Abstract
Prostate cancer poses a public health threat to hundreds of people around the world. p62 has been identified as a tumor suppressor, however, the mechanism by which p62 promotes prostate cancer remains poorly understood. The present study aimed to investigate whether p62 promotes proliferation, apoptosis resistance and invasion of prostate cancer cells via the Kelch-like ECH-associated protein 1/nuclear factor erytheroid-derived 2-like 2/antioxidant response element (Keap1/Nrf2/ARE) axis. Immunohistochemical staining and immunoblotting were performed to determine the protein levels. Rates of proliferation, invasion and apoptosis of prostate cancer cells were assessed using an RTCA system and flow cytometric assays. Levels of reactive oxygen species (ROS) were assessed using Cell ROX Orange reagent and mRNA levels of Nrf2 target genes were detected by qRT-PCR. It was revealed that p62 increased the levels and activities of Nrf2 by suppressing Keap1-mediated proteasomal degradation in prostate cancer cells and tissues, and high levels of p62 promoted growth of prostate cancer through the Keap1/Nrf2/ARE system. Silencing of Nrf2 in DU145 cells overexpressing p62 led to decreases in the rate of cell proliferation and invasion and an increase in the rate of cell apoptosis. p62 activated the Nrf2 pathway, promoted the transcription of Nrf2-mediated target genes and suppressed ROS in prostate cancer. Therefore, p62 promoted the development of prostate cancer by activating the Keap1/Nrf2/ARE pathway and decreasing p62 may provide a new strategy to ameliorate tumor aggressiveness and suppress tumorigenesis to improve clinical outcomes.
Keywords: apoptosis, autophagy, Keap1, Nrf2, p62, prostate cancer
Introduction
Prostate cancer (PCa) is one of the most common types of cancer among men and accounts for 1/5 patients newly-diagnosed with cancer in the USA alone (1). There were at least 180,000 newly-diagnosed cases and 26,000 estimated-deaths in 2016. Current treatments include hormonal therapy, radical prostatectomy and radiation therapy. However, almost all patients with PCa eventually develop incurable metastatic castration-resistant PCa (mCRPC) and generally succumb to this disease 2 years later (2,3). Currently, the mechanism underlying PCa development is still far from being understood, and thus investigating its molecular mechanisms is important and will help improve therapeutic efficacy.
p62 (also known as sequestosome-1, SQSTM-1 or A170) contains 440 amino acid residues, is named based on its molecular weight of 62 kDa, and plays a key role in the regulation of cell proliferation and survival (4). p62 serves as a selective autophagy adaptor protein that links ubiquitinated proteins to autophagy machinery for degradation. p62 harbors multiple functional domains and plays important roles in multiple critical signaling pathways. An increasing amount of evidence has demonstrated that high levels of p62 are associated with multiple diseases including liver disorders, neurodegenerative diseases and cancers (5).
The Kelch-like ECH-associated protein 1/nuclear factor erytheroid-derived-2-like 2/antioxidant response element (Keap1/Nrf2/ARE) system is currently recognized as one of the major defense mechanisms against oxidative stress and xenobiotics (6,7), thus, the dysregulation of the Keap1/Nrf2/ARE system is involved in several human illnesses, particularly cancers (8,9). Nrf2 is a critical leucine zipper (bZIP) transcription factor that serves as a major regulator in cellular defenses against reactive oxygen species (ROS) and chemical detoxification to protect from oxidative damages. Under normal conditions, Keap1 specifically recognizes Nrf2 (10) and constantly carries Nrf2 to proteasomes for degradation (11,12). Conversely, the activity of a Keap1-associated E3 ubiquitin ligase is inhibited so that Nrf2 protein is activated and transfers from the cytoplasm to the nucleus leading to nuclear accumulation under oxidative stress. Nuclear Nrf2 increases the expression of ARE-bearing genes including heme oxygenase-1 (HO-1), NAD(P)H quinine oxidoreductase (NQO-1), glutamate-cysteine ligase (GCL), glutathione (GSH) generation enzymes and GSH peroxidase (GPx), to neutralize ROS and maintain a balance of cellular redox (13–15).
When autophagy is impaired, p62 forms a complex with Keap1 and sequesters Keap1 into inclusion bodies, resulting in a stabilization of Nrf2 protein and an induction of ARE-containing genes. Therefore, p62 emerges as a linker molecule between autophagy and oxidative stress (15).
The crosstalk between autophagy and the Keap1/Nrf2/ARE axis was reported in 2010, and since then great efforts have been made to investigate the underlying molecular mechanisms, particularly in different types of cancers such as hepatocellular carcinoma (16), ovarian cancer (17) and lung cancer (18). A previous study demonstrated that the clinical drug verteporfin induced aggregation of p62 and inhibited autophagy thus suppressing prostate tumorigenesis in a xenograft model (19). However, the interplay between the two signaling pathways remains largely unknown in PCa. Given that p62 acts as a regulator of the Keap1/Nrf2 pathway, the previous study hypothesized that p62 is involved in the Keap1/Nrf2/ARE pathway and plays a critical role in the pathogenesis of mCRPC.
The present study demonstrated that p62 functioned as a molecular regulator in the Keap1/Nrf2/ARE axis to promote the transcription of Nrf2-mediated genes and decrease the levels of ROS in PCa. Oxidative stresses generated under defective autophagy trigger genome instability and tumorigenesis (20). Therefore, inhibiting p62 and Nrf2 activity may suppress carcinogenicity.
Materials and methods
Collection of animal and patient tissue samples
In the present study, PTEN−/− mice and transgenic adenocarcinoma of mouse prostate (TRAMP) mice were obtained from Jackson Laboratory. All animal protocols were approved by the Animal Care and Use Committee of the Texas A&M Health Science Center at Houston (AUP IACUC 2015-0132-IBT). Normal prostate tissues and prostate tumor tissues were collected from three sets of 11-month-old wild-type, PTEN−/− and TRAMP mice after mice were sacrificed by performing cervical dislocation by appropriately trained personnel approved on the animal protocol. A total of 150 patients including 112 patients with PCa and 38 with BPH as controls were evaluated in the present study. As previously described (21), these patients had consented to donate prostate tissue samples from surgery or via biopsy and underwent treatment including radiotherapy, prostatectomy, transurethral resection of the prostate (TURP) and hormone therapy between 1999 and 2003 at The Fifth Affiliated Hospital of Guangzhou Medical University, Sun Yat-Sen Memorial Hospital, and The First People's Hospital of Guangzhou and The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, China.
Immunohistochemistry analyses
All primary samples were fixed in 10% formalin, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Paraffin-embedded, formalin-fixed prostate tissues of patients and mice were immunostained for p62, Keap1 and Nrf2 protein using an immunohistochemical detection kit (GTVision) according to the manufacturer's protocol. Immunostained slides were independently evaluated under a Leica DM3000 microscope by two pathologists in a double-blind manner.
Cell culture, transfection and siRNA. DU145 cells were obtained from the American Type Culture Collection (ATCC); maintained in RPMI-1640 medium (HyClone Laboratories; GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 units/ml penicillin and 100 µg/ml streptomycin in a humidified incubator at 37°C with 5% CO2; and sub-cultured for a passage every 3–4 days. Stable DU145 cell strains expressing different levels of p62 were constructed as previously described: C1 carrying the control plasmid vector, OE overexpressing p62, KO with p62 knocked out by CRISPER technology and C2 negative control of KO (22). Validated siRNAs against Keap1 and Nrf2 were purchased from Guangzhou Ribobio Co., Ltd. Cells were treated with 10 mM bafilomycin A1 (BAF) or 10 mM MG-132 for 6 h when necessary.
Immunoblotting analyses
Whole-cell extracts were prepared with RIPA buffer containing the broad protease inhibitor cocktail (Roche Diagnostics). Nucleoprotein extraction was prepared with NE-PER Nuclear Cytoplasmic Extraction Reagent kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The protein concentration was determined using a BCA kit. Equal amounts of total proteins (25 µg) were separated via 10% SDS-PAGE gel and transferred to PVDF membranes. Then, 5% skimmed milk (Devondale Milk) was used to incubate at room temperature for 2 h. Then the membranes were blotted with primary antibodies against p62 (product code ab56416, dilution 1:2,500), Keap1 (product code ab218815, dilution 1:1,500) and Nrf2 (product code ab194986, dilution 1:1,000; all were from Abcam), Lamin B1 (cat. no. sc-374015, dilution 1:1,000; Santa Cruz Biotechnology, Inc.) or GAPDH (cat. no. AF0911, dilution 1:4,000; Affinity Biosciences), all the primary antibodies were incubated at 4°C overnight. Protein bands were visualized using their respective goat anti-rabbit IgG (H+L) (dilution 1:1,000; incubated at room temperature for an 1 h) or goat anti-mouse IgG (H+L) (dilution 1:1,000; incubated at room temperature for an 1 h) (Wuhan Boster Biological Technology, Ltd.) and ECL reagent (Thermo Fisher Scientific, Inc.). ImageJ® software (v1.52p; National Institutes of Health) was used for densitometric analysis.
Confocal fluorescent microscopy
Confocal fluorescent microscopy was performed as previously described (22). Briefly, cells were grown on glass coverslips in 6-well plates for 6 h, washed three times with PBS, fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and incubated with 2% BSA for 1 h at room temperature. Treated cells were first blotted with an antibody against Keap1 (product code ab139729) or Nrf2 (product code EP1808Y; both from Abcam) for 16 h at 4°C, and subsequently incubated with DYLight649-conjugated goat anti-rabbit IgG (H+L) secondary antibody (cat. no. GAR6492; Multi Sciences) for 1 h at room temperature. After washing with PBS, coverslips were mounted with Fluoromount-G™ (Invitrogen; Thermo Fisher Scientific, Inc.) and visualized under a confocal laser scanning microscope (Zeiss) at an ×20 magnification.
Reverse transcription-quantitative PCR (RT-qPCR) analyses
Total RNA was isolated using the UNlQ-10 Column TRIzol Total RNA Isolation kit (Sangon Biotech Co., Ltd.) according to the manufacturer's protocol, and reverse-transcribed to cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc.). The conditions used were as follows: 30 sec 95°C, followed by 50 cycles of 5 sec 95°C, 34 sec 60°C, then 15 sec 95°C, and 1 min 50°C. The gene-specific primers presented in Table I were obtained from PrimerBank. Differential expression was calculated using the 2−ΔΔCq method as described (23). Data are presented as the mean ± standard deviation of three independent experiments.
Table I.
The sequences of primers used for qPCR experiments.
| Primer | Sequence |
|---|---|
| hKeap1 | F: GCTGATGAGGGTCACCAGTT |
| R: CCAACTTCGCTGAGCAGATT | |
| hNrf2 | F: GCTCATACTCTTTCCGTCGC |
| R: ATCATGATGGACTTGGAGCTG | |
| hNQO-1 | F: GCACTGATCGTACTGGCTCA |
| R: GAACACTCGCTCAAACCAG | |
| hGCLC | F: GGATGATGCTAATGAGTCTGACC |
| R: TCTACTCTCCATCCAATGTCTGAG | |
| HO-1 | F: CCAGGCAGAGAATGCTGAGTTC |
| R: AAGACTGGGCTCTCCTTGTTGC | |
| hGCLM | F: TTGGGAACTCCATTCATTCA |
| R: CGGGAACCTGCTCAACTG | |
| GAPDH | F: GAAGGTGAAGGTCGGAGTC |
| R: GAAGATGGTGATGGGATTTC |
Keap1, Kelch-like ECH-associated protein 1; Nrf2, nuclear factor erytheroid-derived 2-like 2; NQO-1, NAD(P)H quinine oxidoreductase; GCLC, glutamate-cysteine ligase catalytic subunit; HO-1, heme oxygenase-1; GLCM, glutamate-cysteine ligase modifier subunit.
Apoptosis analyses
The Annexin V-APC/7AAD Apoptosis Detection system was used to measure rates of apoptosis according to the manufacturer's protocol. Briefly, cells were cultured in RPMI-1640 medium in 10-cm dishes to a confluence of 80–90%, digested with 0.25% trypsin without EDTA, re-suspended in 500 µl of binding buffer supplied with 5 µl APC-Annexin V and 10 µl 7AAD, and incubated at room temperature for 15 min after mixing. The apoptosis rate was detected using BD Accuri C6 flow cytometer and analyzed with the associated software from BD Biosciences.
Cell proliferation assays
The rates of cell proliferation were measured using the Real Time Cellular Analysis (RTCA) system. Briefly, cells were harvested at 48 h after infection with siRNA, diluted and seeded into a 16-well strip at a density of 1×104 cells/well. Cell proliferation was monitored in real time with sensor devices placed in the incubator with 5% CO2.
Cell invasion assays
Cell invasion was continuously monitored every 15 min for 36 h by real-time monitoring on CIM-Plate 16 pre-coated with Matrigel (356234; Corning Inc.) with an RTCA iCELLigence Analyzer (ACEA Biosciences). Briefly, the infected cells were starved for 6 h in serum-starved medium, diluted and seeded into a 16-well strip at a density of 3×105 cells/well in the upper chamber. The lower chamber was filled with medium containing 10% serum.
ROS detection
The detection of ROS was performed using the Cell ROX Orange reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cell-permeable reagents are non-fluorescent or weakly fluorescent in a reduced state, and emit strong fluorescence signals upon oxidation. DU145 cells in a 6-well plate were incubated in medium containing a final concentration of 5 µM CellROX Orange reagent at 37°C for 30 min and washed three times with pre-warmed PBS after the medium was removed. Levels of ROS of cells were quantified using ImageJ software (v1.52p; National Institutes of Health) based on the fluorescence intensities of cells in captured fields.
Statistical analysis
Experimental data were presented as the mean ± standard error of at least three independent experiments. Differences in results were analyzed by a two-tailed unpaired t-test using the GraphPad Prism version 7 for Windows (GraphPad Software, Inc.). P<0.05, P<0.01, and P<0.001 were considered to indicate a statistically significant result (*P<0.05, **P<0.01 and ***P<0.001 as indicated in the figures).
Results
Expression of P62, Keap1 and Nrf2 in prostate tissues
It has previously been reported that PCa tissues have higher levels of cytoplasmic p62 than benign prostate hyperplasia (BPH) tissues although p62 whose significance remains unknown was also observed to be distributed in nuclei. In addition, PCa tissues from patients with higher grades or metastasis have significantly higher levels of p62 than those from patients with lower grades or no metastasis (20). The present study further demonstrated that p62 was primarily located in the cytoplasm of epithelial cells of human PCa and BPH tissues (Fig. 1A-C). Keap1 was also primarily located in the cytoplasm of epithelial cells of human PCa and BPH tissues (Fig. 1D and E) while Nrf2 was detected in both the nucleus and cytoplasm (Fig. 1F and G). The levels of p62 and Nrf2 were higher in human PCa than those in BPH (Fig. 1A-C, F and G), while the levels of Keap1 were higher in BPH than those in PCa (Fig. 1D and E). Similarly, higher levels of p62 and Nrf2 and lower levels of Keap1 were detected in PCa tissues collected from PTEN−/− and TRAMP mice compared with normal prostate tissues collected from normal wild-type mice (Fig. 1H-P). The results indicated that p62 and Nrf2 may act as oncogenes of PCa.
Figure 1.
The expression of p62, Keap1 and Nrf2. (A and B) Representative images and (C) quantification revealing the levels of p62 in (A) BPH and (B) PCa. A total of 150 patients including 112 patients with PCa and 38 with BPH were evaluated. The relative levels of p62 in prostate glands and stromas and in cytoplasm and nuclei were compared between BPH and PCa. *P≤0.05 and **P≤0.01 Representative images showing (D) high levels of Keap1 in BPH, or (E) low levels of Keap1 in PCa, and (F) low levels of Nrf2 in BPH, or (G) high levels of Nrf2 in PCa. Representative images showing (H) low or (I and J) high levels of p62 expressed in prostate tissues from (H) normal, (I) PTEN−/−, or (J) TRAMP mice; (K) high, or (L and M) low levels of Keap1 expressed in (K) prostate tissues from normal, (L) PTEN−/−, or (M) TRAMP mice; and (N) low, or (O and P) high levels of Nrf2 expressed in (N) prostate tissues from normal, (O) PTEN−/−or (P) TRAMP mice. Normal prostate tissues and prostate tumors were collected from three sets of wild-type, PTEN−/− and TRAMP mice. Scale bar, 50 µm. Keap1, Kelch-like ECH-associated protein 1; Nrf2, nuclear factor erytheroid-derived 2-like 2; BPH, benign prostate hyperplasia; PCa, prostate cancer; TRAMP, transgenic adenocarcinoma of mouse prostate.
p62 suppresses the expression of Keap1 and enhances the activity of Nrf2
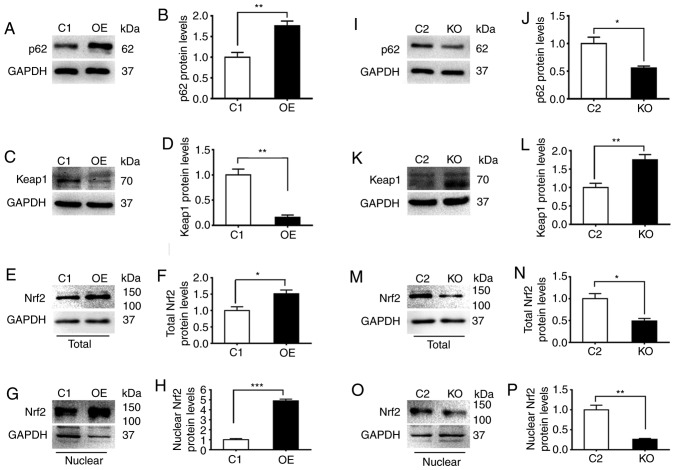
To examine the association between the levels of p62, Keap1 and Nrf2 proteins, the present study prepared extracts from four different DU145 cell strains expressing different levels of p62. Increasing the expression levels of p62 significantly decreased the levels of Keap1 and enhanced the levels of total Nrf2 and nuclear Nrf2 (Fig. 2A-H), respectively. In contrast, decreasing the expression level of p62 significantly increased levels of Keap1 and reduced levels of total Nrf2 and nuclear Nrf2 (Fig. 2I-P), respectively. In addition, the effects of p62 on the levels of Keap1 and total Nrf2 were further confirmed by immunofluorescence microscopy analyses (Fig. 2Q-T). Therefore, p62 suppressed the levels of Keap1 and enhanced the levels of Nrf2.
Figure 2.
p62 suppresses the expression of Keap1 and enhances the activity of Nrf2. C1, DU145 cells carrying the control plasmid vector; OE, DU145 cells overexpressing p62; KO, OE cells with p62 knocked out by CRISPER technology; and C2, OE cells carrying negative control for CRISPR knockout. (A-P) Representative (A, C, E, G, I, K, M and O) immunoblots and (B, D, F, H, J, L, N and P) quantification showing levels of (A, B, I and J) p62, (C, D, K and L) Keap1, and (E, F, M and N) total and (G, H, O and P) nuclear Nrf2 in (A-H) C1 and OE cells and (I-P) C2 and KO cells. (Q-T) Representative images revealing the (Q and R) immunostaining intensities and (S and T) quantification of (Q and S) Keap1 and (R and T) Nrf2 in C1, OE, C2 and KO cells. The nuclei were stained with DAPI (blue). All images were captured under an identical setting. The results represent three independent experiments. *P≤0.05, **P≤0.01, and ***P≤0.001. Keap1, Kelch-like ECH-associated protein 1; Nrf2, nuclear factor erytheroid-derived 2-like 2.
p62 decreases apoptosis and promotes proliferation and invasion of DU145 cells through Nrf2
In order to determine whether aberrant Nrf2 activity was associated with apoptosis, proliferation and invasion in vitro, DU145 cells overexpressing p62 (OE) were transfected with negative control siRNA (Ctl siRNA) and Nrf2-specific siRNA (Nrf2 siRNA). A knockdown efficiency of 80% was confirmed by qPCR and immunoblotting (Fig. 3A and B). Flow cytometric analyses of cell apoptosis revealed that DU145 cells overexpressing p62 (OE) had lower rates of apoptosis than its control C1 (Fig. 3C and D), while silencing the expression of Nrf2 in OE cells induced prominently higher rates of apoptosis (Fig. 3E and F). In contrast, the overexpression of p62 led to increases in cell proliferation and invasion, while further decreasing the increased levels of Nrf2 expression with siRNA led to decreases in cell proliferation and invasion (Fig. 3G and H). Notably, rates of cell proliferation and invasion between OE and Ctl siRNA cells were different potentially due to the impact of control siRNA (Fig. 3G and H). Therefore, p62 decreased apoptosis and promoted proliferation and invasion of DU145 cells through Nrf2.
Figure 3.
p62 decreases apoptosis and promotes proliferation and invasion of DU145 cells through Nrf2. C1, DU145 cells carrying the control plasmid vector; OE, DU145 cells overexpressing p62. (A) Analyses of the levels of Nrf2 mRNA by RT-qPCR in DU145 cells stably expressing p62 (OE as presented in Fig. 2) infected with Nrf2-specific siRNA (Nrf2 siRNA) or negative control siRNA (Ctl siRNA) for 48 h. The mRNA levels were normalized to those of GAPDH and presented as ratios relative to those in cells transfected with control siRNA. (B) Immunoblot analyses of Nrf2 expression in C1 and OE as presented in Fig. 2, and OE cells treated with control siRNA (Ctl siRNA), and OE cells treated with Nrf2-specific siRNA (Nrf2 siRNA). The results represent three independent experiments. (C and E) Flow cytometric analyses and (D and F) quantification of apoptotic rates in (C and D) C1 and OE, and (E and F) OE cells treated with Ctl or Nrf2 siRNA. The results represent three independent experiments. *P≤0.05 and **P≤0.01. (G) A cell proliferation assay revealing the effects of p62 and Nrf2 expression on the rates of cell growth. (H) An invasion assay revealing the effects of p62 and Nrf2 expression on the rates of cell invasion. Nrf2, nuclear factor erytheroid-derived 2-like 2.
p62 increases the expression levels of Nrf2-targeted genes and suppresses ROS
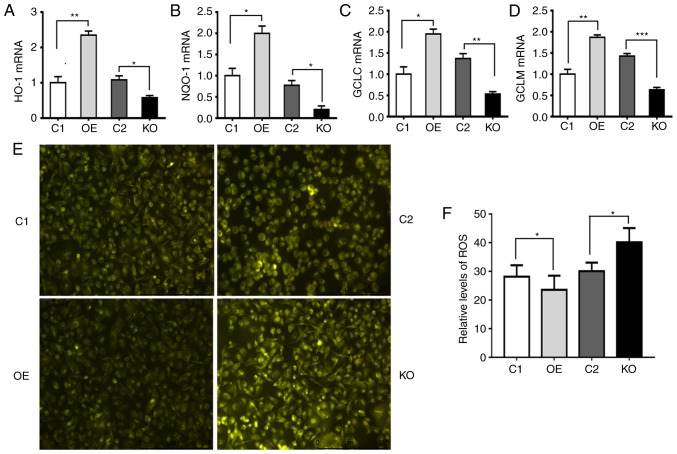
In order to investigate the mechanism by which p62 enhances proliferation, invasion and anti-apoptosis, RT-qPCR was used to assess the mRNA levels of Nrf2 target genes such as HO-1, NQO-1, GCLC and glutamate-cysteine ligase modifier subunit (GCLM). It was revealed that p62 increased the mRNA levels of Nrf2 target genes (Fig. 4A-D). Overexpression of p62 led to a decrease in ROS levels, while suppressing the expression of p62 induced an increase in ROS levels (Fig. 4E and F). Thus, p62 increased the expression of antioxidant-responding genes and suppressed ROS.
Figure 4.
p62 increases the expression of Nrf2-targeted genes and decreases the levels of ROS. C1, DU145 cells carrying the control plasmid vector; OE, DU145 cells overexpressing p62; KO, OE cells with p62 knocked out by CRISPER technology; and C2, OE cells carrying negative control for CRISPR knockout. (A-D) Plots revealing the mRNA levels of (A) HO-1, (B) NQO-1, (C) GCLC and (D) GLCM in C1, OE, C2 and KO cells. Values were normalized to GAPDH. (E and F) Representative (E) images and (F) quantification revealing the levels of cellular ROS reflected by the intensities of fluorescence in C1, OE, C2 and KO cells. Data represent the mean ± standard error of the mean of readings from >8 fields of views. *P<0.05, **P<0.01, ***P<0.001. Nrf2, nuclear factor erytheroid-derived 2-like 2; HO-1, heme oxygenase-1; NQO-1, NAD(P)H quinine oxidoreductase; GCLC, glutamate-cysteine ligase catalytic subunit; GLCM, glutamate-cysteine ligase modifier subunit; ROS, reactive oxygen species.
Nrf2 is degraded through the proteasomal system independent of the p62 levels but the autophagy system in the absence of p62
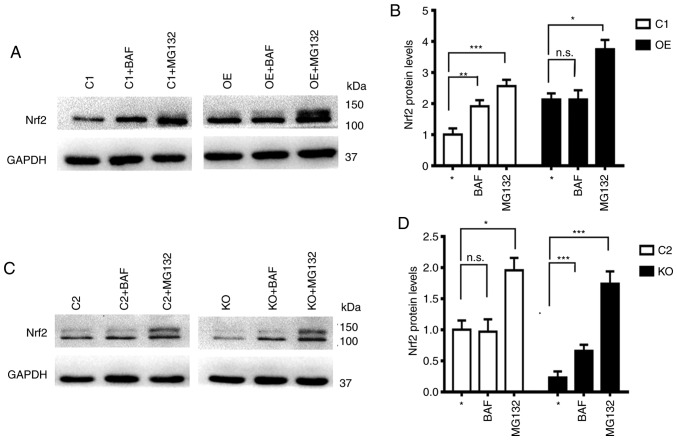
Numerous studies have demonstrated that Nrf2 protein is degraded through the a proteasomal system (13,24,25). It was revealed in the present study that the levels of Nrf2 in the presence of proteasomal inhibitor MG132 were always increased independent of the levels of p62, while levels of Nrf2 in the presence of autophagy inhibitor BAF were only increased in cells with low levels of p62 (C1 and KO), but remained constant in cells with high levels of p62 (C2 and OE) (Fig. 5). These results indicated that some portions of Nrf2 protein are always degraded through the proteasomal system but some portions of the Nrf2 protein that are modified differentially in the absence of p62 are degraded through the autophagy system.
Figure 5.
Nrf2 protein is always degraded through the proteasomal system but the autophagy system in the absence of p62. (A-D) Representative (A and C) immunoblot results and (B and D) quantification revealing the levels of Nrf2 in cells with different levels of p62 in the absence or presence of BAF or MG-132. The results represent three independent experiments. *P≤0.05, **P≤0.01 and ***P≤0.001. Nrf2, nuclear factor erytheroid-derived 2-like 2; BAF, bafilomycin.
p62 regulates Nrf2 through Keap1
In order to investigate the association between Keap1 and Nrf2, the expression of Keap1 in DU145 cells with p62 knocked down was suppressed. The levels of Nrf2 were increased (Figs. 2G and H and 6A). The suppression of Keap1 was confirmed in both mRNA (Fig. 6A) and protein levels (Fig. 6B and C). Such suppression caused a significant increase in the levels of Nrf2 protein (Fig. 6B and D) but had no impact on the levels of Nrf2 mRNA (Fig. 6E). Therefore, p62 increased the level of Nrf2 through Keap1 in a post-transcriptional manner.
Figure 6.
p62 regulates levels of Nrf2 through Keap1. (A) Plots revealing the mRNA levels of Keap1 in p62-deficient DU145 cells infected with negative control siRNA (Ctl siRNA) or Keap1-specific siRNA (Keap1 siRNA) for 48 h. The mRNA levels were normalized to GAPDH and presented as ratios relative to those in cells transfected with Ctl siRNA. Representative (B) immunoblots and plots revealing the levels of (C) Keap1 or (D) Nrf2 protein in the aforementioned cells in A. (E) Plots revealing the mRNA levels of Nrf2 in similarly-treated cells as aforementioned in A. The results represent three independent experiments. *P≤0.05 and **P≤0.01, n.s., not significant. Nrf2, nuclear factor erytheroid-derived 2-like 2; Keap1, Kelch-like ECH-associated protein 1.
Discussion
It has previously been demonstrated that p62 functions as an oncogene in 73% of PCa cases, particularly at the advanced stage of disease (26). However, the mechanism underlying the carcinogenicity of P62 remains unclear in PCa. The present study revealed new evidence that the p62/Keap1/Nrf2 pathway is a crucial regulator of PCa by transcriptional inhibition of genes involved in regulating ROS. p62 was positively correlated with total Nrf2, particularly nuclear Nrf2, and negatively correlated with Keap1. Notably, high levels of p62 were crucial for Nrf2 activity in association with cell proliferation, apoptosis and invasion. Inhibiting the p62/Keap1/Nrf2 pathway significantly suppressed PCa development both in vitro and in vivo.
There are multiple mechanisms that regulate the Keap1/Nrf2/ARE pathway. For example, p62, a substrate and an adaptor for selective autophagy (27) and a linker between autophagy and oxidative stress (15), could be involved in the regulation of this signaling pathway. Autophagy deficiencies cause the accumulation of p62 and the formation of aggregates (28). p62 blocks the interaction between Keap1 and Nrf2 and promotes nuclear translocation of Nrf2 to regulate a battery of genes encoding antioxidant proteins (15,29). Increased levels of both p62 and Nrf2 were observed in PCa cells and PCa tissues from PTEN−/− and TRAMP mice, while decreased levels of p62 led to the accumulation of Nrf2. Collectively, these studies provide evidence to suggest that the accumulation of p62 and prolonged activation of Nrf2 activity play a critical role in the pathogenesis of numerous human diseases including cancers.
The Keap1-Nrf2-ARE pathway is a key mechanism of cellular defense that helps maintain the homeostasis of cellular redox (30). The activation of the Keap1/Nrf2/ARE pathway leads to the activation of diverse antioxidant and phase II detoxifying enzymes, including NQO-1, HO-1, GCLC and GCLM, to cope with oxidative stress. It was reported that the suppression of p62 prevents UVA-induced production of ROS potentially as p62 induction disrupts mitochondrial function leading to increased production of ROS (31). Notably, it was revealed in the present study that decreasing the levels of p62 in DU145 cells resulted in an increase in basal levels of ROS, while high levels of p62 caused a decrease in ROS levels. The role of ROS in cancers has been under debate for decades. Recent studies have revealed that low levels of ROS induced the activation of pathways regulating proliferation and survival of cells (32), moderately increased levels of ROS caused DNA damage and promoted mutagenesis in cells (33), but high levels of ROS could ultimately cause cell senescence or death (34). It was speculated in the present study that the decrease in the levels of ROS caused by p62 overexpression through the Keap1/Nrf2/ARE pathway was more significant than the increase of ROS levels caused by p62 overexpression through the inhibition of the autophagy pathway and the induction of mitochondrial dysfunction. Therefore, p62 may induce low levels of ROS to promote the progression and survival pathways of tumor cells.
It has been reported that overexpressed Keap1 is degraded through autophagy (35) and the degradation of Keap1 depends on a direct physical interaction between Keap1 and p62 (13). In addition, it has been reported that Nrf2 is degraded through the proteasomal system (35,36). The present study confirmed that the degradation of Nrf2 protein is always through the proteasomal system. However, Nrf2 is degraded through the autophagy system in the absence of p62.
It was previously demonstrated that p62 enhances the epithelial-mesenchymal transition (EMT) and promotes development of PCa (22). The present study further revealed that p62 increased the rates of proliferation and invasion and decreased the rates of apoptosis of PCa cells through the p62/Keap1/Nrf2 pathway. Once the activity of Nrf2 was inhibited, the overexpression of p62 led to an enhancement of apoptosis and a decrease in the rates of cell proliferation and invasion. Therefore, Nrf2 was revealed to be a key mediator for p62 to exert its oncogenetic impact.
Epidemiological studies have attributed obesity as an important risk factor of PCa progression and poor outcome of those patients diagnosed, and one recent study demonstrated that the underlying molecular mechanism was associated with p62 activity in adipocytes (37,38). The deletion of p62 in adipocytes was revealed to induce EMT and promote PCA aggressiveness independent of the differences in locomotor activity and food intake in mice. It was previously reported that p62 enhanced EMT of PCa, and the present study further demonstrated that Nrf2 is a key regulator of such a phenomenon. Nrf2 is reported to be involved in obesity-induced oxidative stress and suggested to be targeted to decrease obesity-triggered oxidative stress damage (39). However, obesity has been characterized by significantly-activated oxidative stress and increased levels of autophagy flux, which have been demonstrated to be detrimental for the male reproductive system (40). p62 in combination with Nrf2 was revealed to be beneficial for sustaining anti-oxidation in adipocytes, while p62 deletion in adipocytes leads to an upregulation of oxidative stress that may cause EMT in PCa. Since Nrf2 is degraded through the autophagy system in the absence of p62, the degradation of Nrf2 is accordingly enhanced in p62-deficient adipocytes where autophagy is enhanced due to p62 inactivation. Collectively, these results contribute to the progression of PCa, such as EMT. It has previously been reported that high levels of p62 caused a promotion of EMT, in addition to an impairment of autophagy flux in PCa. The present study has further demonstrated that high levels of p62 suppressed ROS in PCa through the anti-oxidative Keap1/Nrf2/ARE pathway. This is consistent with a previous study which revealed that p62 and Nrf2 could cooperatively regulate antioxidants (41).
In conclusion, p62 interacted with Keap1 to prevent the degradation of Nrf2 through the proteasomal system. In the absence of p62, a portion of Nrf2 was delivered to the autophagy machinery to be degraded. p62 suppressed the Keap1/Nrf2/ARE system and induced low levels of ROS to promote PCa metastasis (Fig. 7). Therefore, p62 inhibitors or Nrf2 inhibitors may be utilized to precisely treat patients with PCa in the future.
Figure 7.
Diagram revealing the impact of p62 on the initiation and development of tumors in prostate tissues through the Keap1/Nrf2/ARE pathway. Keap1/Nrf2/ARE, Kelch-like ECH-associated protein 1/nuclear factor erytheroid-derived 2-like 2/antioxidant response element.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guangzhou Municipal Science and Technology Project (grant no. 1563000448), the National Natural Science Foundation of China (grant no. 81772931) and the National Cancer Institute of National Institutes of Health (grant no. CA142862).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
GJ, XL, YH, ZL, ZZ, ZS, ZF, YL, WY, TL, LH, FW and HH performed the experiments. GJ, XL and YH wrote the original draft of the manuscript. LL and XJ conceived, designed and directed the project and reviewed the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The collection of human prostate samples was approved by the Fifth Affiliated Hospital of Guangzhou Medical University, Sun Yat-sen Memorial Hospital, the Guangzhou First People's Hospital and the First Affiliated Hospital of Sun Yat-sen University in Guangzhou, China. Animal protocols were approved by the Animal Care and Use Committee of the Texas A&M Health Science Center at Houston (AUP IACUC 2015-0132-IBT).
Patient consent for publication
Not applicable.
Competing interests
The authors have declared that they have no competing interests.
References
- 1.DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, Jemal A. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016;66:290–308. doi: 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 2.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr, Saad F, et al. Abiraterone and increased survival in metastatic prostate cancer. New Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. New Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18:3069–3080. doi: 10.1128/MCB.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moscat J, Karin M, Diaz-Meco MT. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Gene Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T, Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Rad Biol Med. 2015;88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell MA, Hayes JD. The Keap1/Nrf2 pathway in health and disease: From the bench to the clinic. Biochem Soc Trans. 2015;43:687–689. doi: 10.1042/BST20150069. [DOI] [PubMed] [Google Scholar]
- 10.Hayes JD, Dinkova-Kostova AT. Oncogene-stimulated congestion at the KEAP1 stress signaling hub allows bypass of NRF2 and induction of NRF2-target genes that promote tumor survival. Can Cell. 2017;32:539–541. doi: 10.1016/j.ccell.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Bol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 13.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 15.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia M, Yu H, Gu S, Xu Y, Su J, Li H, Kang J, Cui M. p62/SQSTM1 is involved in cisplatin resistance in human ovarian cancer cells via the Keap1-Nrf2-ARE system. Intern J Oncol. 2014;45:2341–2348. doi: 10.3892/ijo.2014.2669. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang J, Huang ZH, Huang XH, Zheng WB, Yin XF, Li YL, Li B, He QY. Isodeoxyelephantopin induces protective autophagy in lung cancer cells via Nrf2-p62-keap1 feedback loop. Cell Death Dis. 2017;8:e2876. doi: 10.1038/cddis.2017.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Kim D, Wise JTF, Shi X, Zhang Z, DiPaola RS. p62 as a therapeutic target for inhibition of autophagy in prostate cancer. Prostate. 2018;78:390–400. doi: 10.1002/pros.23483. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X, Zhong W, Huang H, He H, Jiang F, Chen Y, Yue F, Zou J, Li X, He Y, et al. Autophagy defects suggested by low levels of autophagy activator MAP1S and high levels of autophagy inhibitor LRPPRC predict poor prognosis of prostate cancer patients. Mol Carcinog. 2015;54:1194–1204. doi: 10.1002/mc.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X, Li X, Huang H, Jiang F, Lin Z, He H, Chen Y, Yue F, Zou J, He Y, et al. Elevated levels of mitochondrion-associated autophagy inhibitor LRPPRC are associated with poor prognosis in patients with prostate cancer. Cancer. 2014;120:1228–1236. doi: 10.1002/cncr.28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X, Huang Y, Liang X, Jiang F, He Y, Li T, Xu G, Zhao H, Yang W, Jiang G, et al. Metastatic prostate cancer-associated P62 inhibits autophagy flux and promotes epithelial to mesenchymal transition by sustaining the level of HDAC6. Prostate. 2018;78:426–434. doi: 10.1002/pros.23487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Gene Cell. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 26.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Can Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Moscat J, Diaz-Meco MT. p62: A versatile multitasker takes on cancer. Trend Biochem Sci. 2012;37:230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 29.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sample A, Zhao B, Wu C, Qian S, Shi X, Aplin A, He YY. The autophagy receptor adaptor p62 is Up-regulated by UVA radiation in melanocytes and in melanoma cells. Photochem Photobiol. 2018;94:432–437. doi: 10.1111/php.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: Role in inflammatory disease and progression to cancer. Biochem J. 1996;313:17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 35.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proce Nat Acad Sci USA. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerr F, Sofola-Adesakin O, Ivanov DK, Gatliff J, Gomez Perez-Nievas B, Bertrand HC, Martinez P, Callard R, Snoeren I, Cocheme HM, et al. Direct Keap1-Nrf2 disruption as a potential therapeutic target for Alzheimer's disease. PLoS Genet. 2017;13:e1006593. doi: 10.1371/journal.pgen.1006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metabol. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Huang J, Duran A, Reina-Campos M, Valencia T, Castilla EA, Muller TD, Tschop MH, Moscat J, Diaz-Meco MT. Adipocyte p62/SQSTM1 suppresses tumorigenesis through opposite regulations of metabolism in adipose tissue and tumor. Can Cell. 2018;33:770–784.e776. doi: 10.1016/j.ccell.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huo L, Su Y, Xu G, Zhai L, Zhao J. Sulforaphane protects the male reproductive system of mice from obesity-induced damage: Involvement of oxidative stress and autophagy. Intern J Environ Res Public Health. 2019;16(pii):E3759. doi: 10.3390/ijerph16193759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metabol. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Yamada M, Iwata M, Warabi E, Oishi H, Lira VA, Okutsu M. p62/SQSTM1 and Nrf2 are essential for exercise-mediated enhancement of antioxidant protein expression in oxidative muscle. FASEB J. 2019;33:8022–8032. doi: 10.1096/fj.201900133R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.