Abstract
Background. Community-acquired pneumonia (CAP) remains an important cause of morbidity and mortality. Polymerase chain reaction (PCR) has been shown to be more sensitive than current standard microbiological methods and, therefore, may improve the accuracy of microbiological diagnosis for patients with CAP.
Methods. Conventional detection techniques and multiplex real-time PCR for atypical bacteria and respiratory viruses were performed on samples collected from 105 adults enrolled in a prospective study. An infiltrate was visible on each patient's chest radiograph, and a pneumonia severity index score was determined for each patient.
Results. Microbiological diagnoses were determined for 52 (49.5%) of 105 patients by conventional techniques and for 80 (76%) of 105 patients by real-time PCR. The time to obtain the result of real-time PCR could be reduced to 6 h. PCR methodology was significantly more sensitive for the detection of atypical pathogens and viruses (P ⩽ .001). Respiratory viral infections and mixed infections were detected in 15 (14.2%) and 3 (2.8%) of 105 patients, respectively, by conventional methods, but were detected in 59 (56.2%) and 28 (26.5%) of 105, respectively, by real-time PCR. Presence of a mixed infection was significantly associated with severe pneumonia (P = .002). Human rhinoviruses and coronaviruses, OC43 and 229E, were frequently identified pathogens.
Conclusions. The combined real-time PCR assay is more sensitive for diagnosis of the main viruses and atypical bacteria that cause CAP compared with conventional methods, and obtains results in a clinically relevant time period.
Community-acquired pneumonia (CAP) remains an important cause of morbidity and mortality. In the United States and the United Kingdom, CAP is the overall sixth leading cause of death [1–3]. Initial antibiotic management of CAP is empirical and dependent on a clear understanding of the likely pathogens [4].
Many professional societies have promulgated guidelines for diagnosis and treatment to aid clinicians who manage patients with CAP [5]. However, the diagnosis of CAP is controversial, because the findings of routine microbiological investigations have minimal impact on care [6, 7], and some studies question the value of sputum culture findings [8, 9]. In addition, results of blood cultures are only positive for 2%–10% of patients [10, 11], and serologic testing requires convalescent-phase samples and, therefore, is not clinically useful [2].
PCR has been shown to be more sensitive than current microbiological methods [12], and it could help to increase the number of microbiological diagnosis for patients with CAP. Specifically, real-time PCR formats allow results to be available rapidly and, therefore, in a clinically relevant time period.
To date, PCR-based methods have not been used to investigate the etiology of CAP in a prospective study, and, therefore, the impact of PCR on the guidelines for the treatment and diagnosis of CAP is unknown. The aim of the present study was to determine the etiology of CAP using multiplex real-time PCR to detect atypical bacterial and viral causes of CAP that occurred in adults enrolled in a prospective study who resided in a defined geographic area. Each patient had an infiltrate that was visible on a chest radiograph. Samples were collected for diagnostic testing, which included PCR analysis, from adult patients who were enrolled in 2 studies: a study of patients in the community with lower respiratory tract infections (LRTI) and a study of hospitalized patients with CAP. In addition, we investigated the use of multiplex, real-time PCR assays and conventional microbiological assays as means to diagnose CAP.
Materials and Methods
Patients. All patients were aged ⩾18 years and had an infiltrate observed by chest radiography. Patients from 2 studies—a study of patients with LRTI and a study of hospitalized patients—were included. Outpatients were observed from 1 September 2000 until 1 June 2001 in the Leiden area of The Netherlands as part of the LRTI study [13], in which patients with an infiltrate visible on their chest radiograph were included. Inclusion criteria for the outpatient study were observation of any abnormality during pulmonary auscultation and presence of at least 2 of the following 3 criteria: (1) fever (temperature, >38°C) in the past 48 h, (2) dyspnea or cough (productive or nonproductive), and (3) tachypnea, malaise, or confusion. Patients with conditions that would preclude completion of the study were excluded. A standard medical history was obtained for each patient, and an investigator performed a physical examination of each patient and collected information to determine a pneumonia severity index (PSI) score [14].
Hospitalized patients admitted from 5 November 2000 until 30 September 2002 as part of a study of CAP in the province of South Holland were included. Three hospitals participated: 2 community general hospitals and 1 academic teaching hospital. An inclusion criterion for hospitalized patients was the presence of a chest infiltrate on admission that was consistent with an infection. Clinical characteristics of the patients were obtained so that a PSI score could be determined for each patient with CAP, as has been described by Fine et al. [14]. The medical research ethics committee of the Leiden University Medical Center approved both studies, and informed consent for PCR analyses was obtained from all patients.
Samples. In the LRTI study, sputum samples (when available) were collected from patients before antibiotic treatment was administered, throat washes and throat swab specimens were obtained for virus isolation and PCR analyses, and paired serum samples were obtained during acute and convalescent phases of infection. In the study of hospitalized patients, the same types of samples were obtained, and, in addition, 2 sets of blood specimens were obtained for culture with the Bactec 9240 system (Becton Dickinson). Basic blood chemistry testing (for blood urea nitrogen, glucose, sodium, and potassium values) and hematological testing (for hematocrit levels, leukocyte and platelet counts, erythrocyte sedimentation rates, and C-reactive protein levels) were performed. All samples were transported at 4°C and processed in the laboratory on the day of collection. The serum samples were stored at -20°C until serological testing. After inoculation into cell culture, the remainder of the throat swab and throat wash specimens were divided into 3 aliquots and stored at -70°C for PCR testing.
Definitions. CAP was diagnosed if an infiltrate was observed on a chest radiograph. All chest radiographs were obtained either by a general practitioner 5–7 days after inclusion in the study or on admission to the hospital. Samples were obtained by investigators at the patient's home or at a clinic or hospital within 1–2 days after study enrollment. The criteria for the etiological classification of CAP are described in table 1.
Table 1.
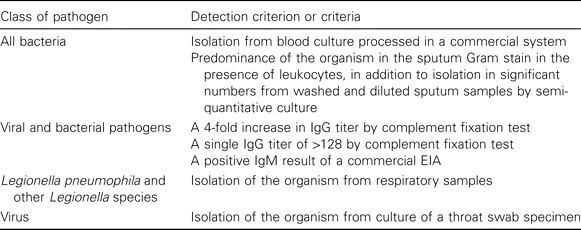
Criteria for the positive detection of pathogens with conventional microbiological methods.
Conventional microbiological methods. Analyses were performed using standard methods and in accordance with criteria described in table 1. For culture, standard methods were used for isolation of aerobic and anaerobic bacteria from sputum and blood samples. Isolation of Legionella pneumophila from sputum was attempted by use of selective medium (buffered charcoal yeast extract). Throat swab specimens were cultured in cell lines for detection of respiratory viruses after centrifugation. Immunofluorescence was used for early detection and confirmation of viral replication. Rhinoviruses were distinguished from enteroviruses by acid-lability testing.
For serologic testing, acute-phase and convalescent-phase samples were analyzed in the same assay. Complement fixation testing with a commercially available antigen (Virion; Ruschikon) was performed to detect adenovirus; influenza A and B viruses; parainfluenza viruses 1, 2, and 3; respiratory syncytial virus; and Mycoplasma pneumoniae. The M. pneumoniae antibody level was also determined by microparticle agglutination testing (Fujirebio). Serologic testing for IgG and IgM antibodies against Coxiella burnetti and IgM and IgG antibodies against L. pneumophila types 1–7 was performed using the serion ELISA (Ruschlikon;Virion). Levels of IgG and IgM antibodies against Chlamydophila species were measured using a commercial EIA (Medac GmbH).
Real-time PCR. Nucleic acids were extracted using the total nucleic acid protocol with the MagNA pure LC nucleic acid isolation system (Roche Diagnostics). Each sample was eluted in 100 µL of buffer, which was sufficient for all PCR analyses. Real-time PCR was performed, as has been described previously, for M. pneumoniae [15], L. pneumophila, Legionella species [16], influenza A virus, influenza B virus, respiratory syncytial virus, parainfluenza viruses 1–4 [17], human rhinovirus, and metapneumovirus [18] and for a modification of the adenovirus assay. An additional molecular beacon of the adenovirus E serogroup, CGTGCGGAGTCYACCCTTCTCTATGT CGCACG (underlined sequence indicates the stem of the molecular beacon), was used, with the 5′ end labeled with Texas red and the 3′ end labeled with black hole quencher 2. In addition, real-time PCR was performed for C. pneumonia and for human coronaviruses OC43 and 299E.
The C. pneumonia assay used primers CPs GCGGAAGGGTTAGTAGTACA and CPas ATCGCATAAACTCTTCCTCA and molecular beacon 5′-GCTGCGGGGATCTTCGGACCTTTCGGCGCAGC-3′, with the 5′ end labeled with hexachloro-6-carboxy-fluorescein (HEX) and the 3′ labeled with dimethylaminoazo-benzene-4-carboxylate (DABCYL), according to the same protocol as that used for M. pneumoniae PCR [15].
The human coronavirus OC43 assay used primers OC43s-CATACTCTGACGGTCACAATAATA, OC43as-ACCTTAGCAACAGTCATATAAGC and the probe 5′-TGCCCAAGAATAGCCAGTACCTAGT-3′ (TaqMan), with the 5′ end labeled with Yakima yellow and the 3′ end labeled with black hole quencher 1.
The human coronavirus 229E assay used primer 229Es-CATACTATCAACCCATTCAACAAG, 229Eas-CACGGCAACTGTCATGTATT, and the probe 5′-ATGAACCTGAACACCTGAAGCCAATCTATG-3′ (TaqMan), with the 5′ end labeled with 6-carboxy fluorescein (FAM) and the 3′ end labeled with black hole quencher–1. Both human coronavirus assays were performed using the same PCR protocol that has been described for assays of other respiratory RNA viruses [17, 18].
Two internal control assays were used to ensure the integrity of PCR, with a spike for phocine herpesvirus [15] and equine arteritis virus [18] for DNA and RNA targets, respectively. Amplification, detection, and data analysis were performed using the iCycler IQ real-time detection system (BioRad).
Statistical methods. Data were analyzed using SPSS software, version 11.0 for Windows (SPSS), and in all instances, a P of <.05 (2-tailed) was considered statistically significant. The differences in category variables between methodologies or patient groups were compared by use of the Fisher's exact test or the χ2 test.
Results
Patient information. The patient's demographic and clinical characteristics are described in table 2. In the outpatient LRTI group, a total of 38 patients gave informed consent, 13 patients had chest radiograph that demonstrated an infiltrate, none of whom required hospitalization. In the hospitalized group, 136 patients were observed, of whom 92 gave informed consent. Therefore, samples from 105 patients with CAP were available for microbiological diagnosis. The 3 patients who died were in the hospitalized group. Seven of 8 patients admitted to the intensive care unit had a PSI score in class IV ; the other patient admitted to intensive care unit had a PSI score in class II. None of the patients with PSI scores in class V were admitted to the intensive care unit.
Table 2.
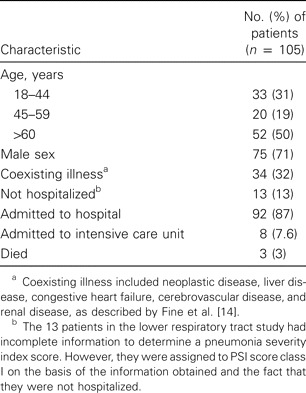
Demographic and clinical characteristics of 105 patients included in the main analysis of the etiology of community-acquired pneumonia in adults in the province of South Holland, The Netherlands.
Microbiological diagnosis. An etiologic agent was detected in 52 (49.5%) of 105 patients by conventional methods, and an etiologic agent was identified by real-time PCR in 80 patients (76%). Tables 3 and 4 show the numbers of patients for which the most common pathogens were detected. Other pathogens detected by conventional methods included Chlamydophila species (2 cases), parainfluenza virus (1 cases), respiratory syncytial virus (2 cases), and influenza B virus (2 cases). In 3 cases, real-time PCR did not detect pathogens that were detected by conventional methods: Chlamydophila was detected by serologic testing in 1 case, and influenza A virus was detected by complement fixation testing in 2 cases. However, real-time PCR identified additional cases of infection with L. pneumophila (1 case), Legionella species (3 cases), Chlamydophila pneumoniae (2 cases), M. pneumoniae (5 cases), parainfluenza virus (7 cases), respiratory syncytial virus (1 case), adenovirus (4 cases), influenza A virus (3 cases), influenza B virus (1 case), and human rhinovirus (16 cases). Metapneumovirus and C. burnetti infections were not detected. of the atypical bacterial and viral pathogens that were detected, 24 isolates were detected by conventional methods and 109 isolates were detected by PCR (P ⩽ .001). Conventional techniques detected >1 causative agent in 5 (10.2%) of 49 cases, and real-time PCR detected >1 causative agent in 28 (35%) of 80 cases.
Table 3.
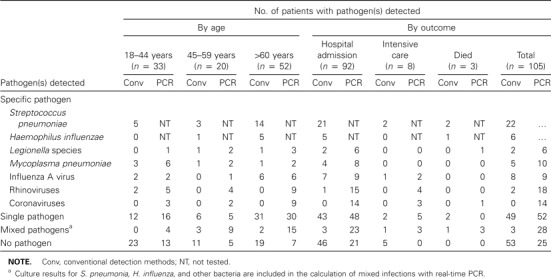
The most frequently identified pathogens detected by conventional methods or real-time PCR, according to patient age and outcome.
Table 4.
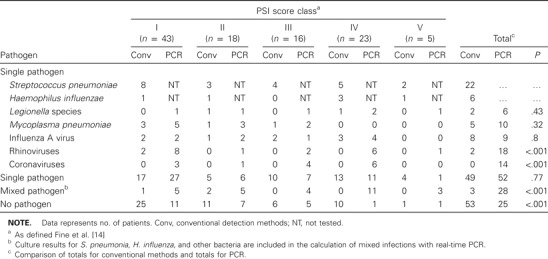
The most frequently identified pathogens detected by conventional methods or real-time PCR, according to pneumonia severity index (PSI) risk status.
Microbiological detection in relation to age and hospital admission. A comparison of detection by conventional assays and real-time PCR is shown in table 3. The patients aged >60 years received the highest number of diagnoses: 33 (65%) of 52 patients had diagnoses determined by conventional methods, and 45 (87%) of 52 patients had diagnoses determined by real-time PCR. Legionella infection was only found in hospitalized patients, and other infections were found in both nonhospitalized and hospitalized patients. All patients in the intensive care unit had an etiologic agent identified by real-time PCR, and 5 patients had no etiologic agent detected by conventional methods. All patients that died had a single bacterial pathogen detected by culture and additional pathogens that were detected by real-time PCR but not by conventional methods.
Microbiological detection according to risk groups. Table 4 shows pathogens detected in relation to PSI score classes, and table 5 shows the presence of pathogens in relation to the incidence of severe pneumonia. For PSI score class IV and V, pathogens were detected in 17 (61%) of 28 patients by conventional methods and in 26 (93%) of 28 patients by real-time PCR. The most significant risk factor for severe pneumonia was an age of >60 years (P < .001). In general, no statistically significant association was observed between the pathogen detected and the infected patient's PSI score class. However, M. pneumoniae was predominantly detected in patients in PSI score classes I and II (P = .05). PCR detected significantly more mixed infections in patients in PSI score classes IV and V (P = .002) than did conventional methods, which did not detect any cases of mixed infection in these risk groups. Human rhinovirus and human coronaviruses OC43 and 229E were identified as the only etiologic agents in 4 patients with PSI scores in classes IV and V. Mixed infection in patients with severe pneumonia was significantly associated with the presence of human rhinovirus (P = .001) and human coronaviruses OC43 and 229E (P = .005).
Table 5.
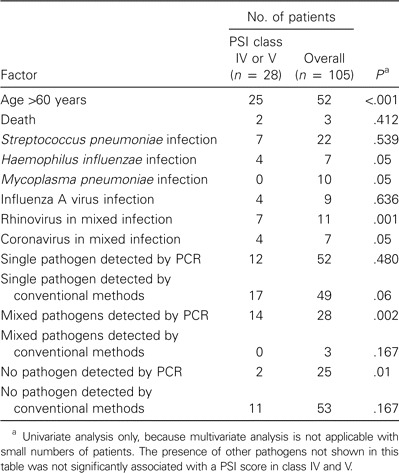
Statistical analysis of the factors associated with severe pneumonia, defined as a pneumonia severity index (PSI) score class of IV or V.
Conclusions
This study demonstrates that real-time PCR can increase the number of microbiological detection of respiratory pathogens in patients with CAP from 49.5% to 76% of patients. This higher number was mainly a result of detection of respiratory viruses, especially human rhinovirus and human coronaviruses, that are poorly detected by conventional techniques. Previous studies using conventional methods have achieved a microbiological detection rate of 2.1%–75% [10, 19, 20, 22]. The reason for these large differences is the diagnostic tests used; Campbell et al. [10] used only blood culture, whereas most studies with detection rates of >40% have used serologic testing of convalescent-phase samples to detect the full range of pathogens [19–22].
In this study, diagnoses were achieved in 87% of elderly patients, which might be related to the fact that diagnosis is more readily achieved because of the severity of disease and the poor immune response in elderly patients. PCR and bacterial culture also detected pathogens in >90% of patients with severe pneumonia. Therefore, a microbiological diagnosis could be made for the majority of elderly patients and patients with severe pneumonia by use of PCR, which would also help in the management of such patients. Rello et al. [23] claimed that microbiological diagnosis was fully justified in cases of severe community-acquired pneumonia because it had an impact on patient outcome, but others have found that diagnosis has had no impact [6, 7]. However, PCR provides the opportunity to identify the etiological agent in a clinically relevant time period. This could result in changes to antimicrobial therapy and could allow the potential use of antivirals. The rapid diagnosis of influenza A and B is especially important because the neuramidiase inhibitors oseltamivir and zanamivir are effective if administered within the 48 h of the initial onset of symptoms.
As in most other studies of series of studies with hospitalized patients with CAP, Streptoccocus pneumoniae was the leading bacterial organism detected (in 21% of cases). M. pneumoniae was detected in 9.5% of patients, and it has been described in 3%–22% of CAP cases [20, 22, 24], and epidemic increases are observed every 3–5 years [25]; so, higher infection rates could be expected in an epidemic year. M. pneumoniae infection has also been described in patients at low risk of pneumonia [19], a finding confirmed in this study, which found M. pneumoniae only in patients in PSI score classes I and II. The incidence of C. pneumoniae infection varies between <1% and 13% [19, 20, 22]. The frequency of Legionella infection in this study (6%) is similar to that in some other studies [19, 20], although rates as high as 15% have been reported as well [26]. Legionella species are regarded as a cause of severe CAP [27]. All cases of Legionella infection occurred in hospitalized patients, and 3 of these cases were in patients in PSI score classes IV and V, and, therefore, could be regarded as severe pneumonia.
Viral infections are increasingly being recognized as important causes of pneumonia; the proportion of cases caused by viruses varies from 2.2%–23% [4, 19, 20], with influenza A virus being the most important pathogen. In this study, influenza A virus was also, by all methods, one of the most frequently detected respiratory viral pathogens. Respiratory virus infections accounted for 14% of cases, with conventional microbiological methods, but that rate increased to 56.2% with PCR methods. The results of this study are in agreement with several reports that describe the increased sensitivity and specificity of PCR for respiratory viruses [12, 17, 28] because the use of PCR resulted in more diagnoses of respiratory virus infections than did conventional methods of detection. Each cell line used in cell culture can only detect 1 pathogen, and conventional techniques are very poor for detection of human rhinovirus and human coronavirus, which could explain the difference. PCR has an increased rate of detection of human rhinovirus [29] and human coronavirus [30], which are well recognized as the causes of common cold. There is increasing evidence that they are also important causes of severe lower respiratory disease. Rhinovirus infections have been reported in elderly adults with LRTI or pneumonia [31, 32] and in immunocompromised patients [33, 34]. Human coronavirus infection has also been reported in cases of pneumonia [30, 32, 35, 36]. In 4 cases in this study, human rhinovirus and human coronavirus were identified as sole etiologic agents, suggesting these viruses were the most likely cause of pneumonia in those cases.
Mixed infections have been identified in 16%–38% of cases of infection [20, 21], and, in this study, >1 causative pathogen was detected in 35% of cases. Previous studies have shown that viral infection may pave the way for bacterial infection [37]. It has also been suggested that dual viral infections, particularly infections with adenovirus and rhinovirus, could enhance the severity of illness [38]. The combinations seen in this study support this possibility, because viral infections were observed in all cases of mixed infection.
As with all studies on the etiology of pneumonia, there was a portion of cases for which no etiological agent was detected. In this study, 24% of cases remained without a diagnosis when PCR was used. This figure could probably be improved if respiratory samples other than throat swab specimens were investigated. Good detection rates have been reported with the use of sputum samples or induced sputum samples [39, 40], and the use of samples from the lower respiratory tract can result in an increased rate of diagnosis [39], but comparative studies using a variety of upper and lower respiratory tract samples still need to be performed. In addition, samples used in this study were frozen and thawed, which can reduce the sensitivity of the PCR. Most of the cases that were left without a diagnosis occurred in the low-risk patient group and among younger patients, suggesting either that there is less pathogen present for detection or that an unidentified organism was present that may cause less severe disease. Respiratory pathogens previously undetected are continuously being discovered; metapneumovirus is an example, and recently, coronavirus NL63 [41, 42] has been described in a child with pneumonia [41]. This coronavirus was not investigated in our patient population, so it may account for some of the unidentified cases.
PCR is increasingly being used for the diagnosis of atypical pathogens causing LRTI and CAP. A multiplex PCR that uses the same extraction procedures simplifies the test, and the results are available within 1 day, improving the speed of diagnosis for patients. In previous studies, the diagnosis of infection due to atypical pathogens and viruses has relied on serologic testing of convalescent-phase samples [19–21]. Serologic testing is only useful for epidemiology because it is too slow to impact clinical management. The application of real-time, multiplex PCR for rapid diagnosis could also have cost benefits and could result in a reduction of rates of antimicrobial resistance, hospital admission, and nosocomial transmission of respiratory viruses.
The real-time PCR assay described in this study is more sensitive for diagnosis of the main viruses and atypical bacteria that cause CAP, compared with conventional methods, and obtains results in a clinically relevant time period. The assay also detected pathogens in 87% of patients >60 years of age and in >90% of patients with severe pneumonia, meaning that this PCR assay may have a role in diagnosis of infections with the highest rates of morbidity and mortality.
Acknowledgments
Financial support. This study was financially supported by European Commission (Framework V) (grant no. QLK2-CT-2000-00294). A.W.G. was funded in part by Pfizer for work on another study.
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Guest JF, Morris A. Community-acquired pneumonia: the annual cost to the National Health Service in the UK. Eur Respir J. 1997;10:1530–4. doi: 10.1183/09031936.97.10071530. [DOI] [PubMed] [Google Scholar]
- 2.Guthrie R. Community-acquired lower respiratory tract infections: etiology and treatment. Chest. 2001;120:2021–34. doi: 10.1378/chest.120.6.2021. [DOI] [PubMed] [Google Scholar]
- 3.Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: global burden of disease study. Lancet. 1997;349:1269–76. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- 4.BTS Guidelines for the management of community acquired pneumonia in adults. Thorax. 2001;56(Suppl 4):1–64. doi: 10.1136/thorax.56.suppl_4.iv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niederman MS. Review of treatment guidelines for community-acquired pneumonia. Am J Med. 2004;117:51–57. doi: 10.1016/j.amjmed.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Pachon J, Prados MD, Capote F, Cuello JA, Garnacho J, Verano A. Severe community-acquired pneumonia: etiology, prognosis, and treatment. Am Rev Respir Dis. 1990;142:369–73. doi: 10.1164/ajrccm/142.2.369. [DOI] [PubMed] [Google Scholar]
- 7.Leroy O, Santre C, Beuscart C, et al. A five-year study of severe community-acquired pneumonia with emphasis on prognosis in patients admitted to an intensive care unit. Intensive Care Med. 1995;21:24–31. doi: 10.1007/BF02425150. [DOI] [PubMed] [Google Scholar]
- 8.Lidman C, Burman LG, Lagergren A, Ortqvist A. Limited value of routine microbiological diagnostics in patients hospitalized for community-acquired pneumonia. Scand J Infect Dis. 2002;34:873–9. doi: 10.1080/0036554021000026967. [DOI] [PubMed] [Google Scholar]
- 9.Bartlett JG, Mundy LM. Community-acquired pneumonia. N Engl J Med. 1995;333:1618–24. doi: 10.1056/NEJM199512143332408. [DOI] [PubMed] [Google Scholar]
- 10.Campbell SG, Marrie TJ, Anstey R, Ackroyd-Stolarz S, Dickinson G. Utility of blood cultures in the management of adults with community acquired pneumonia discharged from the emergency department. Emerg Med J. 2003;20:521–3. doi: 10.1136/emj.20.6.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skerrett SJ. Diagnostic testing to establish a microbial cause is helpful in the management of community-acquired pneumonia. Semin Respir Infect. 1997;12:308–21. [PubMed] [Google Scholar]
- 12.van Elden LJ, van Kraaij MG, Nijhuis M, et al. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis. 2002;34:177–83. doi: 10.1086/338238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graffelman AW, Knuistingh NA, le Cessie S, Kroes AC, Springer MP, van den Broek PJ. Pathogens involved in lower respiratory tract infections in general practice. Br J Gen Pract. 2004;54:15–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Fine MJ, Auble TE, Yealy DM, et al. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336:243–50. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 15.Templeton KE, Scheltinga SA, Graffelman AW, et al. Comparison and evaluation of real-time PCR, real-time nucleic acid sequence-based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae. J Clin Microbiol. 2003;41:4366–71. doi: 10.1128/JCM.41.9.4366-4371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Templeton KE, Scheltinga SA, Sillekens P, et al. Development and clinical evaluation of an internally controlled, single-tube multiplex real-time PCR assay for detection of Legionella pneumophila and other Legionella species. J Clin Microbiol. 2003;41:4016–21. doi: 10.1128/JCM.41.9.4016-4021.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–9. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheltinga SA, Templeton KE, Beersma MF, Claas EC. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex PCR. J Clin Virol (in press) [DOI] [PMC free article] [PubMed]
- 19.Roson B, Carratala J, Dorca J, Casanova A, Manresa F, Gudiol F. Etiology, reasons for hospitalization, risk classes, and outcomes of community-acquired pneumonia in patients hospitalized on the basis of conventional admission criteria. Clin Infect Dis. 2001;33:158–65. doi: 10.1086/321808. [DOI] [PubMed] [Google Scholar]
- 20.Lim WS, Macfarlane JT, Boswell TC, et al. Study of community acquired pneumonia aetiology (SCAPA) in adults admitted to hospital: implications for management guidelines. Thorax. 2001;56:296–301. doi: 10.1136/thorax.56.4.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jokinen C, Heiskanen L, Juvonen H, et al. Microbial etiology of community-acquired pneumonia in the adult population of 4 municipalities in eastern Finland. Clin Infect Dis. 2001;32:1141–54. doi: 10.1086/319746. [DOI] [PubMed] [Google Scholar]
- 22.Luna CM, Famiglietti A, Absi R, et al. Community-acquired pneumonia: etiology, epidemiology, and outcome at a teaching hospital in Argentina. Chest. 2000;118:1344–54. doi: 10.1378/chest.118.5.1344. [DOI] [PubMed] [Google Scholar]
- 23.Rello J, Bodi M, Mariscal D, et al. Microbiological testing and outcome of patients with severe community-acquired pneumonia. Chest. 2003;123:174–80. doi: 10.1378/chest.123.1.174. [DOI] [PubMed] [Google Scholar]
- 24.Bohte R, van Furth R, van den Broek PJ. Aetiology of community-acquired pneumonia: a prospective study among adults requiring admission to hospital. Thorax. 1995;50:543–7. doi: 10.1136/thx.50.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macfarlane JT, Finch RG, Ward MJ, Macrae AD. Hospital study of adult community-acquired pneumonia. Lancet. 1982;2:255–8. doi: 10.1016/s0140-6736(82)90334-8. [DOI] [PubMed] [Google Scholar]
- 27.Benin AL, Benson RF, Besser RE. Trends in legionnaires disease, 1980–1998: declining mortality and new patterns of diagnosis. Clin Infect Dis. 2002;35:1039–46. doi: 10.1086/342903. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg GA, Erdman DD, Edwards KM, et al. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J Infect Dis. 2004;189:706–10. doi: 10.1086/381456. [DOI] [PubMed] [Google Scholar]
- 29.Makela MJ, Puhakka T, Ruuskanen O, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–42. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Elden LJ, van Loon AM, van Alphen F, et al. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J Infect Dis. 2004;189:652–7. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson KG, Kent J, Hammersley V, Cancio E. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. BMJ. 1996;313:1119–23. doi: 10.1136/bmj.313.7065.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002;185:1338–41. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malcolm E, Arruda E, Hayden FG, Kaiser L. Clinical features of patients with acute respiratory illness and rhinovirus in their bronchoalveolar lavages. J Clin Virol. 2001;21:9–16. doi: 10.1016/s1386-6532(00)00180-3. [DOI] [PubMed] [Google Scholar]
- 34.Ison MG, Hayden FG, Kaiser L, Corey L, Boeckh M. Rhinovirus infections in hematopoietic stem cell transplant recipients with pneumonia. Clin Infect Dis. 2003;36:1139–43. doi: 10.1086/374340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folz RJ, Elkordy MA. Coronavirus pneumonia following autologous bone marrow transplantation for breast cancer. Chest. 1999;115:901–5. doi: 10.1378/chest.115.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pene F, Merlat A, Vabret A, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis. 2003;37:929–32. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakaletz LO. Viral potentiation of bacterial superinfection of the respiratory tract. Trends Microbiol. 1995;3:110–4. doi: 10.1016/s0966-842x(00)88892-7. [DOI] [PubMed] [Google Scholar]
- 38.Papadopoulos NG, Moustaki M, Tsolia M, et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002;165:1285–9. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- 39.Drosten C, Chiu LL, Panning M, et al. Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol. 2004;42:2043–7. doi: 10.1128/JCM.42.5.2043-2047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson JL, Moric I, Wark PA, Johnston SL, Gibson PG. Use of induced sputum for the diagnosis of influenza and infections in asthma: a comparison of diagnostic techniques. J Clin Virol. 2003;26:339–46. doi: 10.1016/S1386-6532(02)00084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouchier RA, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Hoek, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]


