Abstract
Background. The diagnosis of severe acute respiratory syndrome (SARS) is difficult early in the illness, because its presentation resembles that of other nonspecific viral fevers, such as dengue. Dengue fever is endemic in many of the countries in which the large SARS outbreaks occurred in early 2003. Misdiagnosis may have serious public health consequences. We aimed to determine simple laboratory features to differentiate SARS from dengue.
Methods. We compared the laboratory features of 55 adult patients with SARS at presentation (who were all admitted before radiological changes had occurred) and 147 patients with dengue. Features independently predictive of dengue were modeled by multivariate logistic regression to create a diagnostic tool with 100% specificity for dengue.
Results. Multivariate analysis identified 3 laboratory features that together are highly predictive of a diagnosis of dengue and able to rule out the possibility of SARS: platelet count of <140 × 109 platelets/L, white blood cell count of <5 × 109cells/L, and aspartate aminotransferase level of >34 IU/L. A combination of these parameters has a sensitivity of 75% and a specificity of 100%.
Conclusions. Simple laboratory data may be helpful for the diagnosis of disease in adults admitted because of fever in areas in which dengue is endemic when the diagnosis of SARS needs to be excluded. Application of this information may help to optimize the use of isolation rooms for patients presenting with nonspecific fever.
An outbreak of severe acute respiratory syndrome (SARS) with rapid international spread occurred from November 2002 until June 2003. Prompt isolation was the mainstay of public health efforts in containing the epidemic, and early diagnosis is therefore paramount. Detection in the first few days of illness relies on clinical acumen combined with a positive contact or travel history. However, the initial symptoms of SARS—commonly, fever, myalgia, and malaise—are nonspecific and similar to those of other viral illnesses. Respiratory symptoms typically do not begin until 2–7 days after onset of illness [1–4]. Chest radiographs may appear to be normal in up to 30% of patients with the clinical diagnosis of SARS at the time when first evaluated [5, 6].
Dengue fever is endemic in many of the countries in which the large SARS outbreaks occurred in early 2003, in particular Singapore [7, 8], Vietnam [9, 10], and South China [11]. Symptoms of dengue fever are nonspecific. A maculopapular rash may appear, but often only at the time of defervescence (day 4–7 of illness), and is absent in >50% of cases [12–14]. Respiratory symptoms may also occur [14]. In the early stages of illness, clinical diagnosis of dengue includes the typical laboratory features, such as thrombocytopenia and lymphopenia—features that have also been reported in patients with SARS, even during the early phases [3, 15]. Dengue may therefore be hard to differentiate from SARS on clinical grounds.
Specific diagnostic tests also have limited use: commonly available diagnostic tests for dengue, such as detection of IgM antibody, do not yield positive results until 4–5 days after onset of illness [16]. SARS coronavirus serology may not accurately detect SARS coronavirus until >28 days after onset of symptoms [17]. A positive test result earlier is reliable, but a negative test result earlier is not definitive. Although quantitative real-time RT-PCR technologies have improved the diagnosis of early SARS, sensitivity in the first week of illness remains poor [18].
Dengue therefore poses a diagnostic dilemma in countries with concurrent outbreaks of SARS or in travelers returning from such countries. Misdiagnosing SARS as dengue may have serious public health consequences, whereas misdiagnosing dengue as SARS will lead to unnecessary alarm, isolation, contact tracing, and quarantine measures. In Singapore, an island state in Southeast Asia where dengue is endemic [7, 19], misclassification of 1 of the patients with SARS as having dengue (and therefore not isolating that patient) led to extensive secondary transmission [20]. The aim of this study was to determine whether laboratory tests, either singly or in combination, could assist in the differentiation of early stages of SARS from dengue.
Methods
Patients. For patients with SARS, we conducted a retrospective case sheet review of the 206 patients with probable SARS, as notified to the Ministry of Health during the period of 14 March through 31 May 2003 on the basis of World Health Organization criteria. We selected those who were admitted to Tan Tock Seng Hospital (the designated SARS hospital) early in the disease before the occurrence of radiological changes and who later developed radiological changes and were confirmed to have SARS either by a positive result of a serological test or a positive result of PCR.
For patients with dengue, we used the data obtained from a prospective study of patients with dengue (confirmed by positive result for dengue IgM by EIA; PanBio) [7, 21] admitted to the same institution during the period of October 1997 through May 2000, by use of standardized forms for data collection.
Data. Age, sex, day of illness at admission (day 1 of illness was counted as the day of onset of fever), and hematologic and biochemical laboratory parameters at presentation were recorded. Only laboratory parameters obtained at the time of admission, before initiation of therapy, were analyzed. We classified laboratory parameters into normal or abnormal, with cutoffs derived from normal laboratory ranges of previously published studies of dengue [12, 22]. The clinical data collection from patients with dengue and SARS was approved by the Tan Tock Seng Hospital Ethics Committee.
Statistical analysis. In the univariate analysis, we compared the mean laboratory values for patients with dengue and with SARS by use of the independent Student's t test. For data that were not normally distributed, we used a natural logarithmic transformation to normalize the distribution. We used Fisher's exact test and/or χ2test to compare the proportions of patients with abnormal readings between the 2 groups of patients.
We then performed a multivariate analysis by use of logistic regression models to determine the predicting and thus distinguishing parameters between patients with SARS and those with dengue. Starting from the most significant categorical predictor identified in the univariate analysis, we used the log-likelihood ratio test to see whether inclusion of a new covariate helped improve the fit of the multivariate model. Then, on the basis of the final model selected, the area under the curve from the receiver operating characteristic curve was used to determine the model's ability to discriminate between SARS and dengue. ORs and their 95% CIs were provided as estimates of the effect sizes. Those parameters identified as independent predictors by multivariate analysis were used for the predicted probability equation. The predicted probability (pi) of dengue was calculated with the following equation: ln [pi/(1 − pi)] = −8.0 + 6.1xi1 + 3.8xi2 + 4.2xi3, for the ith individual; and x1, x2, and x3 are indicator variables taking on the values of 1 for abnormal and 0 for normal variables. By use of the equation for predicted probability, we calculated the sensitivity (i.e., ability to identify dengue) and specificity (i.e., ability to rule out SARS) of the combination of independent laboratory predictors.
Data analysis was done using Stata software, version 7.0 (Stata). The level of significance was set at 5%.
Results
We identified a total of 55 patients with probable SARS who met the selection criteria. The mean age was 35.4 years (range, 14–72 years), and 15 (27.3%) were male. They were admitted a median of 3 days (range, 1–7 days) after onset of fever.
Data for 147 patients with dengue were included for comparative analysis. The mean age was 29.5 years (range, 15–71 years), and 116 subjects (78.9%) were male. They were admitted a median of 4 days (range, 2–8 days) after onset of fever.
The mean laboratory parameters at the time of hospital admission in patients with SARS and dengue are presented in table 2. Patients in both groups had low mean lymphocyte counts, but patients with dengue had significantly lower mean values for WBC, neutrophil, and platelet counts. Patients with dengue had significantly higher mean values for hemoglobin, hematocrit, alanine aminotransferase (ALT), aspartate aminotransferase (AST), urea, protein, and bilirubin than did patients with SARS. There were no differences in mean values of clotting profile or levels of sodium, potassium, creatinine, albumin, and alkaline phosphatase.
Table 2.
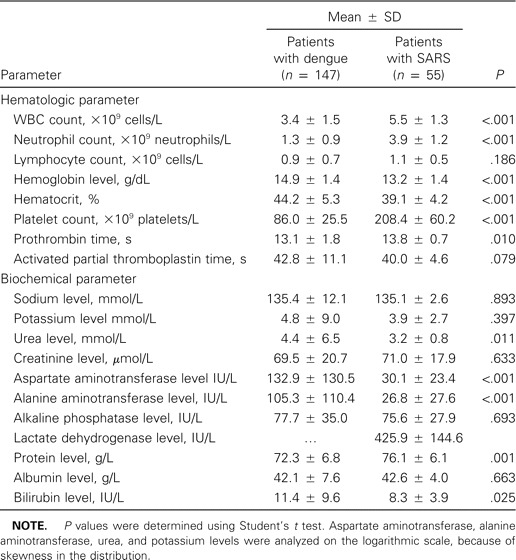
Summary of laboratory parameters on admission for patients with dengue fever or with severe acute respiratory syndrome (SARS).
The proportion of patients with abnormal laboratory findings is shown in table 1. All patients with SARS and dengue had lymphopenia. A significantly higher proportion of patients with dengue fever had elevated liver transaminase levels, with increased AST and ALT levels, and reduced platelet, leukocyte, and neutrophil counts, whereas a significantly higher proportion of patients with SARS had normal leukocyte and neutrophil counts.
Table 1.
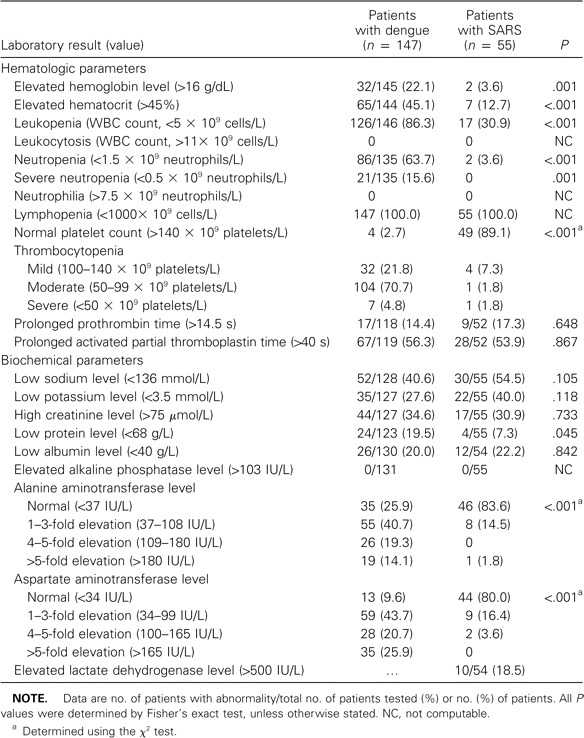
Abnormal laboratory results at hospital admission for patients with dengue fever and patients with severe acute respiratory syndrome (SARS).
Multivariate analysis showed that the only independent discriminating laboratory parameters between SARS and dengue were a platelet count of <140 × 109 platelets/L, a WBC count of <5 × 109 cells/L, and an AST level of >34 IU/L (table 3). The OR of these 3 parameters to predict dengue versus SARS was high (table 3), and the area under the curve was 0.99 (95% CI, 0.98–1.00), thus indicating that the model had a very good ability to discriminate between SARS and dengue. We performed a power calculation to see whether the calculations were adequately powered. For each of the variables in the multivariate analysis, we found that the study had a power exceeding 99% to detect any statistical significance, in terms of testing for differences in proportion.
Table 3.
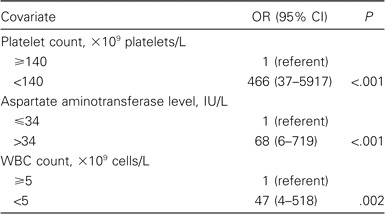
Multivariate predictors of dengue versus severe acute respiratory syndrome.
By use of the equation for predicted probability, the sensitivity and specificity of the diagnosis of dengue for a patient with a combination of these laboratory parameters (platelet count of <140 × 109 platelets/L, a WBC count of <5 × 109 cells/L, and an AST level of >34 IU/L) were 75.4% and 100.0%, respectively.
Discussion
The initial SARS epidemic was effectively contained worldwide, but sporadic cases continue to occur, and there is potential for new epidemics. The current focus is on ruling out SARS in patients presenting with undifferentiated fever—in particular, in travelers from countries previously affected with SARS. For patients without evidence of pneumonia at the initial evaluation, serial observations over time may be needed to identify those for whom isolation precautions can be discontinued safely [23]. Resolution of symptoms and lack of development of radiographic evidence of pneumonia by week 2 of illness argue against the diagnosis of SARS [17]. However, isolating all patients with fever of unknown origin over an extended period of time will overburden any health care system. This study has shown that, with a constellation of basic hematologic and biochemical laboratory parameters, it is possible to differentiate SARS from dengue fever, one of the common differential diagnostic dilemmas in early SARS in Asia.
We identified 3 laboratory parameters independently predictive of distinction between dengue and early SARS: a low platelet count, low WBC count, and elevated AST level. For the analysis, we elected to use a cutoff of <140 × 109 platelets/L for platelet count, because this was the cutoff used in a study of hematologic manifestations in patients with SARS [22], and this cutoff was also shown to be predictive in distinguishing dengue from other infectious diseases in Asia [12]. A WBC count of <5 × 109 cells/L has been reported to have a high positive predictive value for dengue fever in patients presenting with febrile illnesses [12, 24]. The cutoff of 34 IU/L for elevated AST level was based on the upper limit of the normal range in our laboratory. A model with the combination of these 3 parameters and using these cutoff values identifies dengue correctly in 75% (sensitivity of 75%) and rules out dengue in 100% (specificity of 100%) of cases. From a public health point of view, a specificity of 100% is desirable so that no case of SARS will be misdiagnosed as dengue, and thus, the patient not be isolated, leading to secondary transmission. With a sensitivity of 75%, still ∼25% of patients with dengue will need to be isolated, until the results of dengue-specific tests become available.
Our findings also highlight laboratory features that are common in both dengue and SARS and therefore not useful in distinguishing between these 2 viral diseases. Lymphopenia is characteristic and of similar magnitude for both dengue and SARS. However, lymphopenia in dengue was also associated with depletion of total WBC count and neutrophil count, whereas for SARS, it was associated with normal total WBC count and normal-to-high neutrophil count. Neutrophilia in patients with SARS has been described in several case series but may be due to steroid treatment or secondary bacterial infection [15, 22]. This was not seen in our patients, most likely because the values were taken before the institution of any therapy. Elevated lactate dehydrogenase level is a predictor for poor prognosis in SARS [15], but the majority of patients with SARS admitted early in the course of disease had normal values. Although we did not have data for the cohort of patients with dengue, lactate dehydrogenase levels have been reported to be moderately elevated in patients with dengue [25]; therefore, this parameter is unlikely to discriminate between dengue and SARS.
Our study has a number of limitations. The sample size was small, although we were still able to construct a highly discriminatory model, adequately powered at >90% for all variables. However, it would be useful to confirm the findings with data sets from other areas affected by SARS. The study used SARS data from an outbreak, but these may differ in a nonoutbreak situation. We did not include epidemiological factors, but such data will provide important additional information in differentiating dengue from early SARS—in particular, a history of close contact with a patient with laboratory-confirmed SARS or hospital staff during a nosocomial SARS outbreak. We did not consider other viral infections in which the patient may present with lymphopenia [26]. However, dengue is the most common and most difficult viral infection to distinguish from SARS in our setting and in that of many tropical countries.
This study suggests that a simple model of laboratory data may be helpful to differentiate dengue from SARS. Application of this information together with detailed epidemiological clues may help to avoid misdiagnosis of SARS as dengue fever, shorten the time of isolation of patients with fever until full diagnostic evaluation is completed, and optimize the use of isolation rooms and expenses for diagnostic tests. This is of particular importance in low-resource settings in which more elaborate early diagnostic tests for dengue, such as PCR or direct virus isolation, are not readily available.
Acknowledgments
We thank staff from Infectious Diseases Research Centre for assistance with obtaining data for use in this study.
Financial support. The prospective study on patients with dengue was financially supported by the Tan Tock Seng Hospital Health Endowment Fund.
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289:2801–9. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 3.Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 4.Tsang KW, Ho PL, Ooi GC, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–85. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 5.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong KT, Antonio GE, Hui DS, et al. Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology. 2003;228:406–6. doi: 10.1148/radiol.2282030593. [DOI] [PubMed] [Google Scholar]
- 7.Wilder-Smith A, Foo W, Earnest A, Sremulanathan S, Paton NI. Seroepidemiology of dengue in the adult population of Singapore. Trop Med Int Health. 2004;9:305–8. doi: 10.1046/j.1365-3156.2003.01177.x. [DOI] [PubMed] [Google Scholar]
- 8.Goh KT. Dengue—a re-emerging infectious disease in Singapore. Ann Acad Med Singapore. 1997;26:664–70. [PubMed] [Google Scholar]
- 9.Bartley LM, Carabin H, Vinh Chau N, et al. Assessment of the factors associated with flavivirus seroprevalence in a population in Southern Vietnam. Epidemiol Infect. 2002;128:213–20. doi: 10.1017/s0950268801006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran KT, Vazeille-Falcoz M, Mousson L, et al. Aedes aegypti in Ho Chi Minh City (Viet Nam): susceptibility to dengue 2 virus and genetic differentiation. Trans R Soc Trop Med Hyg. 1999;93:581–6. doi: 10.1016/s0035-9203(99)90056-1. [DOI] [PubMed] [Google Scholar]
- 11.Qiu FX, Gubler DJ, Liu JC, Chen QQ. Dengue in China: a clinical review. Bull World Health Organ. 1993;71:349–59. [PMC free article] [PubMed] [Google Scholar]
- 12.Watt G, Jongsakul K, Chouriyagune C, Paris R. Differentiating dengue virus infection from scrub typhus in Thai adults with fever. Am J Trop Med Hyg. 2003;68:536–8. doi: 10.4269/ajtmh.2003.68.536. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz E, Mendelson E, Sidi Y. Dengue fever among travelers. Am J Med. 1996;101:516–20. doi: 10.1016/s0002-9343(96)00278-1. [DOI] [PubMed] [Google Scholar]
- 14.Jelinek T, Muhlberger N, Harms G, et al. Epidemiology and clinical features of imported dengue fever in Europe: sentinel surveillance data from TropNetEurop. Clin Infect Dis. 2002;35:1047–52. doi: 10.1086/342906. [DOI] [PubMed] [Google Scholar]
- 15.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–94. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz E, Mileguir F, Grossman Z, Mendelson E. Evaluation of ELISA-based sero-diagnosis of dengue fever in travelers. J Clin Virol. 2000;19:169–73. doi: 10.1016/s1386-6532(00)00114-1. [DOI] [PubMed] [Google Scholar]
- 17.Jernigan JA-DE, Low DE, Helfand RF. Combining clinical and epidemiologic features for early recognition of SARS. Emerg Infect Dis. 2004;10:327–33. doi: 10.3201/eid1002.030741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon LL, Chan KH, Wong OK, et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. 2003;28:233–8. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ooi EE, Hart TJ, Tan HC, Chan SH. Dengue seroepidemiology in Singapore. Lancet. 2001;357:685–6. doi: 10.1016/S0140-6736(00)04137-4. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. Severe acute respiratory syndrome—Singapore, 2003. MMWR Morb Mortal Wkly Rep. 2003;52:405–11. [PubMed] [Google Scholar]
- 21.Cuzzubbo AVD, Nisalak A, McBride J, Aaskov J, Devine P. Commercial assays for the serological diagnosis of dengue infections. In: Kay BH, Brown MD, Aaskov JG, editors. Arbovirus research in Australia, papers from the Seventh Arbovirus Research in Australia Symposium and the Second Mosquito Control Association of Australia Conference, Surfers Paradise. Vol. 7. Brisbane, Australia: Queensland Institute of Medical Research; 1997. pp. 25–9. [Google Scholar]
- 22.Wong RS, Wu A, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–62. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainer TH, Cameron PA, Smit D, et al. Evaluation of WHO criteria for identifying patients with severe acute respiratory syndrome out of hospital: prospective observational study. BMJ. 2003;326:1354–8. doi: 10.1136/bmj.326.7403.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalayanarooj S, Vaughn DW, Nimmannitya S, et al. Early clinical and laboratory indicators of acute dengue illness. J Infect Dis. 1997;176:313–21. doi: 10.1086/514047. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz E, Mendelson E, Sidi Y. Dengue fever among travelers. Am J Med. 1996;101:516–20. doi: 10.1016/s0002-9343(96)00278-1. [DOI] [PubMed] [Google Scholar]
- 26.Tran TH, Nguyen TL, Nguyen TD, et al. Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med. 2004;350:1179–88. doi: 10.1056/NEJMoa040419. [DOI] [PubMed] [Google Scholar]


