Abstract
The mechanism of systemic spread of H5N1 virus in patients with avian influenza is unknown. Here, H5N1 nucleoprotein and hemagglutinin were identified by immunohistochemistry in the nucleus and cytoplasm of neutrophils in the placental blood of a pregnant woman. Viral RNA was detected in neutrophils by in situ hybridization and enhanced real-time polymerase chain reaction. Therefore, neutrophils may serve as a vehicle for viral replication and transportation in avian influenza.
Human avian influenza (H5N1) infection possesses a great potential threat to human health. However, the pathogenesis of this emerging disease is poorly understood. A previous study demonstrated that extrapulmonary organs, including the placenta, trachea, intestine, and brain, can be infected with human avian influenza H5N1 virus [1]. This virus has been detected in serum [2], but the exact means of viral spread beyond the respiratory system is unclear.
In the present study, we investigated organs obtained at autopsy from an H5N1 virus–infected pregnant woman and from her fetus in an attempt to study the mechanism of systemic dissemination of H5N1 virus. Neutrophils were abundant in the placenta, and therefore, we evaluated blood cells in the placental villi obtained at autopsy to determine whether neutrophils were infected by H5N1 virus.
Materials and methods. Clinical information about the 24-year-old pregnant woman (with a fetus at 4 months gestation) who was investigated in this study is detailed elsewhere [3]. H5N1 virus–infected autopsy tissue samples were fixed in 4% formalin, were embedded in paraffin, and were sectioned at 4 µm. Two H5N1 virus–positive control tissue samples were used. One was a lung tissue sample from the patient, and the other was a brain tissue sample from an H5N1 virus–infected black-headed gull. In addition, in a previous study of the same placenta by our group [1], we observed H5N1 virus–infected macrophages and trophoblastic cells. Therefore, cells from the same placenta were used as additional internal positive control samples when we attempted to determine whether the neutrophils were infected in the same tissue sections [1]. Three H5N1 virus–uninfected placental samples were used as negative control samples. This investigation was performed in accordance with the Helsinki Declaration, and the procedures were approved by the Research Administrative Committee of Peking University (Beijing, China).
Immunohistochemistry was performed as described elsewhere [4]. Monoclonal antibodies against nucleoprotein (NP) and hemagglutinin (HA) were used to identify H5N1 virus–specific NP and HA, respectively. Monoclonal antibodies against cell markers of CD15 (neutrophils), CD20 (B lymphocytes), CD3 (T lymphocytes), and CD68 (macrophages) were used to identify specific cell types, at a dilution of 1:100. Secondary antibodies against the primary antibodies were labeled with horseradish peroxidase, and the color reaction was developed with a horseradish peroxidase reaction kit (3-amino-9-ethylcarbazole; Zymed Laboratories), which resulted in a red signal. When double labeling was performed, alkaline phosphatase–labeled secondary antibodies were used to identify the primary antibodies against NP, and the color reaction was developed with nitro blue tetrazolium/5-bromo-4-choloro-3-indolyl phosphate (Promega), which resulted in a purple-blue signal. Omission of the primary antibodies was also used as a negative control test.
In situ hybridization was performed as described elsewhere [1]. Specific probes against NP and HA RNA sequences were used [1]. The color reaction was developed with nitro blue tetrazolium/5-bromo-4-choloro-3-indolyl phosphate (Promega), which resulted in a purple-blue signal. Positive and negative control tissue samples were identical to those used for immunohistochemistry.
Double labeling was performed to identify cell types with CD15 as a marker for neutrophils. In this case, we used purple-blue for NP signals and red for CD15 signals, to achieve a good contrast to best illuminate the colocalization of the H5N1 virus and the cell marker. In brief, immunohistochemistry or in situ hybridization to detect NP antigen or NP RNA sequence was performed first, and the color reaction was developed to give a purple-blue signal. The sections were then incubated with monoclonal antibodies against CD15 (neutrophil marker), and the color reaction was developed to give a red signal. Additional control reactions were performed to ascertain the specificity of the double labeling, as described elsewhere [1].
Laser microdissection is a visual microdissection technique used under a light microscope to microdissect and collect tissue samples from areas of interest in a tissue section, using the energy of a laser beam. In this case, accumulations of neutrophils in the blood pool in the placental tissue sections were dissected to collect pure populations of neutrophils for further analysis of an H5-specific sequence in those cells by RT-PCR. Laser microdissection was performed, according to the protocol of a Leica Microdissection System, on 8-µm–thick tissue sections prepared on membrane-coated glass slides. Leukocytes dissected from an uninfected placenta served as a negative control group. To establish that viral fragments were originated from leukocytes and not from other elements in the serum, areas of blood pooling that were adjacent to placental villi without leukocytes were microdissected as a blank group from the same tissue sections as those from which the additional negative control groups were obtained.
Enhanced real-time PCR has an increased detecting sensitivity, compared with conventional TaqMan real-time PCR, because of the addition of a target gene preamplification step to precede TaqMan real-time fluorescent PCR [5, 6]. RNA extractions from the test group, the negative control group, and the blank group were performed according to the methods of Masuda et al. [7]. Samples were homogenized in digestion buffer with proteinase K (Merck KGaA). Total RNA was prepared with Trizol (Invitrogen) and was incubated in Tris-EDTA buffer (pH, 7.5) at 70°C for 60 min. The HA gene of H5N1 virus was detected by enhanced real-time PCR, as described elsewhere [6, 7]. The amplification product from the first round of PCR was used as the template for a second round of TaqMan real-time PCR, with the same set of primers (forward primer, 5′-gTg AYA ATg AAT gYA Tgg AA-3′; reverse primer, 5′-CCA IAA AgA YAg ACC AgC TA-3′; probe, FAM-AGT-A-A-A-A-T-T-G-G-A-A-T-C-AATRGGAAT-BHQ).
Results. A subset of leukocytes in placental tissue sections showed positive findings of immunostaining with the HA and NP antibodies, and these cells proved to be neutrophils. First, these cells showed characteristic neutrophil morphology (figure 1A), with multilobed nuclei (figure 1B), demonstrated by secondary hematoxylin and eosin staining of destained immunostained sections. In addition, immunohistochemistry double labeling with antibody to CD15 (neutrophil marker) revealed cellular colocalization with NP. These NP-positive cells were thereby unequivocally established as neutrophils (figure 1C), and the H5N1 virus infection rate among these CD15-positive cells was ∼30%. Immunopositive neutrophils were also observed in the spleen (data not shown).
Figure 1.
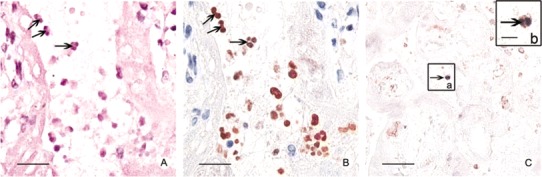
Nucleoprotein (NP) and hemagglutinin detected in neutrophils. A, Hematoxylin and eosin staining after destaining of the positive immunostaining response performed to identify neutrophils (black arrows; bar, 20 µm). B, Immunohistochemistry with antibody against NP (1:300; Abcam) performed to identify the viral proteins in the nuclei of neutrophils (brown-red; black arrows; bar, 20 µm). C, CD15 (brown-red, black arrow)also present in the NP-positive cells (purple-blue, black arrow; bar, 50 µm). Inset b shows enlargement of a cell highlighted by an arrow in inset a, clearly indicating colocalization of NP (viral protein) and CD15 (neutrophil marker) in the same cells (bar, 10 µm).
With the technique of in situ hybridization, H5N1 nucleotide sequences were detected in putative neutrophils. Double labeling of CD15 and viral RNA colocalized them into the same cells, further establishing that these H5N1 virus RNA–containing cells were neutrophils (figure 2A). Positive signals were also detected in cells of the placenta villi; these cells were previously demonstrated to be macrophages and cytotrophoblastic cells (figure 2A) [1].
Figure 2.
H5N1 nucleotide sequences detected in neutrophils by in situ hybridization, microdissection, and enhanced real-time PCR (ERT-PCR). A, Positive in situ hybridization signals of H5N1 (purple-blue; black arrows) detected in CD15-positive cells (red-brown; black arrows) in placental villi (inset a). Macrophages and trophoblastic cells in placental villi were also positive (purple-blue; red arrows; bar, 50 µm). Inset b shows enlargement of inset a, which shows that the same neutrophils contain H5N1 nucleotide sequences (bar, 10 µm). B and C, Three thousand leukocytes, dissected from the infected placenta as the test group. B, Before laser microdissection (bar, 50µm). C, After laser microdissection (bar, 50µm). D and E, Blood preparations with no leukocytes, dissected from the infected placenta as the blank group. D, Before laser microdissection (bar, 50 µm). E, After laser microdissection (bar, 50 µm). Three thousand leukocytes from uninfected placenta were also dissected as a negative control (data not shown). F, Measurement of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) by reamplification of PCR. Lane 1, GAPDH PCR of the positive control group. PCR reamplification of GAPDH of the positive control group (lane 2), on the test group (lane 3), of the negative control group (lane 4), and of the blank group (lane 5). Lane 6, GAPDH PCR of water. Lane 7, PCR reamplification of GAPDH of water. G, Measurements of H5 ERT-PCR for different samples. 1, ERT-PCR measurement of the H5-positive control group. 2, Real-time PCR measurement of the H5-positive control group. ERT-PCR measurement of the test group (3), of the blank group (4), and of the negative control group (5). 6, Real-time PCR measurement of water. 7, ERT-PCR measurement of water. bp, Base pairs.
Laser microdissection and enhanced real-time PCR further confirmed the presence of H5N1 viral sequences in the cell pool of neutrophils (figure 2G) collected from the H5N1 virus–infected placenta (figure2B and 2C). Areas of blood pooling that were adjacent to placental villi without leukocytes (figure 2D and 2E) showed no H5N1 virus RNA signal (figure 2G). Glyceraldehyde 3-phosphate dehydrogenase served as an internal control for confirmation of RNA extraction when results were convincingly positive (figure 2F).
Discussion. There is no known case of neutrophil infection with virus in human influenza in vivo, and the result of our study represents, to our knowledge, the first observation of neutrophil infection due to H5N1 virus in a patient with human avian influenza. In our study, we found that H5N1 viral proteins and nucleotide sequences localized in placental neutrophils and that H5 sequences localized in leukocytes among placenta villi from an H5N1 virus–infected patient. Positive signals were present in both the cytoplasm and the nucleus. NP- and HA-immunopositive neutrophils observed in the spleen indicate that this phenomenon probably occurs throughout the circulation.
One important function of neutrophils is phagocytosis [8]. In vitro, it has been reported that human influenza virus could be engulfed by phagocytosis of neutrophils [9]. Viral hepatitis DNA was detected in neutrophils [10–12], and other viruses could be phagocytized by neutrophils [13–15]. Therefore, H5N1 virus might enter neutrophils via phagocytosis in a similar manner. Recently, we reported the presence of avian influenza receptors on the surface of a number of human cell types, including neutrophils [16]; therefore, the H5N1 virus might enter neutrophils via receptor-mediated endocytosis. Several viruses, including cytomegalovirus and Epstein-Barr virus, were reported to replicate in neutrophils [17–19]. The nuclear localization of viral protein in our study suggests that H5N1 virus might also be capable of replicating in neutrophils.
The unequivocal evidence of H5N1 viral proteins and nucleotide sequences in the nuclei and cytoplasm of neutrophils of patients with avian influenza indicates novel mechanisms of pathogenesis. These cells may serve as a viral carrier in systemic circulation and may cause multiple organ infection, as was reported elsewhere [1]. It was reported that H5N1 virus could replicate in dendritic cells and reduce their survival rate [20]. In addition, it was observed that numerous leukocytes were apoptotic in the lungs of an H5N1 virus–infected patient [21]. Therefore, H5N1 virus may accelerate the destruction of neutrophils by direct killing or apoptosis and may cause the neutropenia that is frequently observed in H5N1 virus infection. Because our study is derived from examination of a unique case of pregnancy, the universality of this discovery warrants further investigation.
Acknowledgments
We thank Yong Guo, Zhengshan Chen, Peng Pan, and Baokai Yang, for their assistance in performing these experiments, and Zhigang Xie, for his assistance in autopsy and tissue collection.
Financial support. The International Collaboration Project (Project 111; B 07001) of the Ministry of Education, China; the Lifu Educational Foundation; and the National High Technology Research and Development Program of China (863 Program; 2006AA02Z452 to A.C.H.Y.).
Potential conflicts of interest. All authors: no conflicts.
Footnotes
Y.Z. and M.L. share equal responsibility as first author.
References
- 1.Gu J, Xie Z, Gao Z, et al. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet. 2007;370:1137–45. doi: 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchy P, Mardy S, Vong S, et al. Influenza A/H5N1 virus infection in humans in Cambodia. J Clin Virol. 2007;39:164–8. doi: 10.1016/j.jcv.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Li Q, Lan Y, Xu CL, et al. Study on a fatal pregnant woman died from by avian influenza (H5N1) [in Chinese] Zhonghua Liu Xing Bing Xue Za Zhi. 2006;27:288–92. [PubMed] [Google Scholar]
- 4.Lin Y, Shen X, Yang RF, et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141–5. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau LT, Fung YW, Wong FP, et al. A real-time PCR for SARS-coronavirus incorporating target gene pre-amplification. Biochem Biophys Res Commun. 2003;312:1290–6. doi: 10.1016/j.bbrc.2003.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu AC, Lau LT, Fung YW. Boosting the sensitivity of real-time polymerase-chain-reaction testing for SARS. N Engl J Med. 2004;350:1577–9. doi: 10.1056/NEJM200404083501523. [DOI] [PubMed] [Google Scholar]
- 7.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–43. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertins SD, Ortona L, Cauda R. Role of polymorphonuclear leukocytes in infection by retroviruses with emphasis on the human immunodeficiency virus. Viral Immunol. 1990;3:173–94. doi: 10.1089/vim.1990.3.173. [DOI] [PubMed] [Google Scholar]
- 9.Ratcliffe D, Migliorisi G, Cramer E. Translocation of influenza virus by migrating neutrophils. Cell Mol Biol. 1992;38:63–70. [PubMed] [Google Scholar]
- 10.Lerat H, Rumin S, Habersetzer F, et al. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood. 1998;91:3841–9. [PubMed] [Google Scholar]
- 11.Catterall AP, Murray-Lyon IM, Zuckerman AJ, Harrison TJ. Southern hybridisation analysis of HBV DNA in peripheral blood leucocytes and of different cell types: changes during the natural history and with interferon-α therapy in patients with hepatitis B virus infection. J Med Virol. 1994;43:269–75. doi: 10.1002/jmv.1890430314. [DOI] [PubMed] [Google Scholar]
- 12.Hoar DI, Bowen T, Matheson D, Poon MC. Hepatitis B virus DNA is enriched in polymorphonuclear leukocytes. Blood. 1985;66:1251–3. [PubMed] [Google Scholar]
- 13.Orenstein JM. In vivo cytolysis and fusion of human immunodeficiency virus type 1-infected lymphocytes in lymphoid tissue. J Infect Dis. 2000;182:338–42. doi: 10.1086/315640. [DOI] [PubMed] [Google Scholar]
- 14.Ratcliffe D, Migliorisi G, Cramer E. Translocation of influenza virus by migrating neutrophils. Cell Mol Biol. 1992;38:63–70. [PubMed] [Google Scholar]
- 15.Gerna G, Baldanti F, Revello MG. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum Immunol. 2004;65:381–6. doi: 10.1016/j.humimm.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Yao L, Korteweg C, Hsueh W, Gu J. Avian influenza receptor expression in H5N1-infected and noninfected human tissues. Faseb J. 2007;22:733–40. doi: 10.1096/fj.06-7880com. [DOI] [PubMed] [Google Scholar]
- 17.Saez-Lopez C, Ngambe-Tourere E, Rosenzwajg M, Petit JC, Nicolas JC, Gozlan J. Immediate-early antigen expression and modulation of apoptosis after in vitro infection of polymorphonuclear leukocytes by human cytomegalovirus. Microbes Infect. 2005;7:1139–49. doi: 10.1016/j.micinf.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Gerna G, Percivalle E, Baldanti F, et al. Human cytomegalovirus replicates abortively in polymorphonuclear leukocytes after transfer from infected endothelial cells via transient microfusion events. J Virol. 2000;74:5629–38. doi: 10.1128/jvi.74.12.5629-5638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savard M, Gosselin J. Epstein-Barr virus immunossuppression of innate immunity mediated by phagocytes. Virus Res. 2006;119:134–45. doi: 10.1016/j.virusres.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Thitithanyanont A, Engering A, Ekchariyawat P, et al. High susceptibility of human dendritic cells to avian influenza H5N1 virus infection and protection by IFN-α and TLR ligands. J Immunol. 2007;179:5220–7. doi: 10.4049/jimmunol.179.8.5220. [DOI] [PubMed] [Google Scholar]
- 21.Uiprasertkul M, Kitphati R, Puthavathana P, et al. Apoptosis and pathogenesis of avian influenza A (H5N1) virus in humans. Emerg Infect Dis. 2007;13:708–12. doi: 10.3201/eid1305.060572. [DOI] [PMC free article] [PubMed] [Google Scholar]


