Abstract
Data on features of Pneumocystis primary infection in infancy are still fragmented. To study Pneumocystis primary infection, 192 infants who were monitored for acute pulmonary disease or fever over a 40-month period were retrospectively investigated. P. jirovecii detection on archival nasopharyngeal aspirates was performed using a qPCR assay. Factors associated with P. jirovecii were assessed using univariate and multivariate analyses. P. jirovecii genotypes in infants and a control group of adults contemporaneously diagnosed with Pneumocystis pneumonia were identified using unilocus, bilocus, and multilocus sequence typing (MLST). P. jirovecii was detected in 35 infants (18.2%). The univariate analysis pointed out four factors: viral infection (P = .035, OR [IC 95], 2.2 [1.1–4.7]), lower respiratory tract infection (P = .032, OR [IC 95], 2.5 [1.1–5.9]), absence of hospital discharge after birth (P = .003, OR (IC 95), 0.1 (0.02–0.5]), and the 63–189-day group (P < .001, OR [IC 95], 42.2 [5.4–332]). The multivariate analysis confirmed these two latter factors (P = .02, OR [IC 95], 0.1 [0.02–0.72]; P = .005, OR [IC 95], 11.5 [2.1–63.5]). Thus, P. jirovecii acquisition mostly takes place in the community. A comparison of these data with those of previously published studies showed that median and interquartile range of positive-infant ages were close to those observed in Chile, Denmark, and Peru, highlighting similar characteristics. Common unilocus or bilocus genotypes were identified in infants and adults, whereas no MLST genotypes were shared. Therefore, a common reservoir made up of infected infants and adults is still hypothetical. Finally, primary infection is a worldwide phenomenon occurring at the same time in childhood regardless of geographical location, rather than an incidental event.
Keywords: Pneumocystis jirovecii, primary infection, genotypes, MLST, infants
Introduction
Pneumocystis pneumonia (PCP) contemporary to Pneumocystis primary infection has been well recognized as an important cause of morbidity and mortality in premature infants or weakened children since at least the 1950s.1 Since the 1980s, PCP has also occurred in immunosuppressed infants and children mostly with human immunodeficiency virus (HIV) infection and specifically in developing countries.2 In contrast, sero-epidemiological surveys conducted in the 1970s have suggested that first contacts with the fungus occurred in 90% of non-immunosuppressed infants before two years of age.3,4 Until the beginning of the 2000s, there were no microbiological data or clinical profiles related to primary infection in nonimmunosuppressed infants which was thought to be essentially asymptomatic.3–5 In fact, studies by Vargas and colleagues and Nevez and colleagues showed that primary infection in non-immunosuppressed infants can be symptomatic, associated or not with viral or bacterial infections.6,7 Later, Larsen and colleagues reported that primary infection occurred contemporary to a self-limiting upper respiratory tract infection (URTI) in infants.8
Available data on P. jirovecii genomic characteristics in infants developing primary infection are limited, whereas P. jirovecii genotypes have mostly been identified in PCP adult patients.9 Totet and colleagues have shown that genotypes of P. jirovecii organisms involved in primary infection in non-immunosuppressed infants and in PCP adults were similar, suggesting that both populations were part of a common reservoir.10,11 Conversely, Beard and colleagues suggested that circulation and transmission of the fungus in the two populations occurred independently.12 Nonetheless, due to method discrepancies these results are still subject of debate.
Be that as it may, clinical profiles, microbiological and P. jirovecii genomic characteristics related to primary infection are still poorly understood. In this context, the objectives of the present study were i) to detect P. jirovecii in a cohort of infants at risk for primary infection, ii) to correlate its presence with infants' characteristics, iii) to identify P. jirovecii genotypes through unilocus, bilocus and multilocus sequence typing (MLST) approach in this infant population, and to compare them to those found in PCP adults; both populations being monitored in the same geographical area.
Methods
Infant population
One hundred and ninety-two nonimmunosuppressed infants (sex ratio M/F 121/71, median age 58 days [limits, 4–729 days]) were retrospectively enrolled. The infants were initially admitted to the Brest University Hospital, France, between April 10, 2013 and July 26, 2016. They were submitted to nasopharyngeal aspirates (NPAs) (192 infants, 233 NPAs) for viral diagnosis within the framework of investigation of pulmonary symptoms or fever. Nucleic acids from the NPAs were extracted using NucliSens® easyMag® (bio-Mérieux) and stored at −80°C for further examination or control. P. jirovecii detection was retrospectively performed in NPAs using a real-time quantitative polymerase chain reaction (qPCR) assay targeting mitochondrial large subunit ribosomal RNA (mtLSUrRNA) as described elsewhere on 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA).13,14
Primary infection was defined as detection of P. jirovecii in a NPA sample from an infant, an infant who was a priori immune-naive to P. jirovecii.
Adult control population
Twenty-one adults with PCP (sex ratio M/F 13/8, median age 65 years [limits, 34–84 years]) were retrospectively enrolled. They were also monitored at Brest University Hospital and were submitted to bronchoalveolar lavage (BAL) procedure for PCP diagnosis. Twenty-one BAL specimens were examined contemporaneously with infant NPA examination between January 31, 2013, and October 7, 2016. P. jirovecii was detected using microscopic examination (indirect immunofluorescence assay, Bio-Rad, Marnes-La-Coquette, France) and the aforementioned qPCR assay as described elsewhere.13,14 This population represented a control population for Pneumocystis genotyping.
To avoid contamination, each step of the PCR assays was performed in different areas of the laboratory with different sets of micropipettes. Mix reagents were prepared in a laminar flow cabinet. To monitor for possible contamination, negative controls were included in each experiment and PCR round.
Pneumocystis genotyping
Extracted DNAs of positive NPAs from infants and BAL samples from adults were examined for Pneumocystis genotyping based on unilocus, bilocus, and multilocus sequence typing (MLST) methods. Three loci, mtLSUrRNA, cytochrome b (CYB) and superoxide dismutase (SOD) genes were analyzed, as we previously described.15 First, a single-round PCR was performed for each locus, as described elsewhere.16–19 A nested PCR was performed for SOD locus, as previously described on samples that failed to give amplification with a single-round PCR. The discriminatory power was determined using Hunter index (H).20,21 Discriminatory power was considered good when H > 0.95.21
Prevention of cross contamination during PCR and sequencing assays of genotyping was done as described above. Moreover, these assays were performed in a different laboratory than the one where P. jirovecii infection diagnosis was initially performed, the two laboratories being located at different hospital sites.
Data collection and analysis
Infant and adult sociodemographic and clinical data were collected using Brest University Hospital online software “Portail d’informatisation du processus de soin” (IPS).
Statistical analyses were performed using Prism software (version 7.0, GraphPad Software, San Diego, CA, USA) and XLSTAT software (version 2018.2.50731, Addinsoft, Brooklyn, NY, USA). Qualitative variables were compared using Fisher exact test or χ2 test when appropriate. Quantitative variables were compared using Mann-Whitney test. A two-sided P-value <.05 was considered significant. Logistic regression was used for univariate and multivariate analyses.
The study was noninterventional, which did not require inform consents and ethical approval according to French laws and regulations (CSP Art L1121e1.1).
Results
P. jirovecii was detected in 35 out of 192 infants, 18.2% (40 out of 233 NPAs, 17.2%). Results of P. jirovecii detection and infants’ characteristics are shown in Table 1. The first approach of qualitative data analysis using χ2 or Fisher exact test pointed out the following characteristics: infant age (P < .001), underlying diseases (P = .009), hospital discharge or not after birth (P < .001). Indeed, some newborns and infants were hospitalized after birth and discharge of their mothers. Univariate and multivariate analyses through a logistic regression are shown in Table 2. The univariate analysis pointed out the following factors positively associated with P. jirovecii detection: distribution of age groups, specifically the 63-189-day group (third quartile) (P < .001, OR [IC 95], 42.2 [5.4–332]) and the 90–120-day group (P = .023, OR [IC 95], 51 [1.7–1498]), congenital diseases (P = .037, OR [IC 95], 2.4 [1.1–5.6]), viral infection (P = .035, OR [IC 95], 2.2 [1.1–4.7]), lower respiratory tract infection (P = .032, OR [IC 95], 2.5 [1.1–5.9]). The univariate analysis pointed out the following factors negatively associated with P. jirovecii detection: hyaline membrane disease and/or bronchopulmonary dysplasia (P < .019, OR [IC 95], 0.3 [0.1–0.8]), absence of hospital discharge after birth (P = .003, OR [IC 95], 0.1 [0.02–0.5]). Among these aforementioned factors, the multivariate analysis retained two factors, the 63–189-day group (P = .005 OR [IC 95], 11.5 [2.1–63.5]), absence of hospital discharge after birth (P = .02, OR [IC 95], 0.1 [0.02–0.72]). The results of genotyping are detailed in Tables 3 and 4.
Table 1.
Pneumocystis jirovecii detection on nasopharyngeal aspirate samples in non-immunosuppressed infants at risk for Pneumocystis primary infection: results and infants’ characteristics.
| No. of positive infants (n = 35) | No. of negative infants (n = 157) | P-value | ||
|---|---|---|---|---|
| Sex ratio, M/F | 21/14 | 100/57 | .70 (κ2 test) | |
| Median age (IQR) (days) | 114 (75–173) | 44 (22–192) | <.001 (Mann-Whitney test) | |
| Birth term | Term birth | 25 (71.4%) | 84 (53.5%) | .068 (Fisher exact test) |
| Moderate to late preterma1 | 7 (20%) | 25 (15.9%) | ||
| Very preterma2 | 1 (2.9%) | 25 (15.9%) | ||
| Extremely preterma3 | 2 (5.7%) | 21 (13.4%) | ||
| Underlying diseases | Congenital diseasesb | 12 (34.3%) | 24 (15.3%) | .009 (Fisher exact test) |
| Hyaline membrane disease and/or bronchopulmonary dysplasia | 4 (11.4%) | 52 (33.1%) | ||
| Cystic fibrosis | 0 | 1 (0.6%) | ||
| Absence of underlying diseases | 19 (54.3%) | 85 (54.1%) | ||
| Hospital discharge after birth c | No | 2 (5.7%) | 55 (35. %) | <.001 (Fisher exact test) |
| Yes | 33 (94.3%) | 100 (63.7%) | ||
| Unknown data | 0 | 2 (1.3%) | ||
| Clinical presentation | Upper respiratory tract infection | 4 (11.4%) | 15 (9.6%) | .079 (Fisher exact test) |
| Lower respiratory tract infection | 22 (62.9%) | 68 (43.3%) | ||
| Absence of respiratory symptomsd | 9 (25.7%) | 70 (44.6%) | ||
| Unknown data | 0 | 4 (2.5) | ||
| Pathogens e | Virusf | 21 (60%) | 63 (40.1%) | .06 (κ2 test) |
| Bacteriag | 7 (20%) | 22 (14%) | ||
| Absence | 11 (31.4%) | 82 (52.2%) |
IQR, interquartile range; F, female; M, male
a P. jirovecii was detected in 10 preterm infants (weeks of amenorrhea, a1 32 -35, a2 28–32, a3 < 28);
bIntrauterine growth restriction (17), unlabeled malformation syndrome (11), Tetralogy of Fallot (3), CHARGE syndrome (coloboma, heart defect, atresia choanae, retarded growth and development, genital hypoplasia, ear anomalies/deafness) (2), Pierre Robin syndrome (2), spinal muscular atrophy (1), central hypoventilation syndrome (1), VACTERL association (1), Down syndrome (1), unbalanced t(8, 20) (1), Menkes disease (1).
c P. jirovecii was detected in only 2 out of 57 infants who had been hospitalized since birth whereas it was mostly detected in infants who were discharged after birth (33 out of the 133 infants); indeed, some newborns and infants were hospitalized after birth and discharge of their mothers;
dInfants who presented fever in absence of overt respiratory symptoms,
e P. jirovecii was detected in a context of co-infections with viruses and/or bacteria in 28 infants whereas it was detected alone in 11 infants.
fRhinovirus, Respiratory Syncytial Virus, Parainfluenza 3 virus, Metapneumovirus, Influenza A virus, Influenza B virus, Enterovirus, Coronavirus, Cytomegalovirus, Adenovirus, Bocavirus, Parechovirus.
g Branhamella catarrhalis, Bordetella pertussis, Haemophilus influenza, Mycoplasma pneumoniae, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus haemolyticus, Enterobacter cloacae, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae
Table 2.
Univariate and multivariate analysis of factors associated with Pneumocystis jirovecii detection in non-immunosuppressed infants at risk for Pneumocystis primary infection.
| Variable | Adjusted odds ratio | 95% CI | P-value |
|---|---|---|---|
| Univariate analysis | |||
| Age quartile | |||
| no. 3 (n = 47; 63–189 days) | 42.2 | 5.4-332 | <.001 |
| no. 4 (n = 48; 190–729 days) | 9.6 | 1.15-80 | .037 |
| 90 ≤ age <120 days | 51 | 1.7-1498 | .023 |
| Hyaline membrane disease and/or bronchopulmonary dysplasiaa | 0.3 | 0.1-0.8 | .019 |
| Congenital disease | 2.4 | 1.1-5.6 | .037 |
| Viral coinfection | 2.2 | 1.1-4.7 | .035 |
| Lower respiratory tract infection | 2.5 | 1.1-5.9 | .032 |
| Absence of hospital discharge after birtha,b | 0.1 | 0.02-0.5 | .003 |
| Multivariate analysis | |||
| Age quartile no. 3 (n = 47; 63–189 days) | 11.5 | 2.1-63.5 | .005 |
| Absence of hospital discharge after birtha,b | 0.1 | 0.02-0.72 | .02 |
CI, confidence interval.
anegatively associated with P. jirovecii; bsome newborns and infants were hospitalized after birth and discharge of their mothers.
Table 3.
Results of Pneumocystis jirovecii genotyping at three loci in non-immunosuppressed infants developing primary Pneumocystis primary infection.
| MLST genotype | ||||
|---|---|---|---|---|
| Bilocus genotype | ||||
| Infant Code | Sample code | MtLSU alleles | CYB alleles | SOD alleles |
| E007 | E007-2a | mtLSU2, mtLSU4 | Allele mixc | SOD2 |
| E007-3 | mtLSU2, mtLSU3, mtLSU4 | Allele mixc | Allele mixc | |
| E008 | E008-1 | ...b | CYB3 | ...b |
| E017 | E017-1 | ...b | CYB1 | SOD1 |
| E029 | E029-3 | mtLSU4 | CYB2 | SOD2 |
| E032 | E032-1 | ...b | CYB1 | ...b |
| E044 | E044-1a | ...b | CYB1 | Allele mixc |
| E044-2 | mtLSU2 | CYB1 | ...b | |
| E069 | E069-1 | Allele mixc | Allele mixc | SOD1 |
| E075 | E075-1 | mtLSU2 | ...b | ...b |
| E076 | E076-1 | mtLSU2 | CYB1 | Allele mixc |
| E082 | E082-1 | ...b | CYB6 | SOD2 |
| E089 | E089-1 | mtLSU3 | CYB1 | SOD2 |
| E103 | E103-1a | mtLSU2, mtLSU4 | CYB6 | ...b |
| E103-2 | mtLSU2 | CYB1, CYB6 | SOD2 | |
| E114 | E114-1 | mtLSU4 | CYB5 | SOD1 |
| E125 | E125-1 | mtLSU3 | ...b | ...b |
| E130 | E130-1 | mtLSU3 | CYB2 | Allele mixc |
| E138 | E138-1 | mtLSU3 | CYB5 | Allele mixc |
| E143 | E143-2 | mtLSU2 | ...b | ...b |
| E154 | E154-1 | ...b | CYB1, CYB6 | ...b |
| E156 | E156-1 | mtLSU2 | CYB2 | SOD2 |
| E159 | E159-1 | mtLSU2 | ...b | ...b |
| E165 | E165-1 | mtLSU2, mtLSU3 | CYB1 | Allele mixc |
| E169 | E169-1 | mtLSU2 | CYB2 | ...b |
| E171 | E171-1 | ...b | CYB1 | ...b |
| E177 | E177-1 | mtLSU2 | CYB1 | ...b |
| E190 | E190-1 | mtLSU3 | CYB1 | ...b |
| E206 | E206-1 | mtLSU3, mtLSU4 | CYB1 | SOD2 |
The alleles were identified considering changes at nucleotide positions described elsewhere by Beard et al.23, Esteves et al.16, and Maitte et al.17.
CYB, cytochrome b gene; mtLSU, mitochondrial large subunit ribosomal RNA gene; SOD, superoxide dismutase gene.
aThe infants underwent nasopharyngeal aspirates two times, only one sample was considered for statistical analysis.
b“…”, typing failure.
cAllele mix, presence of several different alleles that were not identified.
Table 4.
Results of Pneumocystis jirovecii genotyping at three loci in immunosuppressed adults developing Pneumocystis pneumonia.
| MLST genotype | |||
|---|---|---|---|
| Bilocus genotype | |||
| Adult and sample code | MtLSU alleles | CYB alleles | SOD alleles |
| A001 | mtLSU2 | CYB2 | SOD1 |
| A002 | mtLSU4 | CYB1 | SOD1 |
| A003 | mtLSU1 | CYB5, CYB8 | SOD2 |
| A004 | mtLSU1 | CYB2 | SOD2 |
| A005 | mtLSU1, mtLSU4 | CYB2 | SOD1 |
| A006 | mtLSU1 | CYB8 | SOD2 |
| A007 | mtLSU2 | Allele mixa | Allele mixa |
| A008 | mtLSU2 | CYB8 | SOD1 |
| A009 | mtLSU2 | CYB1, CYB7 | Allele mixa |
| A010 | mtLSU3 | CYB1 | SOD1 |
| A011 | mtLSU4 | CYB1 | SOD1 |
| A012 | mtLSU4 | CYB2 | SOD1 |
| A013 | Allele mixa | CYB1, CYB2 | SOD2 |
| A014 | mtLSU1 | CYB1 | Allele mixa |
| A015 | mtLSU3 | CYB6 | SOD1 |
| A016 | Allele mixa | CYB1, CYB6 | SOD2 |
| A017 | mtLSU3, mtLSU1 | CYB1 | SOD1, SOD4 |
| A018 | mtLSU2 | CYB1 | Allele mixa |
| A019 | mtLSU2 | CYB2 | Allele mixa |
| A020 | mtLSU1, mtLSU3 | CYB1 | SOD1 |
| A021 | mtLSU3 | CYB5 | SOD1 |
The alleles were identified considering changes at nucleotide positions described elsewhere by Beard et al.23, Esteves et al.16, and Maitte et al.17. Multilocus genotypes are named following the pattern [x-y] (two-loci genotypes) or [x-y-z] (three-loci genotypes) where x = mtLSU allele number, y = CYB allele number and z = SOD allele number.
CYB, cytochrome b gene; MLST, multilocus sequence typing; mtLSU, mitochondrial large subunit ribosomal RNA gene; N/A, not applicable; SOD, superoxide dismutase gene.
Not typed: Typing failure due to amplification and/or sequencing and/or sequence analysis issues.
Undetermined mix: Detection of several undetermined alleles or multilocus genotypes in the same sample.
aallele mix, presence of several different alleles that were not identified.
Taking the results of P. jirovecii typing in infants and adults together, the Hunter indexes of the unilocus approach based on each mtLSU, CYB, and SOD genes were ≤0.75 each. The Hunter index of the bilocus genotyping method and of MLST was 0.94 and 0.99, respectively.
In the infant population, considering the mtLSU, CYB, and SOD alleles previously described elsewhere, three mtLSU alleles were identified; mtLSU2 being the most frequent (11 infants).16,17,22 Five CYB alleles were identified; CYB1 being the most frequent (12 infants). Two SOD alleles were identified, SOD2 being the most frequent allele (seven infants). The bilocus typing system, based on analysis of mtLSU locus combined with CYB locus, enabled us to identify seven mtLSU-CYB genotypes in 11 infants; mtlLSU2-CYB1 being the most frequent (three infants). Using the MLST, seven genotypes were identified in six infants.
In the adult population, four mtLSU alleles were identified; mtLSU1 being the most frequent (seven adults). In addition, six CYB alleles were identified; CYB1 being the most frequent (10 adults). Three SOD alleles were identified; SOD1 being the most frequent (11 adults). The bilocus typing locus permitted to identify 14 mtLSU-CYB genotypes, mtLSU3-CYB1, and mtLSU1-CYB1 being the most frequent (three adults respectively). Twelve MLST genotypes were also identified. Three of them were found in two adults each, the nine others were found in one adult each.
Among the four mtLSU alleles, mtLSU1 was identified only in the adults, whereas the three others were both identified in infants and adults. Among the seven CYB alleles, CYB3 was identified only in the infants, whereas CYB8 and CYB7 were identified only in the adults. The two major CYB alleles, CYB1 and CYB2, were shared by the two populations. Among the three SOD alleles, SOD4 was only identified in adults, whereas the two others, SOD2 and SOD1, were shared by the two populations. Five bilocus genotypes were common in both populations, whereas two other genotypes were found only in infants and nine other genotypes were exclusively found in adults. A total of 17 MLST genotypes were identified, but none were shared by the two patient populations.
Discussion
We detected P. jirovecii in 35 out of 192 infants (18.2%), while in our former study it was detected in 45 out of 178 infants (25.3%).6 These results, obtained 17 years apart in two similar infant populations from two French regions 700 km apart, do not differ significantly (P = .09, χ2). P. jirovecii presence and its detection in an infant by definition a child less than 2 years old, was considered to be related to first contacts with the fungus and consequently to be contemporary with primary infection. This definition of primary infection could be discussed because putative P. jirovecii reinfection could not be strictly ruled out in some cases. It is noteworthy that P. jirovecii was mainly detected in young infants and in only four infants over 1 year old. Thus, the concept of reinfection with P. jirovecii remains speculative. The infants enrolled in the study may have presented with pulmonary colonization or PCP. Nonetheless, the hypothesis of PCP development was improbable since they recovered without specific treatment.
Infection due to viruses as an associated factor to the presence of P. jirovecii was expected since infants enrolled in the present study mostly presented with bronchiolitis, a pulmonary syndrome specifically related to viruses. Moreover, we have previously described and reported this microbial association.6
In the present study, P. jirovecii was detected in the apparent absence of other pathogens in 11 infants, suggesting that the fungus may play a role in pulmonary syndrome pathogenesis. Nonetheless, all positive infants recovered, despite the absence of specific treatment for the fungus. We can hypothesize that the presence of P. jirovecii followed by clinical improvement could be explained by the “immune-naive” status of infants on the one hand, and the role of symptomatic treatment and physiotherapy in the absence of overt immunosuppression on the other hand. Contrary to Larsen and colleagues’ study in which P. jirovecii detection was specifically associated with URTI, in our study lower respiratory tract infections (LRTI) appear as an associated factor.8 This result is consistent with the fact that P. jirovecii infection effectively takes place deep within the lungs and specifically in alveoli in close contact with type I pneumocytes.2Pneumocystis primary infection contemporary to first contacts with the fungus may present as a mild LRTI.
We did not identify prematurity as a factor associated with P. jirovecii. Nonetheless, P. jirovecii was detected in 10 out of 81 preterm infants (12.3%), whereas Rojas et al. in Seville, Spain, have recently detected the fungus in 33 of 128 preterm infants (25.7%).23 These discrepant results, 12.3% versus 25.7% (P = .02), are probably due to the difference in sampling methods and difference in hospital locations, Brest, Brittany, France versus Seville, Andalusia, Spain, geographical differences in P. jirovecii epidemiology in Europe having previously been reported in patients with chronic pulmonary diseases.24,25 Be that as it may, our results combined with those of Rojas and colleagues obtained from French and Spanish populations, respectively, show that P. jirovecii is still involved in non-immunosuppressed preterm infants in Europe, as it was described in the 1950s.1,23 These important findings have already been published separately as a correspondence and should arouse the interest of pediatricians.26
Congenital diseases appear as an associated factor (Table 1). To the best of our knowledge none of them are associated with overt immunosuppression. Nonetheless, this association provides additional arguments to the well-recognized links between P. jirovecii and infant frailty.1
Hyaline membrane disease or bronchopulmonary dysplasia is a factor negatively associated with P. jirovecii detection. This negative association remains unclear, but it may be explained by the fact that we did not identify prematurity as an associated factor, whereas it is assumed that there is a relationship between prematurity and hyaline membrane disease or bronchopulmonary dysplasia.27
The univariate analysis pointed out the absence of hospital discharge after birth as a factor negatively associated with P. jirovecii detection. This result was confirmed by multivariate analysis. Thus, P. jirovecii is mostly acquired in the community by infants who have left the hospital after birth. In contrast, P. jirovecii was only found in two out of the 57 infants who had not left the hospital after birth. These results were expected since the infants who stayed at the hospital after birth were younger than those who were discharged (median age, 22 days; interquartile range [IQR], 14.5–36 vs. median age, 118 days; IQR, 43–283). Nonetheless, P. jirovecii acquisition can happen within the hospital premises, the potential human sources such as health care workers, patients or mothers remaining undetermined in this case. To sum up, our results support the possibility of both community and nosocomial acquisitions of the fungus also in the context of primary infection in infants as previously shown for PCP in immunosuppressed adults.28,29
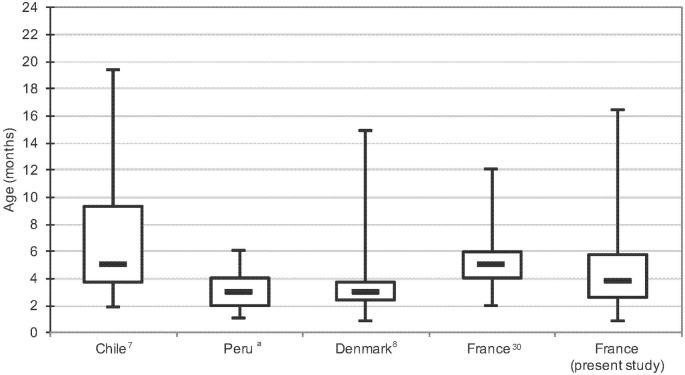
The multivariate analysis also identified the 63-189-day group as a factor associated with P. jirovecii detection. The median age of positive infants was 3 months and 24 days (IQR, 75–173 days), whereas it was 5 months (IQR, 3.7–9.3 months), 3 months (IQR, 73–112 days), 3 months (2–4 months) in Chile, Denmark, and Peru, respectively (Ochoa TJ, Bustamante B, Garcia C al., manuscript in preparation).7,8 In our study reported 17 years ago, the median age of positive infants was 5 months (IQR, 4–6 months) (Figure 1).6,30 Comparing the results of the aforementioned studies must be done cautiously, nonetheless the values of median and IQR ages of positive infants are close. These common characteristics suggest that first contacts with P. jirovecii, and consequently primary infection in infants, is a common process occurring at the same time in life, regardless the geographical location, rather than an incidental event.
Figure 1.
Boxplots of age distribution of infants positive for Pneumocystis jirovecii in Chile, Peru, Denmark, and France (two geographical locations). Lines within each box represent the median values. Ends of whiskers mark the lowest and highest diversity values in each instance. aOchoa TJ, Bustamante B, Garcia C al., manuscript in preparation.
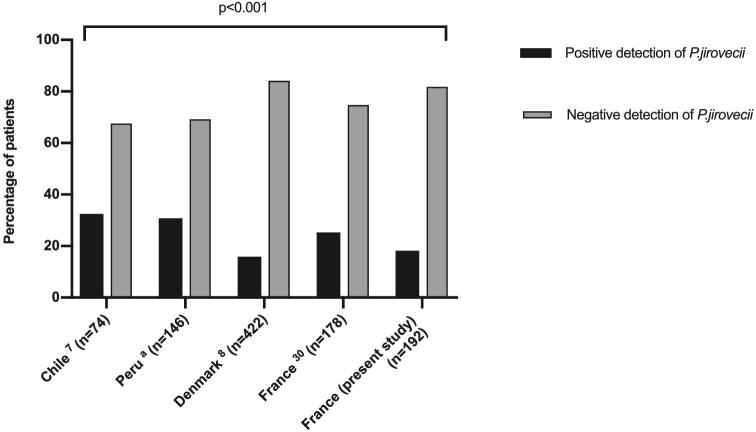
The analysis of P. jirovecii detection results in infants in Chile, Denmark, Peru, and France, pointed out significant differences of prevalence (P < .001) with a higher prevalence in South America than in Europe (Figure 2) (Ochoa TJ, Bustamante B, Garcia C al., manuscript in preparation).7,8 Beyond differences in methods, these results may be related to the genuine geographical specificity of Pneumocystis epidemiology. This hypothesis was previously assumed to explain the difference of Pneumocystis colonization prevalence in patients with chronic pulmonary diseases.24,25
Figure 2.
Prevalence of Pneumocystis jirovecii in non-immunosuppressed infants at risk for Pneumocystis primary infection in Chile, Peru, Denmark, and France (two geographical locations). aOchoa TJ, Bustamante B, Garcia C al., manuscript in preparation. Foot notes. The analysis pointed out significant differences of prevalence (P < .001, κ2 test) with a higher prevalence in South America than in Europe.
Considering Maitte and colleagues’ proposal for P. jirovecii genotyping and our skill in this area, we chose to analyze the mtLSU, CYB and SOD loci.15,17 The discriminatory power of the typing system increased from the unilocus approach through the bilocus approach to the MLST method. These results were expected, considering recommendations of MLST for microorganism typing methods published elsewhere.21 The apparent diversity of P. jirovecii grew according to the increase in discriminatory power of the typing method used. Consequently, the unilocus and bilocus approaches showed that genotypes were common in infants and adults, whereas no MLST genotype was shared. These apparently contradictory results due to technological constraints make it impossible to conclude whether the acquisition/transmission cycle of P. jirovecii in primary infected infants and PCP adults are independent or not.
Our results of MLST suggested a putative correlation between specific genotypes and clinical presentation, that is, primary infection in infants versus PCP in adults. The hypothesis can be ruled out, since only one MLST genotype (mtLSU3-CYB1-SOD2) identified in our infant population was also identified by Maitte and colleagues among adults developing PCP and monitored in another French University Hospital Centre.17 It is noteworthy that the genotypes in the two positive infants who had not left the hospital after birth (E029, E130) were different than those in infants who were discharged after birth (Table 3). This observation is consistent with a different source of P. jirovecii in this two-infant population. However, genotyping results should not be too conclusive considering the low numbers of patients for whom MLST was successful (13 PCP adults and six infants) (Tables 3 and 4).
The study presents some limitations such as the retrospective approach, the enrolment of only symptomatic infants, the examination of NPAs from infants and BAL specimens in adults. Nonetheless, our investigation of a 192 infant population in Brittany, France, combined with the results of a meta-analysis of available data from another French region, and from Chile, Denmark, and Peru, shows that, despite putative geographical specificities, Pneumocystis primary infection in nonimmunosuppressed infants appears to be a common event occurring worldwide.
Acknowledgments
The authors would like to thank V Abiven, P Bargain, C Cam, E Guillotel, C Le Guen, K Quinaou, C Roger, and D Roué for their contribution and Doctor Hans Henrik Larsen for providing additional data on Pneumocystis primary infection in Denmark.
The results were reported in part at ASM Microbe meeting, New Orleans, LA, USA, June 1–5, 2018. This study was supported by the European Commission (ERANet-LAC) within the framework of the project “Recognition of the primary infection by Pneumocystis in infants: a silent threat to public health” (CAPRI-PC HID-0254).
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1. Gajdusek DC. Pneumocystis carinii-Etiologic agent of interstitial plasma cell pneumonia of premature and young infants. Pediatrics. 1957; 19: 543–565. [PubMed] [Google Scholar]
- 2. Walzer PD, Cushion MT.. Pneumocystis Pneumonia, 4th edn. New York: Marcel Dekker, 2005. [Google Scholar]
- 3. Meuwissen JH, Tauber I, Leeuwenberg AD, Beckers PJ, Sieben M. Parasitologic and serologic observations of infection with Pneumocystis in humans. J Infect Dis. 1977; 136: 43–49. [DOI] [PubMed] [Google Scholar]
- 4. Pifer LL, Hughes WT, Stagno S, Woods D. Pneumocystis carinii infection: evidence for high prevalence in normal and immunosuppressed children. Pediatrics. 1978; 61: 35–41. [PubMed] [Google Scholar]
- 5. Djawe K, Daly KR, Vargas SL et al.. Seroepidemiological study of Pneumocystis jirovecii infection in healthy infants in Chile using recombinant fragments of the P. jirovecii major surface glycoprotein. Int J Infect Dis. 2010; 14: e1060–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nevez G, Totet A, Pautard JC, Raccurt C. Pneumocystis carinii detection using nested-PCR in nasopharyngeal aspirates of immunocompetent infants with bronchiolitis. J Eukaryot Microbiol. 2001; Suppl: 122S–123S. [DOI] [PubMed] [Google Scholar]
- 7. Vargas SL, Hughes WT, Santolaya ME et al.. Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin Infect Dis. 2001; 32: 855–861. [DOI] [PubMed] [Google Scholar]
- 8. Larsen HH, von Linstow ML, Lundgren B, Hogh B, Westh H, Lundgren JD. Primary pneumocystis infection in infants hospitalized with acute respiratory tract infection. Emerg Infect Dis. 2007; 13: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matos O, Esteves F.. Pneumocystis jirovecii multilocus gene sequencing: findings and implications. Future Microbiol. 2010; 5: 1257–1267. [DOI] [PubMed] [Google Scholar]
- 10. Totet A, Pautard JC, Raccurt C, Roux P, Nevez G. Genotypes at the internal transcribed spacers of the nuclear rRNA operon of Pneumocystis jiroveci in nonimmunosuppressed infants without severe pneumonia. J Clin Microbiol. 2003; 41: 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Totet A, Respaldiza N, Pautard JC, Raccurt C, Nevez G. Pneumocystis jiroveci genotypes and primary infection. Clin Infect Dis. 2003; 36: 1340–1342. [DOI] [PubMed] [Google Scholar]
- 12. Beard CB, Fox MR, Lawrence GG et al.. Genetic differences in Pneumocystis isolates recovered from immunocompetent infants and from adults with AIDS: epidemiological implications. J Infect Dis. 2005; 192: 1815–1818. [DOI] [PubMed] [Google Scholar]
- 13. Meliani L, Develoux M, Marteau-Miltgen M et al.. Real time quantitative PCR assay for Pneumocystis jirovecii detection. J Eukaryot Microbiol. 2003; 50: 651. [DOI] [PubMed] [Google Scholar]
- 14. Le Gal S, Robert-Gangneux F, Pepino Y et al.. A misleading false-negative result of Pneumocystis real-time PCR assay due to a rare punctual mutation: a French multicenter study. Med Mycol. 2017; 55: 180–184. [DOI] [PubMed] [Google Scholar]
- 15. Vindrios W, Argy N, Le Gal S et al.. Outbreak of Pneumocystis jirovecii infection among heart transplant recipients: molecular investigation and management of an interhuman transmission. Clin Infect Dis. 2017; 65: 1120–1126. [DOI] [PubMed] [Google Scholar]
- 16. Esteves F, Gaspar J, Tavares A et al.. Population structure of Pneumocystis jirovecii isolated from immunodeficiency virus-positive patients. Infect Genet Evol. 2010; 10: 192–199. [DOI] [PubMed] [Google Scholar]
- 17. Maitte C, Leterrier M, Le Pape P, Miegeville M, Morio F. Multilocus sequence typing of Pneumocystis jirovecii from clinical samples: how many and which loci should be used? J Clin Microbiol. 2013; 51: 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chabe M, Khalife S, Gantois N, Even G, Audebert C. An improved single-round PCR leads to rapid and highly sensitive detection of Pneumocystis spp. Med Mycol. 2014; 52: 841–846. [DOI] [PubMed] [Google Scholar]
- 19. Monroy-Vaca EX, de Armas Y, Illnait-Zaragozi MT et al.. Genetic diversity of Pneumocystis jirovecii in colonized Cuban infants and toddlers. Infect Genet Evol. 2014; 22: 60–66. [DOI] [PubMed] [Google Scholar]
- 20. Hunter PR. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990; 28: 1903–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Struelens MJ. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996; 2: 2–11. [DOI] [PubMed] [Google Scholar]
- 22. Beard CB, Carter JL, Keely SP et al.. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg Infect Dis. 2000; 6: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rojas P, Friaza V, Garcia E et al.. Early acquisition of Pneumocystis jirovecii colonization and potential association with respiratory distress syndrome in preterm newborn infants. Clin Infect Dis. 2017; 65: 976–981. [DOI] [PubMed] [Google Scholar]
- 24. Morris A, Norris KA.. Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev. 2012; 25: 297–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nevez G, Robert-Gangneux F, Pougnet L et al.. Pneumocystis jirovecii and cystic fibrosis in Brittany, France. Mycopathologia. 2018; 183: 81–87. [DOI] [PubMed] [Google Scholar]
- 26. Nevez G, Guillaud-Saumur T, Cros P et al.. Pneumocystis is still involved in nonimmunosuppressed preterm infants in Europe. Clin Infect Dis. 2018; 67: 645–646. [DOI] [PubMed] [Google Scholar]
- 27. Hart M. Current concepts on respiratory distress syndrome in the newborn (hyaline membrane disease). Can Fam Physician. 1970; 16: 64–67. [PMC free article] [PubMed] [Google Scholar]
- 28. Nevez G, Chabe M, Rabodonirina M et al.. Nosocomial Pneumocystis jirovecii infections. Parasite. 2008; 15: 359–365. [DOI] [PubMed] [Google Scholar]
- 29. Yiannakis EP, Boswell TC.. Systematic review of outbreaks of Pneumocystis jirovecii pneumonia: evidence that P. jirovecii is a transmissible organism and the implications for healthcare infection control. J Hosp Infect. 2016; 93: 1–8. [DOI] [PubMed] [Google Scholar]
- 30. Totet A. Molecular Biology and Epidemiology of Pneumocystis jirovecii in Infants. PhD thesis, Université de Picardie Jules Verne (France), 2004[in French]. [Google Scholar]