from microarray datasets and protein expression from human cancer tissue arrays. Our profile indicates that OGA expression is extensively elevated in human cancers, supporting the notion that OGA may function to enhance tumor progression. While previous studies have provided a correlative link between OGA and cancer, our mouse xenograft studies reveal a causative role for OGA in tumor growth. Our results are corroborated by Yang et al., who showed that OGA heterozygosity attenuates tumorigenesis in a mouse model of colorectal cancer39.
The ability of OGA to promote tumor growth appears to contradict previous studies that have established a direct role for OGT in tumor progression8, 40. However, we demonstrate that OGA drives aerobic glycolysis and tumor growth primarily through its cryptic lysine acetyltransferase activity and not through its O-GlcNAc hydrolase activity, resolving this potential contradiction. This finding is unexpected because, outside of a few studies showing that OGA can modulate histone acetylation12, 30, the acetyltransferase activity associated with OGA has been largely ignored. Recent structural studies suggest that OGA itself may be a catalytically incompetent “pseudo-acetyltransferase”13, 23. Thus, it is likely that OGA is associated with additional proteins that confer acetyltransferase activity. Although the identity of the bona fide acetyltransferase remains to be determined, our study suggests a potential causative link between OGA-associated acetyltransferase activity and human disease.
Using proteomics, biochemical, cell biological, and in vivo approaches, we demonstrate that OGT and OGA target PKM2. PKM2 is known to be subject to extensive post-translational regulation, including serine/threonine phosphorylation by ERK35, tyrosine phosphorylation by FGFR114, and acetylation by PCAF and p30018, 19. These modifications have been shown to modulate PKM2 activity, degradation, oligomerization, and subcellular localization. Our study reveals two additional mechanisms of PKM2 post-translational regulation: O-GlcNAcylation by OGT and acetylation induced by OGA. Similar to phosphorylation at Y105 by FGFR1 and acetylation at K433 by p30014, 19, O-GlcNAcylation of PKM2 at S362 and T365 blocks its tetramerization and inhibits its catalytic activity. OGA-induced acetylation of PKM2 is also associated with reduced PKM2 activity, though the precise mechanism through which this occurs is not clear. In future studies, it will be important to determine whether these post-translational regulatory mechanisms are specific to PKM2 and cancer cells or whether they also apply to PKM1 and normal cells.
Although the specific PKM2 amino acid residues targeted by OGA-associated acetyltransferase activity have not yet been identified, our study elucidates one potential mechanism through which OGA may regulate PKM2 function. Specifically, OGA-induced acetylation facilitates OGT-mediated O-GlcNAcylation of PKM2, which blocks its tetramerization and inhibits its catalytic activity. OGT and OGA have previously been shown to associate and form an O-GlcNAczyme complex34, but the crosstalk between PKM2 acetylation and O-GlcNAcylation that we have described represents the first specific example of cooperation between these two classically opposing enzymes. Moreover, our study demonstrates that this crosstalk is sensitive to glucose availability. Specifically, under low glucose conditions, OGA O-GlcNAcase activity dominates, leading to reduced PKM2 O-GlcNAcylation; however, when glucose is abundant, OGA-associated acetyltransferase activity becomes more prominent, facilitating enhanced O-GlcNAcylation of PKM2 by OGT (Figure 7B). These results suggest that OGT and OGA are glucose-sensitive regulators of metabolism that allow cancer cells to allocate their nutritional resources according to glucose availability, a function that is critical for tumor growth.
Though our finding that OGA-associated acetyltransferase activity promotes aerobic glycolysis is consistent with a role for PKM2, it does not necessarily implicate PKM2 directly. Previous studies have demonstrated that OGT reprograms cancer cell metabolism through PFK1 and HIF-1α8, 40; since OGA cooperates with OGT in our model, it is possible that the mechanisms elucidated in these studies also play a role in OGA-mediated metabolic reprogramming22. Nonetheless, we show that O-GlcNAcylation of PKM2, which is enhanced by OGA-induced acetylation at high glucose, inhibits PKM2 activity and thereby drives aerobic glycolysis and tumor growth. While our finding that reduced PKM2 activity increases glycolytic flux and lactate production is paradoxical according to canonical glycolysis, it is consistent with previous studies and supported by evidence for the existence of an alternative glycolytic pathway6, 7, 14, 18, 32.
In conclusion, our study reveals that glucose availability can regulate cancer cell metabolism through the interplay between acetylation and O-GlcNAcylation of PKM2. This process is mediated by the cooperative action of OGT and OGA. Our study also provides a mechanistic explanation for the cooperative action of these two classically opposing enzymes. Our findings suggest that the crosstalk between various post-translational modifications on metabolic enzymes may integrate nutritional and hormonal signals to control intermediary metabolism. Targeting specific activities of the O-GlcNAc cycling enzymes to reset cellular metabolism may provide an attractive anticancer strategy.
Materials and Methods
Cell culture
HeLa, H1299, A549, and HEK293T cells were cultured in DMEM plus 10% FBS and 1% Pen-Strep (Life Technologies). MCF7 and MD-MBA-231 cells were cultured in RPMI plus 10% FBS and 1% Pen-Strep (Life Technologies). MCF10A cells were cultured in DMEM/F12 plus 5% horse serum and 1% Pen-Strep (Life Technologies) supplemented with EGF (Peprotech, 20 ng/mL), hydrocortisone (Sigma, 0.5 mg/mL), cholera toxin (Sigma, 100 ng/mL), and insulin (Sigma, 10 μg/mL). Cells were transfected with the indicated plasmids using FuGENE HD (Promega). Cells were treated with the indicated glucose concentrations by supplementing no glucose medium with glucose and 10% dialyzed FBS (Life Technologies). PUGNAc (Tocris, 10 μM), nicotinamide (Sigma, 5 mM), and trichostatin A (Sigma, 2 μM) were added to the treatment medium as indicated.
Plasmids and adenovirus preparation
Flag-OGTwt, Flag-OGAwt, and HN-PKM2 were constructed through subcloning of PCR products. Flag-PKM2 was kindly provided by Dr. Jinbo Yang. OGT E899A/E900A (OGTcd)16, OGA D177N (OGAcd) and Y891F (OGAhd)4, and PKM2 S362A and T365A point mutations were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent). OGA shRNA was purchased from Origene and OGA shRNA adenovirus was generated as described in the literature17. PKM2 shRNA lentivirus was kindly provided by Dr. Jinbo Yang.
Antibodies, immunoprecipitation, and Western Blotting
Cells and tumors were lysed in a 1% Nonidet P-40, 50 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA buffer with protease, protein phosphatase, and O-GlcNAcase inhibitors. Proteins were immunoprecipitated by incubating whole cell lysates/tumor homogenates with the indicated antibodies for 2 hours and precipitating with Protein A/G agarose beads. Whole cell lysates/tumor homogenates and immunoprecipitation samples were subjected to 8% SDS-PAGE gel electrophoresis and transferred to PVDF membranes. Membranes were incubated with the indicated primary antibodies overnight at 4°C. Western blots were visualized using HRP-conjugated secondary antibodies and ECL. The following antibodies were used: anti-actin (Sigma), anti-acetyl-lysine (AcK, Cell Signaling), anti-Flag (Sigma), anti-HN (Clontech), anti-OGT (Cell Signaling), anti-OGA (Novus), anti-O-GlcNAc (RL2, Abcam), and anti-PKM2 (Cell Signaling).
Generation of stable cell lines
To generate HeLa cells with stable expression of Flag-OGA or HN-PKM2, the HeLa Tet-Off Advanced Cell Line (Clontech) was transfected with a pTRE2pur vector expressing the protein of interest and selected for stable expression using culture medium supplemented with puromycin. The pTRE2pur-Flag-OGA/HN-PKM2 expression vectors were constructed through subcloning of PCR products. Stable cell lines were maintained in doxycycline (2 μg/mL) and passaged three times to wash out doxycycline and induce transgene expression. MD-MBA-231 and H1299 stable cells were generated by transfection with a pcDNA3 vector expressing Flag-OGA followed by G418 selection.
Cell proliferation assay
Cells were seeded in 6-well plates and cultured in 10 mL culture medium. Culture medium was changed every 16 hours to maintain the indicated glucose concentration. Live cells were counted at the indicated time points using trypan blue dye exclusion staining.
Clonogenic assay
Cells were seeded in 6-well plates and allowed to grow for 14 days. Culture medium was changed every 48 hours to prevent nutrient depletion. After 14 days, cells were fixed and stained with methanol:acetic acid (3:1, v/v) containing 0.5% (w/v) crystal violet as described in the literature9. Images were taken and analyzed using Adobe Photoshop. Stained colonies were de-stained with methanol:water (9:1, v/v) and relative dye incorporation was measured using a UV-Vis spectrophotometer.
Xenograft tumor assay
10 × 106 cells were suspended in 100 μL PBS, mixed with 100 μL matrigel, and injected subcutaneously into the flanks of 5-week-old male nude mice (Charles River Laboratories) on a normal chow diet. Tumor size measurements were taken over the course of 25–30 days. Tumor volumes were calculated using the ellipsoid formula (Pi/6)*L*W*H. Tumors were surgically removed to take final measurements and images. BrdU incorporation was analyzed by immunohistochemistry (detailed description in Extended Procedures). All procedures have been approved by the Institutional Animal Care and Use Committee of Yale University.
Glucose consumption assay
Cells were seeded in 6-well plates at low density and cultured in 10 mL culture medium. After 72 hours, 200 μL culture supernatant was collected and live cells were counted. Residual glucose in the culture supernatant was measured using a glucose assay kit (Sigma) and used to calculate residual glucose in the culture medium. Residual glucose was subtracted from initial glucose in the culture medium and the difference was normalized to cell number.
Measurement of basal ECAR
60,000 cells were seeded in each well of a Seahorse XF24 Cell Culture Microplate. After 24 hours, cells were washed with warm PBS, culture medium was changed to unbuffered DMEM, and basal ECAR was measured using the Seahorse XF24 Analyzer.
Quantification of 13C-lactate enrichment
Cells were incubated with 25 mM uniformly labeled 13C-glucose for 4 hours to achieve steady-state labeling of glucose metabolites. After flash freezing in liquid nitrogen, metabolite extracts were prepared for 13C-NMR isotopomer analysis using HCl/methanol as described in the literature21. Metabolite concentrations and 13C-enrichments were measured using 1H-[13C] NMR (POCE) at 11.7T on a Bruker AVANCE vertical bore spectrometer and referenced to 13C-glycine as an internal control.
Quantification of glycolysis, PPP, and TCA cycle metabolites
Cells were seeded in 10-cm dishes and cultured in 10 mL culture medium for 24 hours. Metabolites were extracted and subjected to LC-MS analysis by METABOLON. Metabolite levels were normalized to total cellular protein as measured by the Bradford method.
Pyruvate kinase activity assay
A lactate dehydrogenase (LDH) coupled-enzyme reaction was used to measure pyruvate kinase activity of whole cell lysate (10 μg total cellular protein per well) or purified PKM2 (8.5 nM). Negative slope of UV-Vis absorbance for NADH per minute was calculated as relative pyruvate kinase activity. The reagents (ADP, FBP, LDH, and NADH) were purchased individually from Sigma and reconstituted as specified.
Sucrose density gradient ultracentrifugation
Cells transiently transfected with Flag-PKM2wt, S362A, or T365A were lysed in a hypotonic lysis buffer containing 100 μM FBP and centrifuged at 10,000 RPM at 4°C. A sucrose density gradient containing 100 μM FBP was prepared by layering less dense sucrose solutions over more dense sucrose solutions in a Beckman ultracentrifuge tube. Whole cell lysate was layered on top of the sucrose density gradient and centrifuged at 50,000 RPM for 16 hours. Fractions were manually collected and analyzed by Western blotting. In parallel, native protein molecular weight markers were loaded onto the sucrose density gradient and protein in the fractions was quantified using the Bradford method.
Statistical analysis
Sample sizes (numbers of mice or wells of cells) were determined by power analyses (Alpha = 0.05, Power = 0.80) based on our previous studies22, 24. Samples were not randomized to experimental groups. Experiments and analyses were not performed in a blinded fashion. For immunohistochemistry, statistical analyses were performed using IBM SPSS 19.0. For all other experiments, statistical analyses were performed using GraphPad Prism 6. The specific statistical analyses performed for each experiment are described in the figure legends. No samples were excluded from the reported analyses. All values represent mean ± SEM. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Jinbo Yang for providing the Flag-PKM2 plasmid and shPKM2 lentivirus. We thank Dr. Julie T. Feldstein for helping with immunohistochemical analysis of human cancer tissues. We thank Dr. Neeraj Tiwari for helping with sucrose density gradient ultracentrifugation. This work was supported by NIH R01 DK089098, P01 DK057751, Yale Comprehensive Cancer Center Pilot Grant, and American Cancer Society Research Scholar Grant to X.Y.
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 2012; 8: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem 1991; 266: 16842–16846. [PubMed] [Google Scholar]
- 3.Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr 2013; 33: 205–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowe DB, Sadlonova A, Toleman CA, Novak Z, Hu Y, Huang P et al. O-GlcNAc integrates the proteasome and transcriptome to regulate nuclear hormone receptors. Mol Cell Biol 2006; 26: 8539–8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 2010; 29: 2831–2842. [DOI] [PubMed] [Google Scholar]
- 6.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008; 452: 230–233. [DOI] [PubMed] [Google Scholar]
- 7.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008; 452: 181–186. [DOI] [PubMed] [Google Scholar]
- 8.Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell 2014; 54: 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc 2006; 1: 2315–2319. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 11.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007; 446: 1017–1022. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa K, Hirosawa M, Tabei Y, Arai D, Tanaka S, Murakami N et al. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J Biol Chem 2013; 288: 17099–17110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Roth C, Turkenburg JP, Davies GJ. Three-dimensional structure of a Streptomyces sviceus GNAT acetyltransferase with similarity to the C-terminal domain of the human GH84 O-GlcNAcase. Acta Crystallogr D Biol Crystallogr 2014; 70: 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2009; 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krzeslak A, Forma E, Bernaciak M, Romanowicz H, Brys M. Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin Exp Med 2012; 12: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP et al. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell metabolism 2013; 17: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2007; 2: 1236–1247. [DOI] [PubMed] [Google Scholar]
- 18.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell 2011; 42: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell 2013; 52: 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong Q, Han W, Yang X. O-GlcNAc as an Integrator of Signaling Pathways. Front Endocrinol (Lausanne) 2018; 9: 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel AB, de Graaf RA, Mason GF, Rothman DL, Shulman RG, Behar KL. The contribution of GABA to glutamate/glutamine cycling and energy metabolism in the rat cortex in vivo. Proc Natl Acad Sci U S A 2005; 102: 5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian K, Wang S, Fu M, Zhou J, Singh JP, Li MD et al. Transcriptional regulation of O-GlcNAc homeostasis is disrupted in pancreatic cancer. J Biol Chem 2018; 293: 13989–14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao FV, Schuttelkopf AW, Dorfmueller HC, Ferenbach AT, Navratilova I, van Aalten DM. Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol 2013; 3: 130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell metabolism 2012; 16: 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab 2013; 24: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan HB, Dietrich MO, Liu ZW, Zimmer MR, Li MD, Singh JP et al. O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell 2014; 159: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh JP, Zhang K, Wu J, Yang X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett 2015; 356: 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer 2011; 11: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 2008; 118: 3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem 2004; 279: 53665–53673. [DOI] [PubMed] [Google Scholar]
- 31.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 2010; 329: 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, Kudlow JE. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology 2006; 16: 551–563. [DOI] [PubMed] [Google Scholar]
- 35.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 2012; 14: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett 2013; 339: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008; 451: 964–969. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017; 18: 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang YR, Jang HJ, Yoon S, Lee YH, Nam D, Kim IS et al. OGA heterozygosity suppresses intestinal tumorigenesis in Apc(min/+) mice. Oncogenesis 2014; 3: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012; 337: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K, Yin R, Yang X. O-GlcNAc: A Bittersweet Switch in Liver. Front Endocrinol (Lausanne) 2014; 5: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.