Abstract
Diagnosis of invasive aspergillosis (IA) is challenging, particularly in high-risk patients with lung lesions other than typical according to 2008-EORTC/MSG criteria. Even if microbiology is positive, they still remain unclassified according to 2008-EORTC/MSG. Quantitative polymerase chain reaction (qPCR) provides new mycological documentation of IA. This retrospective study assessed Aspergillus fumigatus real time qPCR (MycoGENIE®) in BAL to diagnose IA and identify azole-resistant strains. Clinical, radiological, and microbiological data from 114 hematology patients (69% HSCT recipients; 29% on mould active agents) from years 2012-2017 were collected; and 123 BAL samples were tested with qPCR (cutoff: Ct < 40) and galactomannan (GM, Platelia®, cutoff: 0.5 ODI). Patients were classified as proven/probable, possible, and no-IA. "Atypical-IA" referred to patients with lesions other than typical according to 2008-EORTC/MSG and positive mycology. Proven IA was diagnosed in two cases (1.6%), probable in 28 (22.8%), possible in 27 (22%), atypical in 14 (11.4%). qPCR was positive in 39 samples (31.7%). Sensitivity and specificity of qPCR for proven/probable IA (vs no-IA; atypical-IA excluded) were 40% (95% confidence interval [CI]: 23–59) and 69% (95%CI: 55–81), respectively. Sensitivity of qPCR was higher when combined with GM (83%, 95%CI: 65–94) and in those receiving mould-active agents at BAL (61%, 95%CI: 32–86). One sample had TR34/L98H mutation. In conclusion, in high-risk hematology patients with various lung lesions, A. fumigatus qPCR in BAL contributes to diagnosing IA, particularly if combined with GM and in patients receiving mould-active agents might allow detecting azole-resistant mutations in culture negative samples.
Keywords: invasive aspergillosis, galactomannan, Aspergillus fumigatus PCR, BAL, HSCT
Introduction
Invasive fungal disease (IFD), and particularly invasive aspergillosis (IA), is an infectious complication affecting mainly patients with haematological disorders and prolonged neutropenia, long-term high dose corticosteroid treatment or allogeneic stem cell transplantation (SCT).1–3 IA in this setting is associated with high morbidity and mortality, particularly when not promptly diagnosed and treated.4 Unfortunately, the diagnosis of IA in hematology patients is challenging because of nonspecific clinical manifestations, low yield of fungal cultures, and difficulty in performing invasive diagnostic procedures due to thrombocytopenia or poor general conditions. In addition, azole resistant A. fumigatus strains have been increasingly frequent in several geographical regions, and given low rate of positive cultures, these cases risk to remain undetected, compromising the outcome of IA.5
Diagnostic criteria for IFD in the immunocompromised patients have been developed by EORTC/MSG in 2002 and subsequently revised in 2008.6 They established three levels of certainty of diagnosis: proven, probable, and possible. In particular, for probable IA, a combination of a host factor (presence of a predisposing condition) plus a clinical criterion plus a mycological criterion are required. Clinical criteria in cases of pulmonary IA include one of the following typical radiological lesions in lung computed tomography (CT) scan: (1) dense, well-circumscribed lesions(s) with or without a halo sign, (2) air-crescent sign, or (3) cavity. Mycological criteria for pulmonary aspergillosis are detection of galactomannan (GM) in serum or bronchoalveolar lavage fluid (BAL) or direct tests positive in sputum or BAL.6 Possible IA is diagnosed in the presence of host and clinical criteria but in the absence of mycological documentation.
Although these criteria revolutionized the clinical research and epidemiological studies in IA, they do not cover numerous possible clinical situations.7 Among them, the most frequent is the presence of host criteria with positive mycological criteria and lung lesions, which are different from the aforementioned typical ones. These patients remain “unclassified” according to EORTC/MSG criteria, but they are usually treated for IA, and several studies showed that they truly have IA.8 In fact, in a recent observational EORTC study, they were classified as those in whom IFD cannot be excluded, thus not suitable for being considered certain negative controls (European prospective invasive mould disease audit [PIMDA] protocol).9 Additionally, the performance of GM might be suboptimal in certain settings, for example, in patients receiving mould active agents, in whom breakthrough IA is suspected.
Molecular methods such as polymerase chain reaction (PCR) are able to detect Aspergillus DNA in BAL samples with good sensitivity (77.2%) and specificity (93.5%).10 Although in recent years many publications11–13 focused on the diagnostic role of Aspergillus PCR, its use is not yet recommended in the 2016 update of IDSA Guidelines on the diagnosis and management of Aspergillosis, mostly because of the lack of standardised and validated assays.2,3 Their advantages include the possibility to detect fungal DNA also if it is no longer viable, such as after antifungal treatment has been started, and to confirm the presence of Aspergillus with higher sensitivity than culture, similarly to GM. Additionally, certain molecular methods offer the possibility to detect resistance mutations in Aspergillus fumigatus even in the absence of strain's growth.
The aim of the study is to evaluate the performance of a commercially available Aspergillus fumigatus real-time quantitative PCR (qPCR), alone and in combination with GM, in BAL samples form patients at high risk of IA, and with various radiological lesions, including those receiving mould active antifungals. Additionally, the rate of mutations conferring azole resistance in high risk patients in our center was also evaluated.
Methods
Samples and patients
The study was conducted in Ospedale Policlinico San Martino, a tertiary care center in Genoa, Italy, with active allogeneic SCT center. We retrospectively identified all available BAL samples from years 2012–2017 from patients with SCT and/or haematological malignancies. All patients had pulmonary lesions reported on CT scan. BAL samples from the same patient were excluded if performed within 4 weeks.
All patients underwent at least two determinations of GM (PlateliaTM Bio-Rad-Inc.) and 1,3-beta-d-glucan (BDG) (Fungitell® Assay) in serum, with cutoff values for positivity of 0.5 optical density index (ODI) and 80 pg/ml, respectively, according to manufactures’ recommendations.
All BAL samples were subject to the following analyses: culture for bacteria, filamentous fungi and mycobacteria, GM, molecular testing for pneumocystosis, Mycobacterium tuberculosis, cytomegalovirus, herpes simplex virus, and respiratory viruses (influenza, parainfluenza, respiratory syncytial virus, metapneumovirus, enterovirus, rhinovirus, coronavirus) and bacteria (Legionella, Bordetella, Streptococcus pneumoniae, Haemophilus influenzae, Chlamydia, Mycoplasma pneumoniae). Direct research for hyphae and antifungal susceptibility testing were not performed routinely.
For BAL GM testing, samples were centrifuged and 300 μl of supernatant was further treated according to manufacturer's instruction. Residual sample after GM testing was stored at −20°C. for GM the cutoff value of 0.5 ODI was considered positive according to manufacturer's instruction.
For each patient, general data, clinical characteristics, data on the underlying disease, including SCT, administration of mould active antifungals, and outcome were collected.
All patients gave informed consent for data collection and research purposes at the hospital admission and BAL performance. The study was approved by local ethics committees under the reference number PR001REG2016.
Classifications and definitions
For all patients, microbiological results and full medical records were reviewed. CT lesions were revalued by two radiologists (I.P. and G.C.) with an expertise in pulmonary fungal infections and classified as typical IFD lesions according to 2008 EORTC/MSG revised criteria,6 or other (atypical) lesions.
Considering also mycological criteria, patients were stratified according to 2008 EORTC/MSG criteria into four groups: (1) proven/probable IA, (2) possible IA, (3) atypical lung lesions but positive mycological criteria, considered as atypical IA, (4) no IA with atypical lung lesions and negative mycological criteria. These groups were mutually exclusive but grouped together for the purposes of analyses.
The evaluations of qPCR performance were carried out considering as cases patients with proven/probable/possible IA (group 1 and 2) or proven/probable IA (group 1) or proven/probable IA/atypical IA (atypical lesions with mycology positive, group 1 and 3), and considering as controls patients from group 4.
The performance of qPCR was evaluated separately for those receiving and not receiving mould active antifungals (either as prophylaxis or treatment) at the time of BAL.
Molecular analyses of BAL
BAL samples were stored at −20°C and subsequently sent to Institute of Microbiology at Università Cattolica del Sacro Cuore in Rome where molecular diagnostic methods have been performed.
They were tested with Aspergillus fumigatus real-time qPCR assay MycoGENIE® (AdemTech, Pessac, France). This is a multiplex commercially available CE-IVD marked assay approved for testing respiratory samples which detects DNA by targeting the 28S rRNA multicopy gene and specific TR34/L98H mutations in the cyp51A single copy gene of A. fumigatus.14 DNA was extracted with a MycoGENIE® DNA extraction kit Automag solution indicated for the isolation and purification of fungal DNA. An amount of 500 microliters of stored BAL fluid was centrifugated at 12,000 rpm for 10 min, after removing 400 microliters of the supernatant, the pellet was resuspended with 125 microliters of Tissue Lysis Buffer and 200 microliters were used to perform the DNA extraction in an AutoMag Solution Instrument equipped with a magnetic particle processor for DNA purification kits (Ademtech). Samples were eluted in 60 microliters. An internal extraction control was added together with the samples during the extraction process as indicated in the manufacturer procedure assay. Positive and negative PCR controls were added for each PCR experiment.
The kit performance data fixed the limit of detection (LOD) of monocopy sequences (TR34 and L98H cyp51A mutations) was determined at six copies. For multicopy genes (aspergillus rRNA gene), the LOD is below one copy. The specificity of primers and probes was verified by the manufacturer and indicated as highly specific (100%).
The cutoff for positivity was considered as < 40 (Cycle threshold) Ct. The performance of cutoff ≤ 35 Ct was also evaluated.
Statistical analyses of BAL
The variables were reported as median value with range. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated where applicable and reported with 95% confidence intervals (95% CI). Distribution of continuous and categorical variables were evaluated with, respectively, Mann-Whitney and χ2 test or Fisher exact test if applicable.
Statistical analysis was performed using MedCalc's Diagnostic test evaluation calculator (© 2018 MedCalc Software bvba).
Results
Patients
A total of 123 BAL samples from 114 patients were collected. In 9 patients, data from a second BAL procedures, performed in median 123 days after the first one (range, 28–406 days), were also included in the study. The patients’ characteristics are reported in Table 1. Briefly, 82 (66.7%) of them were male, median age was 54 years (range, 20–81 years) and the most frequent underlying disease was acute myeloid leukaemia (37.4%). In 85 (69.7%) cases, BAL was performed after SCT, allogeneic in 94% of cases, in median 165 days after SCT (range, 15–8543 days); and in 12 cases as diagnostic work up in pre-SCT evaluation, in median 17 days before SCT.
Table 1.
Main characteristics of baseline and outcome data of patients undergoing 123 BAL procedures.
| Characteristics | N = 123 (%) |
|---|---|
| Baseline variables | |
| Sex, male | 82 (66.7) |
| Median age, years (range) | 54 (20–81) |
| Underlying disease | |
| Acute myeloid leukaemia | 46 (37.4) |
| Non-Hodgkin lymphoma | 16 (13.0) |
| Acute lymphoblastic leukaemia | 13 (10.6) |
| Hodgkin's lymphoma | 11 (8.9) |
| Myelodysplastic syndromes | 10 (8.1) |
| Idiopathic myelofibrosis | 9 (7.3) |
| Multiple myeloma | 6 (4.9) |
| Chronic lymphocytic leukaemia | 4 (3.3) |
| Severe aplastic anaemia | 4 (3.3) |
| Chronic myeloid leukaemia | 2 (1.6) |
| Others | 2 (1.6) |
| SCT | |
| No | 38 (30.9) |
| BAL performed as pre-SCT evaluation | 12 (9.8) |
| Yes | 85 (69.1) |
| Autologous | 5 (5.8) |
| Allogeneic | 81 (94.2) |
| HLA-identical | 16 (18.8) |
| Matched unrelated | 12 (14.1) |
| Haploidentical | 49 (57.6) |
| Cord blood | 3 (3.5) |
| Time from SCT to BAL, days, median (range) | 165 (15–8543) |
| Neutropenia at BAL, neutrophils < 500 | 15 (12.2) |
| Neutropenia at BAL, neutrophils < 1000 | 28 (22.8) |
| Radiological findings of lung computed tomography | |
| Compatible with IA (according to EORTC/MSG) | 57 (46.3) |
| Atypical lesions | 66 (53.7) |
| Mycology | |
| GM serum > 0.5 | 6 (4.9) |
| BDG serum > 80 pg/mL | 15 (12.2) |
| Culture BAL positive for filamentous fungi | 7 (5.7)* |
| GM BAL > 0.5 | 35 (28.5) |
| GM BAL > 1.0 | 26 (21.1) |
| GM positivity in BAL, ODI, median (range) | 2,497 (0.519–9.386) |
| Aspergillus fumigatus qPCR positive | 39 (31.7) |
| Cycles of positivity. Median (range) | 35.5 (26.0–38.3) |
| Diagnosis of IA according to 2008 EORTC/MSG criteria | |
| Proven | 2 (1.6) |
| Probable | 28 (22.8) |
| Possible | 27 (22.0) |
| No IA according to 2008 EORTC/MSG criteria | 66 (53.7) |
| Atypical lesions and mycology positive = atypical IA | 14 (11.4) |
| Atypical lesions and mycology negative = controls | 52 (42.3) |
| Not receiving mould-active agents at BAL | 87 (70.7) |
| Diagnosis of IA according to 2008 EORTC/MSG criteria | |
| Proven | 1 (1.1) |
| Probable | 16 (18.4) |
| Possible | 18 (20.7) |
| No IA according to 2008 EORTC/MSG criteria | |
| Atypical lesions and mycology positive = atypical IA | 11 (12.6) |
| Atypical lesions and mycology negative = controls | 41 (47.1) |
| Receiving mould-active agents at BAL | 36 (29.3) |
| Diagnosis of IA according to 2008 EORTC/MSG criteria | |
| Proven | 1 (2.8) |
| Probable | 12 (33.3) |
| Possible | 9 (25) |
| No IA according to 2008 EORTC/MSG criteria | |
| Atypical lesions and mycology positive = atypical IA | 3 (8.3) |
| Atypical lesions and mycology negative = controls | 11 (30.6) |
| Other microbiological results | |
| Bacterial growth detected | 28 |
| Pneumocystis PCR positive | 8 |
| CMV DNA positive | 9 |
| Respiratory viruses detected | 27 |
| HSV DNA detected | 9 |
| Outcome variables | |
| Mould active treatment after BAL | 80 (65.0) |
| Alive at 12 weeks after BAL | 104 (84.6) |
| Alive at the last follow-up | 66 (53.7) |
| Median follow up after BAL, days (range) | 386 (10–1843) |
*3 A. flavus, 1 A. fumigatus, 1 A. niger, 1 Fusarium spp., 1 Paecilomyces spp.
BAL, broncoalveolar lavage; BDG, beta-d-glucan; GM, galactomannan; IA, invasive aspergillosis; SCT, stem cell transplant.
At the time of BAL, 15 patients (12.2%) were neutropenic and 36 (29.3%) were receiving mould active agents. CT scan was performed in median 2 days before BAL.
Diagnosis of IA
According to 2008 EORTC/MSG criteria, proven IA was diagnosed in two cases (1.6%), probable IA in 28 (22.8%), and possible in 27 (22%). Overall, 21/28 probable cases had positive GM in BAL, while in seven cases, all receiving antifungal therapy, the diagnosis of IA was made with serum GM in median 30 days before BAL, which was performed mainly due to suspected failure or breakthrough infection.
Among the remaining 66 patients without 2008 EORTC/MSG typical lesions, 14 had a positive mycological result: GM in BAL in 13 cases, with median ODI of 1.9 (range, 0.7–8.3) and serum GM in one case, and were considered cases of atypical IA.
In seven cases (5.7%) BAL cultures were positive for filamentous fungi: five Aspergillus (three A. flavus, one A. fumigatus, one A. niger), one Fusarium spp., and one Paecilomyces spp. Patients with growing Aspergillus were diagnosed with proven/probable IA in four cases, and atypical IA in one.
In 35 BAL samples GM was positive, with median ODI of 2.50 (range, 0.52–9.39).
Aspergillus fumigatus qPCR was positive in 39 samples with median cycle to positive of 35.4 (range, 26–38).
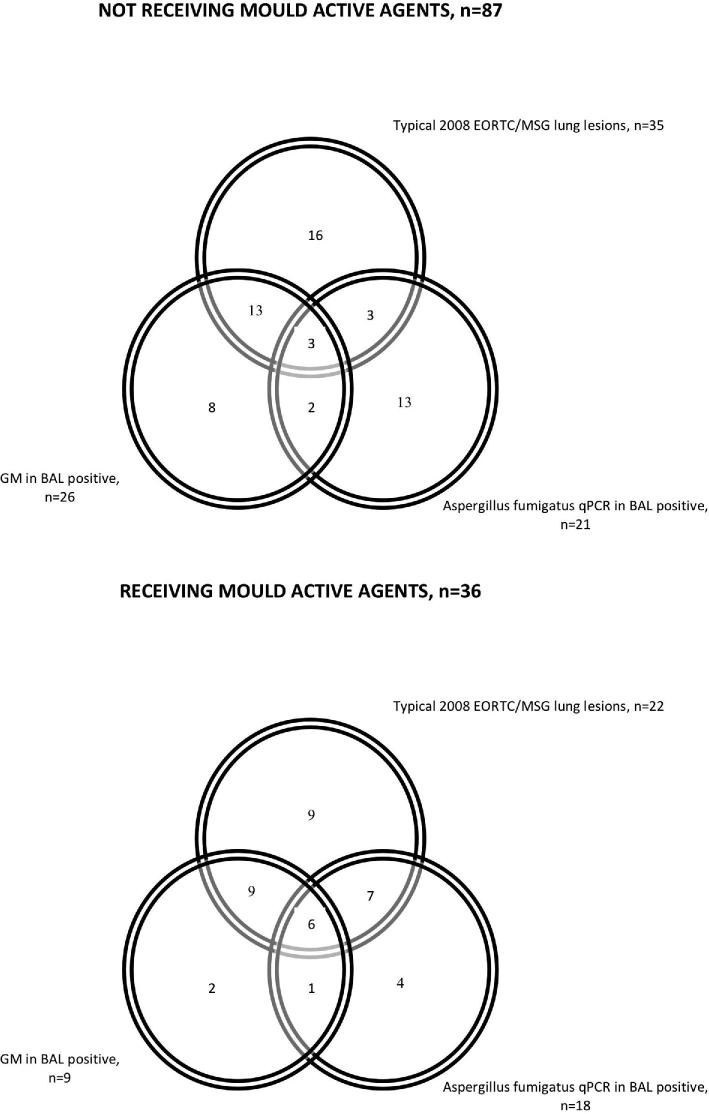
The concordance between typical radiological lesions, GM positivity in BAL and qPCR positivity was limited, as shown in Figure 1, both in patients receiving mould active agents and in patients without ongoing therapy. Also irrespective of radiological lesions, there was poor concordance between BAL GM and qPCR, even if BAL cutoff for positivity of 1 was applied (Supplement Fig. S1). There was no correlation between BAL GM positivity and qPCR Ct values and between PCR positivity and BAL GM ODI values (data not shown).
Figure 1.
Number of patients with results positive for invasive aspergillosis according to radiological criteria, BAL galactomannan (GM) and BAL Aspergillus fumigatus qPCR.
The classification of IA in those receiving and not antifungal agents is outlined in Table 1.
Performance of Aspergillus fumigatus qPCR
The prevalence of positive and negative results of BAL GM and Aspergillus fumigatus qPCR in four different IA diagnostic categories, divided also into patients receiving and not mould active agents at the time of BAL, is reported as supplement in Table S1.
The performance of A. fumigatus qPCR is reported in Table 2. The sensitivity was 33% (95%CI: 21–47) and specificity of 69% (95%CI: 55–81) when patients with proven/probable/possible IA were considered as cases, and, respectively, 40% (95%CI: 23–59) and 69% (95%CI: 55–81) considering as cases only those with proven/probable IA. Sensitivity and specificity were similar also when patients with atypical IA were considered together with proven/probable IA as cases (Table 2).
Table 2.
Diagnostic performance of A. fumigatus qPCR in all cases, divided into those receiving and not receiving mould active agents at the time of broncoalveolar lavage (value with 95% CI).
| Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|
| All cases, n = 123 | ||||
| Proven/probable/possible vs no IA (mycology negative). Atypical IA excluded | ||||
| PCR | 33 (21–47) | 69 (55–81) | 54 (41–67) | 49 (42–55) |
| PCR or GM | 54 (41–68) | 69 (55–81) | 66 (55–76) | 58 (50–66) |
| PCR and GM | 14 (6–26) | 100 (93–100) | 100 | 51 (49–54) |
| Proven/probable vs no IA (mycology negative). Atypical IA excluded | ||||
| PCR | 40 (23–59) | 69 (55–81) | 43 (29–58) | 67 (59–74) |
| PCR or GM | 83 (65–94) | 69 (55–81) | 61 (50–71) | 88 (76–94) |
| PCR and GM | 27 (12–46) | 100 (93–100) | 100 | 70 (66–75) |
| Proven/probable/atypical IA vs no IA (mycology negative) | ||||
| PCR | 36 (22–52) | 69 (55–81) | 50 (36–54) | 56 (49–63) |
| PCR or GM | 89 (75–96) | 69 (55–81) | 71 (62–79) | 88 (76–94) |
| PCR and GM | 25 (13–40) | 100 (93–100) | 100 | 61 (57–65) |
| Not receiving mould active agents, n = 87 | ||||
| Proven/probable/possible vs no IA (mycology negative). Atypical IA excluded | ||||
| PCR | 17 (7–34) | 71 (55–84) | 33 (17–54) | 50 (44–56) |
| PCR or GM | 54 (37–71) | 71 (54–84) | 61 (47–74) | 64 (55–73) |
| PCR and GM | 9 (2–23) | 100 (91–100) | 100 | 56 (54–59) |
| Proven/probable vs no IA (mycology negative). Atypical IA excluded | ||||
| PCR | 24 (7–50) | 71 (55–84) | 25 (11–47) | 69 (62–76) |
| PCR or GM | 100 (81–100) | 71 (54–84) | 59 (47–70) | 100 |
| PCR and GM | 18 (4–43) | 100 (91–100) | 100 | 75 (70–78) |
| Proven/probable/atypical IA vs no IA (mycology negative) | ||||
| PCR | 25 (11–45) | 71 (54–84) | 37 (21–56) | 58 (51–65) |
| PCR or GM | 100 (88–100) | 71 (54–84) | 70 (59–79) | 100 |
| PCR and GM | 18 (6–37) | 100 (91–100) | 100 | 64 (60–68) |
| Receiving mould active agents, n = 36 | ||||
| Proven/probable/possible vs no IA (mycology negative). Atypical IA excluded | ||||
| PCR | 59 (36–79) | 64 (31–89) | 76 (58–88) | 44 (28–60) |
| PCR or GM | 55 (32–76) | 64 (31–89) | 75 (56–88) | 41 (27–57) |
| PCR and GM | 27 (11–50) | 100 (72–100) | 100 | 41 (35–47) |
| Proven/probable vs no IA (mycology negative). Atypical IA excluded | ||||
| PCR | 61 (32–86) | 64 (31–89) | 67 (45–83) | 58 (38–76) |
| PCR or GM | 61 (32–86) | 64 (31–89) | 67 (45–83) | 58 (38–76) |
| PCR and GM | 46 (19–75) | 100 (72–100) | 100 | 61 (49–72) |
| Proven/probable/atypical IA vs no IA (mycology negative) | ||||
| PCR | 56 (30–80) | 64 (3189) | 69 (48–85) | 50 (33–67) |
| PCR or GM | 69 (41–89) | 64 (31–89) | 73 (54–87) | 58 (37–77) |
| PCR and GM | 44 (20–70) | 100 (72–100) | 100 | 55 (44–65) |
GM, galactomannan; ODI for GM in BAL and serum 0.5 ODI.
When considering the influence of antifungal treatment, the sensitivity of qPCR was higher in those receiving mould active agents at the time of BAL (61%, 95%CI: 32–86) compared to those not receiving antifungals (24%, 95%CI: 7–50), while the specificity was similar (respectively, 64%, 95%CI: 31–89 and 71%, 95%CI: 55–84) (Table 2). Among 36 patients receiving mould active agents, nine had positive BAL GM, and qPCR was positive in seven of them, compared to five among 26 of those not receiving antifungals (P < .05).
In all cases, the performance was much better when qPCR was used together with GM, and was worse if positivity of both qPCR and GM was required. The sensitivity values were similar irrespective of the definition used for cases (only those with proven/probable IA, those with proven/probable/possible IA, or those with proven/probable/atypical), being only slightly higher in case for proven/probable IA cases (Table 2).
If a cutoff for qPCR of ≤ 35 Ct was used, sensitivity was lower and specificity higher than for the cutoff of < 40 Ct. For proven/probable IA, the sensitivity was 23% (95%CI: 10–42) and the specificity was 88% (95%CI: 76–96), with higher sensitivity in those receiving mould active agents compared to those not in treatment, respectively, 31% (95%CI: 9–61) versus 18% (95%CI: 4–43).
qPCR was positive in three of five BAL samples with culture growth of Aspergillus with Ct of 25.88 and 37.10 for A. flavus (respectively, probable IA, BAL GM ODI 8.2, and proven resistant IA under treatment with BAL GM ODI 7.6) and 36.83 for A. niger (probable IA under treatment, BAL GM ODI 0.08) and in case of growth of Fusarium (35.30 Ct, BAL GM ODI 0.1) and Paecilomyces (33.33 Ct, BAL GM ODI 0.44), while it was negative in one case of A. flavus (BAL GM ODI 2.5) and one case of A. fumigatus (BAL GM ODI 0.99).
Overall, 16 out of 52 patients with negative mycology and atypical lesions had a positive A. fumigatus qPCR (31%); eight of them received mould active drugs after BAL (either as treatment or prophylaxis, including four patients who started mould-active agents before BAL), while eight patients did not received any mould active agents within 3 months after BAL, and they were all alive at 12 weeks follow-up. Thus, considering high mortality of IA in patients with hematological malignancy if untreated, they were considered as false positives for qPCR or colonized.
Antifungal resistance
TR34/L98H mutation was detected in one case, in a SCT recipient previously exposed to azoles for prolonged treatment of IA after the first SCT. After the second SCT, GM in BAL was positive but culture negative. Initially after BAL, azole therapy was started but after a clinical diagnosis of treatment failure, the therapy was changed to liposomal amphotericin B. GM became negative and radiological lesions improved, but the patient deceased 3 months after the transplant due to a relapse of acute leukemia.
Discussion
This retrospective study showed a poor sensitivity of this qPCR for A. fumigatus, with better results in patients receiving mould active agents at the time of BAL (sensitivity 61% vs 24%, respectively) and moderate specificity (64% and 71%, respectively). The combined performance of qPCR and GM was significantly better than the use of qPCR alone.
Our cohort included very selected patients, all at high risk for IA, with various radiological lesions, in whom BAL was performed mainly because non-invasive results were negative for IA and diagnosis was not reached with other tests. Moreover, 12% of them had lung lesions atypical according to EORTC/MSG criteria and positive mycological results. These patients are “unclassified” according to EORTC, and they are usually considered as having IA and treated.8 Therefore, they could not be included as controls for the assessment of diagnostic performance. However, even patients included in the control group (with non-typical lesions and negative mycology), belonged to a high risk population, and the positivity of PCR cannot be easily interpreted as false positive results. Indeed, 50% of patients in this group with a positive PCR did receive empirically mould active agents, so only in remaining 50% of them, positive PCR could be confidently considered as false positive results or colonisation. Moreover, even the presence of an alternative diagnosis should not serve a criterion for the absence of IA, since fungal and bacterial or viral co-infections are frequent in this setting.15
Good diagnostic performances have reported in meta-analyses, which included mainly studies of in-house methods, with sensitivity and specificity reported of >75% and >93%, respectively.16 However, in some recent studies, the sensitivity was markedly lower (approximately 30%).11,17 The commercial assays allow standardization, high reproducibility and have a validated quality control. However, they do not contain a control of DNA extraction, which may differ for hyphae and for free DNA, and quality control for the BAL itself. Additionally, their performance depends on the assay used and the clinical setting. The low value obtained in our study is not comparable with most of the performances described to date in the literature using MycoGenie®. However, most other studies included patients with high rate of culture-positive respiratory samples. Indeed, the sensitivity was 92.9% in a cohort with 59 of 88 respiratory samples positive in culture for Aspergillus,14 71% in 31 cases of probable IA with 45% of positive BAL culture rate,18 and 77.4% in case of fungal rhinosinusitis in which a high concentration of fungus DNA is present.19 Also a recent study comparing various diagnostic methods, which reported the sensitivity of MycoGenie® of 73.7% in 38 patients with proven/probable IA, had a very high (22/38, 58%) rate of positivity of culture for Aspergillus.12 In addition, in that cohort, the sensitivity was lower in 41 hematological patients than in those from intensive care unit (ICU) suggesting a lower fungal burden sufficient to cause IFD in highly immunocompromised hosts.12 Indeed, also the low yield of fungal cultures and rather low median BAL GM ODI confirm the low burden of viable moulds in this study. Therefore, the performance of MycoGenie® in a cohort of with low fungal burden remains to be established. Another explanation for low sensitivity of this qPCR, even in a subgroup of BAL GM positive patients with probable IA, might be the infection with species other than A. fumigatus which are not detected by this assay.
A very interesting factor which influenced sensitivity in our cohort was the presence of mould active treatment at the time of BAL, which increased the sensitivity from 24% to 61%. One possible explanation is that mould active agents caused lysis of the fungal wall with a higher percentage of free DNA detectable in the respiratory tree, particularly in the supernatant of a centrifuged BAL sample, which was the part used in this study.20 The availability of free fungal DNA would result in much better sensitivity of DNA extraction, which is a critical process for the performance of fungal PCR.16 This observation is indirectly confirmed by cohorts reporting higher performance of PCR, both in BAL and serum, in those receiving mould active agents.21–23 Also in our study, the rate of qPCR positivity in GM positive BAL samples was higher form patients receiving mould active drugs compared to those not treated. Better specification which portion of BAL fluid should be used might be warranted, since by testing supernatant, DNA still within the organism or phagocytic host cells might not be detected.
The specificity of qPCR in our study was 70% which is lower than in the abovementioned meta-analyses16 and other studies using the same assay.14,18 However, in this study there were no heathy controls, as all the patients were at high risk of IA and had lung lesions, which are referred to in a recent prospective EORTC study as those in whom IA cannot be excluded.9
When considering the cases in which PCR represented the only positive mycological criterion, that is, with BDG and GM negative, half of these patients (8/16) survived > 3 months without antifungal treatment and without developing IA, confirming that these were either false positives or cases of colonization. Obviously, this could be established only due to a retrospective nature of our study in which PCR results were not available at the time of diagnosis, but it documents a high rate of clinically irrelevant false positive results.
Furthermore, considering the cross-reactivity with other species, in our study the qPCR, which should be specific for A. fumigatus, was positive in four cases in which different species grew in culture (two A. flavus, both with BAL GM > 7.5 ODI; one Fusarium spp.; one Paecilomyces). Although in case of fungi other than Aspergillus co-infection might be present, it is unlikely based on negative BAL GM. On the contrary, cases of cross-reactivity of the method with strains of A. flavus have already been described in the literature,23 and such a false positive result might be particularly likely in case of high fungal burden, as demonstrated in our two cases. From the clinical point of view, a qPCR should detect all species of Aspergilllus, since species other than A. fumigatus might be more prevalent, particularity in some geographical zones, for example, A. flavus in Mediterranean.24
Although triazole-resistant strains of A. fumigatus are increasing in several geographical regions, fortunately in our cohort only one patient had the TR34/L98H mutation, accounting for 2.6% rate among PCR positive samples. Such a low incidence is in line with what reported for Italy;25 however, it should be considered with caution given overall poor sensitivity of this qPCR assay. Although molecular methods are important tools, in addition to culture, to monitor the changes in resistance patters, also in this case, detection of more than one resistance mutation might be useful. Finally, the absence of detected mutations does not exclude antifungal resistance in case of clinical failure since various mutation patterns can occur, particularly in case of previous azole exposure.
The possibility of overestimating the performance of BAL GM, which was the most frequent mycological criterion in this cohort, should be acknowledged. However, it could not be avoided in a population with mostly negative serum GM and BDG results, low culture yield, and lung biopsy frequently not feasible.
In conclusion, our study demonstrated that the Aspergillus fumigatus qPCR in BAL should be performed together with GM, and it may offer clinical contribution particularly in patients receiving mould active agents, in whom GM is usually negative but PCR had better sensitivity. However, assays detecting most of the common species should be preferred and testing materials other than supernatant might result in higher sensitivity. PCR might be a valuable tool for detecting antifungal resistance both in case of infection or colonization, which may have significant implications for treatment and prophylaxis of IA.
Supplementary Material
Acknowledgements
Financial support. This study was supported by the Fellowship Program Award Gilead 2015.
Declaration of interest
M.M. has received speaker and advisory board fees from Gilead, Pfizer, Biotest, Janssen and MSD. C.V. has received research support to his institution from Pfizer and MSD, speaker, and advisory board fees from Gilead, Pfizer, and MSD. Other authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1. Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax. 2015; 70: 270–277. [DOI] [PubMed] [Google Scholar]
- 2. Patterson TF, Thompson GR, 3rd, Denning DW et al.. Practice guidelines for the diagnosis and management of aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016; 63: e1–e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ullmann AJ, Aguado JM, Arikan-Akdagli S et al.. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin Microbiol Infect. 2018; 24: e1–e38. [DOI] [PubMed] [Google Scholar]
- 4. Bongomin F., Gago S. Global and multinational prevalence of fungal diseases-estimate precision. J Fungi. 2017; 3: E57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meis JF, Chowdhary A. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc London B Biol Sci. 2016; 371. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Pauw B, Walsh TJ, Donnelly JP et al.. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008; 46: 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maertens JA, Nucci M, Donnelly JP. The role of antifungal treatment in hematology. Haematologica. 2012; 97: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nucci M, Nouer SA, Grazziutti M et al.. Probable invasive aspergillosis without prespecified radiologic findings: proposal for inclusion of a new category of aspergillosis and implications for studying novel therapies. Clin Infect Dis. 2010; 51: 1273–1280. [DOI] [PubMed] [Google Scholar]
- 9. Observational study to determine the rate of occurrence of invasive mould disease and treatment outcomes in at-risk patients: a European prospective invasive mould disease audit (PIMDA) [Internet] 2011ECMM. available at http://www.pimda.eu/.
- 10. Avni T, Levy I, Sprecher H et al.. Diagnostic accuracy of PCR alone compared to galactomannan in bronchoalveolar lavage fluid for diagnosis of invasive pulmonary aspergillosis: a systematic review. J Clin Microbiol. 2012; 50: 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boch T, Spiess B, Cornely OA et al.. Diagnosis of invasive fungal infections in haematological patients by combined use of galactomannan, 1,3-beta-D-glucan, Aspergillus PCR, multifungal DNA-microarray, and Aspergillus azole resistance PCRs in blood and bronchoalveolar lavage samples: results of a prospective multicentre study. Clin Microbiol Infect. 2016; 22: 862–868. [DOI] [PubMed] [Google Scholar]
- 12. Guegan H, Robert-Gangneux F, Camus C et al.. Improving the diagnosis of invasive aspergillosis by the detection of Aspergillus in broncho-alveolar lavage fluid: Comparison of non-culture-based assays. J Infect. 2018; 76: 196–205. [DOI] [PubMed] [Google Scholar]
- 13. Eigl S, Hoenigl M, Spiess B et al.. Galactomannan testing and Aspergillus PCR in same-day bronchoalveolar lavage and blood samples for diagnosis of invasive aspergillosis. Med Mycol. 2017; 55: 528–534. [DOI] [PubMed] [Google Scholar]
- 14. Dannaoui E, Gabriel F, Gaboyard M et al.. Molecular diagnosis of invasive aspergillosis and detection of azole resistance by a newly commercialized PCR kit. J Clin Microbiol. 2017; 55: 3210–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alshabani K, Haq A, Miyakawa R, Palla M, Soubani AO. Invasive pulmonary aspergillosis in patients with influenza infection: report of two cases and systematic review of the literature. Expert Rev Respir Med. 2015; 9: 89–96. [DOI] [PubMed] [Google Scholar]
- 16. White PL, Wingard JR, Bretagne S et al.. Aspergillus polymerase chain reaction: systematic review of evidence for clinical use in comparison with antigen testing. Clin Infect Dis. 2015; 61:1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buess M, Cathomas G, Halter J et al.. Aspergillus-PCR in bronchoalveolar lavage for detection of invasive pulmonary aspergillosis in immunocompromised patients. BMC Infect Dis. 2012; 12: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denis J, Forouzanfar F, Herbrecht R et al.. Evaluation of two commercial real-time PCR kits for Aspergillus DNA detection in bronchoalveolar lavage fluid in patients with invasive pulmonary aspergillosis. J Mol Diagn. 2018; 20: 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morio F Dannaoui E, Chouaki T et al.. PCR-based detection of Aspergillus fumigatus and absence of azole resistance due to TR34 /L98H in a french multicenter cohort of 137 patients with fungal rhinosinusitis. Mycoses. 2018; 61: 30–34. [DOI] [PubMed] [Google Scholar]
- 20. Springer J, White PL, Kessel J et al.. A Comparison of Aspergillus and mucorales PCR testing of different bronchoalveolar lavage fluid fractions from patients with suspected invasive pulmonary fungal disease. J Clin Microbiol. 2018; 56:e01655–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Postina P, Skladny J, Boch T et al.. Comparison of two molecular assays for detection and characterization of Aspergillus fumigatus triazole resistance and cyp51A mutations in clinical isolates and primary clinical samples of immunocompromised patients. Front Microbiol. 2018; 9: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. White PL, Barnes RA, Springer J et al.. Clinical performance of Aspergillus PCR for testing serum and plasma: a Study by the European Aspergillus PCR Initiative. J Clin Microbiol. 2015; 53: 2832–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guegan H, Chevrier S, Belleguic C et al.. Performance of molecular approaches for Aspergillus detection and azole resistance surveillance in cystic fibrosis. Front Microbiol. 2018; 9: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zarrinfar H Mirhendi H, Fata A, Khodadadi H, Kordbacheh P. Detection of Aspergillus flavus and A. fumigatus in bronchoalveolar lavage specimens of hematopoietic stem cell transplants and hematological malignancies patients by real-time PCR, nested polymerase chain reaction and mMycological assays. Jundishapur J Microbiol. 2015; 8: e13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prigitano A, Venier V, Cogliati M et al.. Azole-resistant Aspergillus fumigatus in the environment of northern Italy, May 2011 to June 2012. Euro Surveill. 2014; 19: 20747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.