Abstract
Site-specific recombinases (SSRs) such as the Cre/loxP system are useful genome engineering tools that can be repurposed by altering their DNA-binding specificity. However, SSRs that delete a natural sequence from the human genome have not been reported thus far. Here, we describe the generation of an SSR system that precisely excises a 1.4 kb fragment from the human genome. Through a streamlined process of substrate-linked directed evolution we generated two separate recombinases that, when expressed together, act as a heterodimer to delete a human genomic sequence from chromosome 7. Our data indicates that designer-recombinases can be generated in a manageable timeframe for precision genome editing. A large-scale bioinformatics analysis suggests that around 13% of all human protein-coding genes could be targetable by dual designer-recombinase induced genomic deletion (dDRiGD). We propose that heterospecific designer-recombinases, which work independently of the host DNA repair machinery, represent an efficient and safe alternative to nuclease-based genome editing technologies.
INTRODUCTION
Over the past few years, genome-engineering techniques have rapidly advanced with the use of genome editing enzymes such as zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs) and clustered regulatory interspaced short palindromic repeats (CRISPR) with the CRISPR-associated protein 9 (Cas9) (1,2). The CRISPR/Cas9 system has especially become a powerful tool for genome manipulation due to its simplicity and efficacy. Through the straightforward design of suitable guide (g)RNAs the Cas9 protein can be programmed for efficient DNA cleavage at defined genomic sites (3). Hence, the Cas9 protein itself does not need to be modified.
Nuclease-based gene editing tools generate double-strand breaks, consequently making them dependent on cellular DNA repair mechanisms, namely non-homologous end-joining (NHEJ), microhomology-mediated end joining (MMEJ) or homologous recombination (HR) (4–6). The most common of these DNA repair mechanisms is the error-prone NHEJ pathway, which can lead to sequence alterations after DNA repair such as nucleotide insertions and deletions (Indels). Indels can be useful in certain experimental settings but can be detrimental in others, such as many clinical applications (7–11). To address this challenge, development of genome editing methods that do not induce DNA double-strand breaks have become an active field of research (12–15).
Site-specific recombinases (SSRs) from the tyrosine recombinase family can be used as flexible genome engineering tools to perform a variety of DNA rearrangements in heterologous hosts (16). Importantly, SSRs work without the introduction of DNA double-strand breaks and do not require the host DNA repair machinery to rearrange genomic DNA. Furthermore, recombinase-based approaches are also efficient in post mitotic cells, where precision genome editing has been difficult to accomplish with current nuclease-based technologies (17–19). The most commonly used tyrosine recombinase system is the Cre/loxP system. This system has been used for decades to achieve precise genomic manipulations in vitro and in vivo, even though, in certain experimental settings, overexpression of the recombinase has been reported to cause inadvertent Cre-mediated effects (16).
Cre-mediated recombination is highly dependent on the presence of a 34 bp target sequence (loxP) consisting of two 13 bp inverted repeats flanking the same 8 bp spacer sequence (20,21). Each inverted repeat is bound by one Cre molecule and forms a homodimer on the target sequence. Two homodimers assemble the recombination synapse in a homotetramer complex and recombine two loxP target sites without the need of any accessory factors (22). Because of its efficiency and specificity, the Cre/loxP system is a widely used tool for precise genetic manipulations in numerous model organisms (23–27).
In order to employ the Cre/loxP system in organisms, the loxP sites need to be introduced beforehand at the anticipated genomic position, limiting its use to basic research. To expand the usefulness of the Cre/loxP system, efforts have focused on altering the DNA-binding specificity of the recombinase protein. By applying different directed molecular evolution strategies, various Cre mutants have been generated that recombine artificial loxP-like target sequences (28,29). More recently, Cre-based SSRs have been developed to target DNA sequences found in the long terminal repeats of primary HIV-1 isolates (30,31). The developed SSRs showed potent antiviral activity in HIV-1 mouse-models suggesting that these enzymes constitute a promising approach to excise the integrated HIV-1 provirus from infected patient cells to reverse the infection (31,32). These studies have demonstrated that efficient and specific designer-recombinases can be generated to target a variety of loxP-like sequences, including therapeutically relevant sequences.
Based on these successes, we reasoned that SSRs could be potentially repurposed to excise naturally occurring sequences in the human genome. We therefore explored possible strategies to identify suitable SSR target sites in the human genome and developed designer-recombinases for these sites.
MATERIALS AND METHODS
Plasmid construction
All cycling programs and oligos can be found in Supplementary Tables S1 and S2. More details of the cloning procedure can be found in the Supplementary Materials and Methods section. In short, the evolution plasmids pEVO-loxHEX-L, pEVO-loxHEX-R, pEVO-loxHEX-R1 and pEVO-loxHEX-R2 were generated in two-steps from the previously described pEVO-loxP (30,33) using the Cold Fusion Cloning Kit (SystemBiosciences). The additional ribosome binding site and appropriate restriction sites to express two recombinases from one plasmid were introduced while preparing the pEVO-loxHEX1+2. All PCR reactions were performed with a high-fidelity polymerase following the manufacturer's manual (Herculase II Fusion DNA Polymerase, Agilent).
The expression plasmid pIRES-NLS-EGFP was generated in two steps using the previously described pIRESneo-Cre vector and standard restriction and ligation cloning (30).
The loxHEX reporter plasmid was generated by a four-step cloning using the plasmids pCAG-loxPSTOPloxP-ZsGreen (Addgene #51269) and pCAGGS-loxP-mCherry-loxP-EGFP (31,34)
Substrate linked directed evolution (SLiDE)
New recombinases were evolved using the experimental principals as described previously (Supplementary Figure S4) (28,30,31). An optimized substrate linked directed evolution protocol was developed to speed up the generation of evolved enzymes (Supplementary Figure S1). The experimental details to reduce the time for one evolution cycle to one day are described in the Supplementary Materials and Methods section.
Identification of critical residues and targeted mutagenesis (ISOR)
Previous publications identified mutation hotspots in the recombinase coding sequence that frequently change during the adaptation to a new target-site (28–31,35–37). However, it is challenging to predict these changes in order to bootstrap the evolution process by skipping subsites (for example loxHEX-R1 and loxHEX-R2, Supplementary Figure S2A).
To rationalize the directed evolution process, we identified a set of critical residues by retrospectively comparing the starting library (loxBTR1A) to the final loxHEX-R library (Supplementary Figure S2B). The nomination of critical residues was based on their frequency in the final loxHEX-R library and known interactions of these residues with the DNA target site (Supplementary Figure S2B). Twelve amino acid changes were predicted to be critical and were introduced into the loxBTR1A library using an adapted version of the protocol of ISOR from Herman et al. (see Supplementary Figure S3) (38).
By using this protocol, it was possible to start the evolution procedure without using subsites (see Supplementary Figure S2C). Experimental details can be found in the Supplementary Materials and Methods section.
Recombinase activity assays
In order to test the recombination activity of a library or single clones, the pEVO plasmid with the respective target sites and libraries was cultivated in E. coli XL-1 Blue (Agilent) and the expression of the recombinases was induced with different concentrations of l-arabinose (Sigma Aldrich). After growing the cells for 14–16 h in 12 ml LB and in the presence of chloramphenicol (25 mg/ml), plasmid DNA was extracted using the GeneJET Plasmid Miniprep Kit (ThermoFisher). The recombined plasmid is smaller in size compared to the nonrecombined plasmid. Therefore, a restriction digest with BsrGI-HF and XbaI will show a slower migrating band (∼5 kb) for the nonrecombined plasmids and a faster migrating (∼4.3 kb) band for recombined plasmids after gel electrophoresis. Recombination efficiencies were calculated by the ratio of band intensities using Fiji for image processing.
In order to test multiple single clones at the same time, single colonies were picked and grown in 50 μl LB in the presence of chloramphenicol (25 μg/ml) in a standard 96-well cell culture plate and induced with different l-arabinose concentrations for 14–16 h. A colony PCR (cycling program 8) for each clone was performed according to the manufacturer's manual (MyTaq™ Polymerases, Bioline) using 1 μl cell suspension as template and primers c1 (oligo 30), c2 (oligo 31) and c3 (oligo 32). The primers c1 and c3 will generate a PCR product of 475 bp for the nonrecombined plasmids, whereas the primers c1 and c2 will generate a PCR product of 400 bp for the recombined plasmids. Recombination efficiencies were calculated by the ratio of band intensities using Fiji for image processing.
Deep sequencing of the recombinase libraries
In order to deep sequence the recombinase libraries, 5 μg pEVO plasmids for loxHEX-R, loxHEX-L and loxHEX-R (ISOR) were digested with the restriction enzymes BsrGI-HF and XbaI (NEB) and the 1.2 kb fragment harboring the recombinase DNA was isolated from a 0.8% stain free TBE agarose gel using the Isolate II PCR and Gel Kit (Bioline). Further purification was performed with a QIAquick PCR Purification Kit (QIAGEN) and 1× Agencourt AMPure XP beads (Beckman Coulter). The DNA was quantified with a Qubit dsDNA HS Assay Kit on a Qubit 2.0 Fluorometer (ThermoFisher) and sent to the CRTD Deep Sequencing facility, where it was sequenced with a Sequel System 6.0 using the SMRT CCS method.
Circular consensus sequence data were generated with PacBio's pbccs v3.1.0. Using pbccs's filtering criteria, only sequences of length 1034–1200 bp with a minimum predicted accuracy of 0.997 were kept. After converting the data to FASTA format using SAMtools 1.9 the recombinase sequences were aligned using the protein2dna:bestfit alignment model of exonerate v2.3.0 (31,39,40).
Further processing was performed in R v3.5.1 (R Core Team 2018), where the amino acid frequencies where calculated. Using the R package dplyr (R package version 0.7.8) the highest amino acid frequencies of the HexR sequences differing from the starting library consensus where filtered out and plotted using the R package ggplot2 (R package version 0.7.8).
Expression of the recombinases in human cells
HeLa cells were seeded with a density of 200,000 cells/well in a standard 12-well cell culture dish and transfected the next day. For each transfection 1.2 μg plasmid DNA was mixed with 75 μl Opti-MEM I Reduced Serum Medium (ThermoFisher, prewarmed at RT). In addition, 3 μl Lipofectamine® 2000 Transfection Reagent (ThermoFisher) was mixed with 75 μl Opti-MEM I Reduced Serum Medium (prewarmed at RT). The DNA mix and Lipofectamine® 2000 Transfection Reagent reaction were mixed and incubated for 5 min at RT. During the incubation time, the medium (DMEM—Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% Penicillin-Streptomycin (10 000 U/ml), ThermoFisher) was removed from the cells and they were washed with 1 ml of PBS. Then, 1.5 ml of prewarmed fresh medium was added to each well and the transfection mixture was added dropwise to the wells. The cells were transfected with combinations of the reporter plasmid (recombined and nonrecombined), the empty recombinase expression plasmid and the recombinase expression plasmid containing the single D12 recombinases. Forty-eight hours after transfection cells were analyzed with the FACSCanto II SORP (BD) or with the EVOS FL Cell Imaging System (ThermoFisher) for EGFP and mCherry expression.
Additionally, gDNA was extracted 48 h after transfection using the QIAamp DNA Blood Mini Kit (Qiagen). In order to detect the genomic deletion, PCR reactions (cycling program 9) were performed with primers flanking the loxHEX target sites gloxHEX-F (oligo 33) and gloxHEX-R (oligo 34) and 200ng of genomic DNA as a template (using MyTaq™ Polymerases, Bioline). To test for possible chromosomal translocations between the loxHEX target sites on chromosome 7 and the loxHEX-Off1 off-target site on chromosome 8, two PCR reactions were performed (see Supplementary Figure S8). The primers were used in different combinations: chr7_8_trans_F (oligo 35—primer a in scheme), chr7_8_trans_R (oligo 36—primer c in scheme), chr8_7_trans_F (oligo 37—primer d in scheme) and chr8_7_trans_R (oligo 38—primer b in scheme).
Quantification of the excision frequency in human cells
In order to calculate the excision efficiency in the pool of cells transfected with the D12 recombinase heterodimer, we developed a semi-quantitative PCR based assay. We amplified the nonrecombined and the recombined loxHEX locus from gDNA (oligo 33 and 34, cycling program 9) and mixed them in defined molar ratios with 50–10% of recombined fragments. A subsequent PCR (oligo 33 and 34, cycling program 9) of these mixtures resulted in two amplicons with different band intensities based on the molar ratio of the templates (Supplementary Figure S10A). Next, we performed the same PCR on the gDNA of D12 transfected HeLa cells (n = 3). By plotting the intensity ratio (nonrecombined compared to the recombined) against the relative excision, we calculated a standard curve and deduced the recombination efficiency in the D12 transfected cells (Supplementary Figure S10B–D).
Identification of pairs of target sites in the human genome
In order to identify pairs of potential target sites, we wrote a Python script (available upon request) that uses a single DNA sequence as input and generates a list of positions and sequences of all pairs of target sites containing identical 8 bp spacers, within a defined distance range. The script traverses the sequence, shifting by one base pair at a time and, for each substring of eight characters located at the current position, searches all other occurrences of the same substring located downstream.
For each pair of identical spacers, the script extracts 13 bp long sequences of the flanking half-sites and calculates their similarity. If the target sites are supposed to be recombined by a homotetramer, all four half-sites are allowed to differ at no more than two positions. If the target sites are supposed to be bound by two different recombinases, the half-sites are split into two sets, where half-sites belonging to the same set are not allowed to have more than two mismatches. Pairs of target sites that did not fulfill this criterion are discarded from further analysis.
To find all pairs of target sites that could be recombined with a homotetramer, we executed the script on all top-level chromosome sequences from the human genome (release hg38 from December 2013). Sub-sequences for searches of pairs of sites targetable by two recombinases were generated with the BEDTools suite (version 2.27.1) (41) where transcript coordinates and annotations were obtained from a GTF file downloaded from the GENCODE database (release 28 from November 2017) (42).
An overlap between putative target sites and repetitive elements was performed with intersectBed from the BEDTools suite, enforcing an overlap of at least 13 bp per target site. Genomic coordinates of interspersed repeats and low complexity DNA sequences were obtained from the UCSC Genome Browser (43). Specifically, from the ‘RepeatMasker Viz. (Current Dataset)’ track.
Off-target site prediction
An exhaustive, genome-wide prediction of potential off-target sites was performed using an in-house developed Python application employing Nvidia's CUDA parallel computing platform via PyCUDA API (version 2018.1.1) (44) and the NumPy package version 1.16.2 (45). The process was performed in two stages. Initially, a dense hash index of the complete genome was generated, where each hash is a two-bit encoding of a seed sequence of 13 nucleotides. Hash codes were generated for each position that did not contain DNA ambiguity codes in the seed sequence.
In the second stage, a set of four on-target half-sites was converted into their two-bit hash representation, with the right half-sites reverse-complemented before the encoding. Next, every hash-code from the genome index was compared to each of the half-site codes and a number of mismatches was calculated. If this number, for at least one half-site, did not exceed 2 mismatches, the second half-site of the potential off-target was evaluated. A hash-code located 21 positions downstream in the genome index was reverse-complemented and compared to each of the half-site codes. Similarly, up to two mismatches were tolerated and, if a match existed, an 8 bp genomic sequence located between the half-sites was extracted and saved in a list. After the whole genome scan was complete, the program counted occurrences of individual spacers or their reverse-complement variants. If a spacers sequence appeared at least twice in the genome, all target sites containing this sequence were labelled as potential off-targets.
mRNA production
D12 mRNA was produced using the HiScribe™ T7 ARCA mRNA Kit (NEB) following the manufacture's manual. The mRNA was purified using the Monarch® RNA Cleanup Kit (NEB).
Induced pluripotent stem cells (iPSCs) culture and mRNA transfection
IPSCs were maintained in the DEF-CS™ Culture System (Takara) according to the manufacture's manual in a six-well format. One day after seeding (1 × 106 cells/well) the iPSCs were transfected with D12 mRNA (one mRNA for each monomer). For each well 4 μl Lipofectamine™ MessengerMAX™ Transfection Reagent (ThermoFisher) and 1.5 μg mRNA was mixed with 125 μl Opti-MEM I Reduced Serum Medium. Next, the Lipofectamine and mRNA mixtures were combined, vortexed and incubated for 15 min. After changing the medium of the iPSCs, the transfection mix was gently added to each well. The medium was changed again after 4 h. Analysis of the samples was performed 24 or 48 h after transfection.
RESULTS
Target site nomination
A prerequisite for employing Cre-like SSRs as tools for custom genome editing, is the presence of suitable pairs of unique loxP-like sequences. For the HIV-1 genome, the identification of suitable loxP-like sequences was relatively straightforward due to the long-terminal repeats (LTR) that flank the viral genome. Because these sequences occur as unique direct repeats that are not found elsewhere in the human genome, they offer numerous possibilities to identify 34 bp sequences with similarities to loxP.
In a random DNA sequence, one would expect an 8 bp repeated sequence that could serve as a spacer sequence only every 65 000 bp. It is also highly unlikely that this spacer is flanked by nearly perfect symmetric, inverted 13 bp repeats (binding sites for the recombinase). Hence, suitable loxP-like sequences would be extremely rare in random DNA sequence and strategies to find suitable pairs of unique loxP-like sequences would most certainly fail.
However, genome sequences are not random and indeed many repeated DNA sequences, such as microsatellites and minisatellites are found throughout the human genome (46–48). We therefore decided to search for naturally occurring candidate SSR target-sites in the human genome by employing a similar computational pipeline used to identify loxP-like sites in the HIV-1 LTRs (Figure 1A). Intriguingly, the work on the HIV-1 recombinases revealed that suitable target sites do not have to comprise a perfect 13 bp symmetry and that a small amount of asymmetry can be tolerated by SSRs (30,31). Therefore, our search algorithm was modified to allow an asymmetry at up to two positions in the otherwise symmetric loxP-like target site. The initial search was followed by an exhaustive off-target analysis (see Materials and Methods for details). In addition, we limited the search to sequences located on the same chromosome and within a 20 kb distance from each other (Figure 1A). This limitation was imposed, because recombination efficiency progressively declines for target sites at increasing distances (49,50).
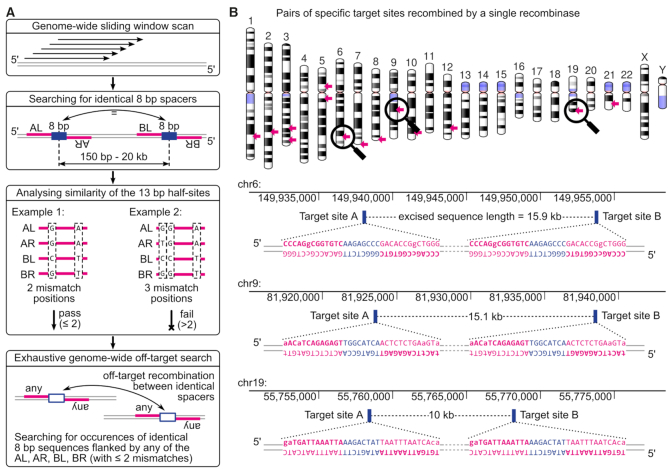
Figure 1.
Identification of loxP-like target sites in the human genome. (A) Workflow of the computational pipeline used for the genome-wide search. First, a sliding window approach is used for identifying positions of identical spacer sequences (blue bars) located within a range of 150 bp – 20 kb. For each pair of matching spacers, their surrounding half-site sequences (in magenta, labelled as AL, AR, BL, BR) are aligned and their nucleotide composition at respective positions is analyzed. Pairs of target sites having more than two mismatch positions are discarded. The remaining pairs undergo an off-target analysis, which defines an off-target as any other pair of identical spacers flanked by any of the on-target half-sites, with up to two mismatches each. (B) Chromosomal distribution of specific pairs of target sites (marked with magenta arrows) that qualify to be recombined with a single SSR. Magnification symbols highlight three regions shown in the bottom panel. The three examples are presented with their chromosomal position, the distance between the two target-sites and the actual sequence. Spacer sequences are highlighted in blue and half-sites sequences are shown in magenta, with asymmetries depicted in lower case.
The genome-wide analysis for a single recombinase detected 85 600 pairs of target sites fulfilling the distance and the sequence composition criteria. However, stringent off-target filtering reduced the number of acceptable sequences to only 14 candidate target-sites, of which eight overlapped with annotated repetitive elements (Figure 1B, Supplementary Table S3). The reason for this massive reduction was that most of the DNA repeats occur multiple times in the genome, rendering them unsuitable. The two loxP-like sequences must be unique enough to ensure specificity and avoid off-target recombination. Therefore, very few suitable target site pairs for evolved single SSRs could be identified in the human genome.
In order to extend the potential of Cre-like SSRs for excising DNA fragments from the human genome, we decided to examine the possibility of using two different recombinase monomers, each specific towards a different target half-site. In an artificial setting a heterodimer of wild type Cre and a Cre mutant was able to cooperatively recombine a hybrid loxP/loxM7 target site (51). If one could apply this principle for two separately evolved recombinases, such SSRs could cooperatively bind a target site composed of two different half sites. In this setting, both target sites would share the same spacer sequence, but they would be flanked by half-sites with high similarity towards one or the other recognition sequence of the two SSRs. Allowing the half sites left and right from the spacer to be different should expand the number of possible target sites. Additionally, the arrangement of two different half-sites around a given spacer allows for more possibilities compared to a single half-site which gives only one combination. Therefore, this flexibility should further increase the number of potential target sites.
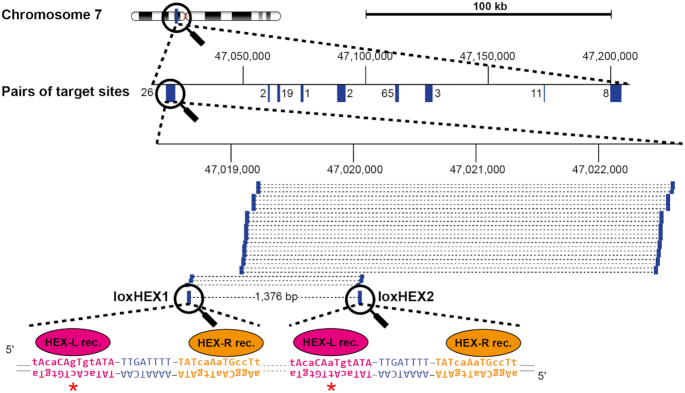
We modified the search algorithm accordingly, and scanned for all possible pairs of target sites suitable for recombination by a dual recombinase system. Remarkably, the independent half-site analysis increased the number of putative recombinase target sites to 155 732 476. Many of these candidate target-sites again occur several times in the human genome, rendering them unacceptable as high-quality SSR target sites. Nevertheless, we anticipated that the massive increase in total number of sequences might also provide a much-increased number of acceptable and unique candidate target-sites. As a proof of concept, and because off-target calculations are computationally extensive, we limited the genome-wide off-target filtering to a randomly chosen 200 kb region from human chromosome 7. While no target site for a single recombinase could be identified within the chosen 200 kb window, 137 candidate target sites were found for dual recombinase systems (Figure 2, Supplementary Table S4), confirming that this approach substantially increased the number of suitable SSR target sites. We inspected the 137 sequences individually and chose two target sites (hereinafter referred to as loxHEX1 and loxHEX2, Figure 2) for further analyses, because of their remote similarity to target sites of previously evolved SSRs (30,31). Hence, recombinase libraries from these previous evolution intermediates could be utilized to accelerate the directed evolution process. LoxHEX1 and loxHEX2 share the same 8 bp spacer sequence, flanked by 13 bp half-sites that are asymmetric in 6 of the 13 positions. In addition, half-sites located left of the spacer differ at one position (red asterisk), whereas half-sites located right of the spacers are identical (Figure 2).
Figure 2.
Nomination of target sites for heterospecific Cre-like recombinases. Bioinformatic analysis of a randomly chosen 200 kb region on chromosome 7 to nominate target sites for dDRiGD. Magnification symbols indicate a zoom-in for the regions of interest. 137 potential target sites cluster in nine different regions, depicted in blue. A zoom into one of these clusters is shown that harbors the selected target sites loxHEX1 and loxHEX2. All pairs of potential target sites located in the cluster are represented as blue bars connected with dashed lines. The selected pair is emphasized by thicker bars, target site names and the distance in base pairs (bp) between them are shown. Numbers above indicate genomic coordinates. The actual DNA sequence of loxHEX1 and loxHEX2 is presented with individual half-sites shown in magenta and orange, with the spacer sequence highlighted in blue. Asymmetric positions are shown in lower case and the red asterisk indicates the single nucleotide difference of the loxHEX1 site compared to the loxHEX2 site. Recombinase monomers (HEX-L rec and HEX-R rec) binding to the individual half-sites are illustrated in the corresponding colors.
Directed evolution of a pair of site-specific recombinases targeting a unique human genomic site
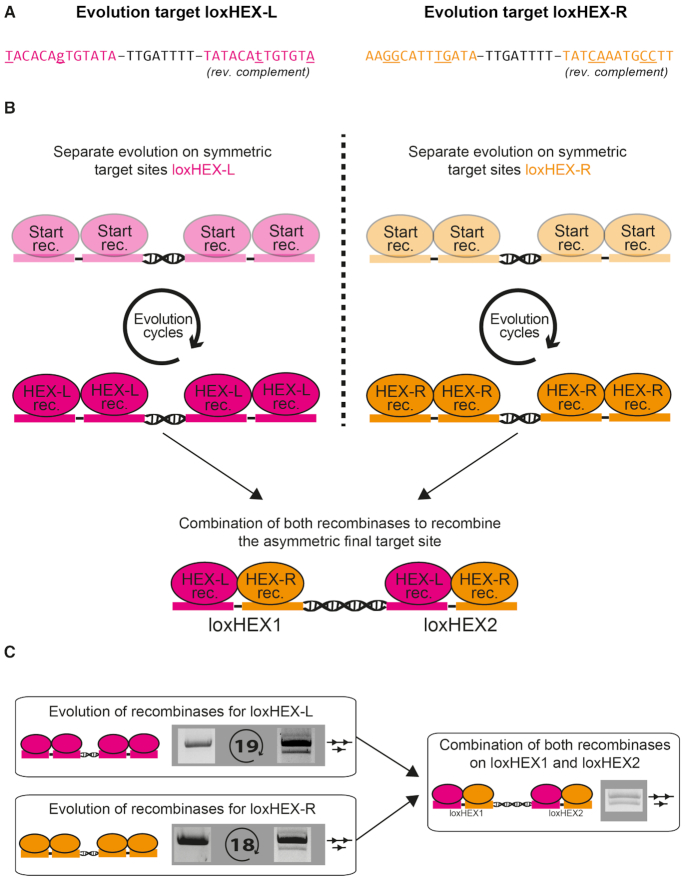
To investigate whether the ∼1.4 kb sequence flanked by loxHEX1 and loxHEX2 could be excised from the human genome, we evolved two SSRs utilizing an optimized substrate linked directed evolution protocol (Supplementary Figure S1). The evolution target-sites (loxHEX-L and loxHEX-R) were composed of either the left or right loxHEX half-sites and their reverse complement sequence (Figure 3A). The evolution process was separated in two steps. In the first step, we evolved recombinases targeting loxHEX-L and loxHEX-R independently. In the second step, we combined the two evolved recombinase libraries to test if they can cooperatively recombine the final asymmetric target site as heterodimers (Figure 3B).
Figure 3.
Substrate linked directed evolution of a recombinase heterodimer. (A) Sequences of the evolution sites loxHEX-L and loxHEX-R. The same color code as in Figure 2 is used, with asymmetric positions shown in lower case. The evolution sites are composed of either the left or the right half site of the loxHEX1/2 sites and the reverse complement sequence. Mismatches of the evolution sites compared to the starting library target site are underlined. (B) Scheme of the evolution process. Two independent evolutions were performed on the target sites loxHEX-L and loxHEX-R (left and right half site of the final loxHEX target site). The light color indicates a starting library with weak activity whereas the dark color represents a new recombinase library with good recombination activity for loxHEX-L or loxHEX-R. After the evolution process, the recombinases were expressed together as a heterodimer to recombine the final asymmetric loxHEX target sites. (C) Substrate linked directed evolution of recombinase libraries. The recombination efficiency of the first and last cycle is shown. Recombinases from the last cycle were coexpressed to recombine the final loxHEX target sites as a heterodimer. The upper band represents the nonrecombined plasmid (represented by a line with two triangles) the lower band represents the recombined plasmid (a line with one triangle). The number of evolution cycles is shown between the gel images. Combination of both libraries resulted in active heterodimers recombining the asymmetric loxHEX target site.
Evolution on loxHEX-L and loxHEX-R was initiated using the existing loxBTR1A or a modified loxBTR1A recombinase library (Supplementary Figure S2) (31). After the first cycle, recombination activity was very low for both loxHEX-L and –R, but over the course of 19 and 18 evolution cycles, respectively, libraries with high activity at low recombinase expression levels were obtained on the symmetric target sites (Figure 3C).
To study whether the recombinases evolved for loxHEX-L or loxHEX-R could recombine the full loxHEX site or show cross activity, we cloned the respective libraries into the pEVO-loxHEX vectors and grew transformed E. coli cells overnight in the presence of l-arabinose to induce recombinase expression. Plasmid DNA isolated from these cultures did not show the recombination specific band patterns. This indicates that the single recombinase libraries were neither active on the final loxHEX target site nor had cross reactivity on the loxHEX-L or loxHEX-R target site (Supplementary Figure S5A). To allow co-expression of both recombinase libraries, we added another ribosome binding site to the pEVO plasmid (Supplementary Figure S5B) and cloned the library of recombinases targeting loxHEX-R in the first position and recombinases targeting loxHEX-L in the second position. This resulted in random pairing of the recombinases but guaranteed that each heterodimer consisted of a recombinase for either the left or right half site of the final loxHEX target site. In contrast to the single recombinase libraries, the co-expression of both (loxHEX-L and loxHEX-R) libraries resulted in recombination on the final target site (Figure 3C), suggesting that individual monomers can form heterodimer complexes to recombine loxHEX. This result demonstrates that it is possible to evolve two different Cre-like recombinases, for either the left or the right half-site of a given target, and combine them to form active heterotetramers. Moreover, our data show that both recombinase libraries are specific to their target sites and do not recombine the final loxHEX sites alone, nor do they recombine the sites of the respective counterparts (Supplementary Figure S5A).
Evolved recombinases show activity as heterodimers in bacteria
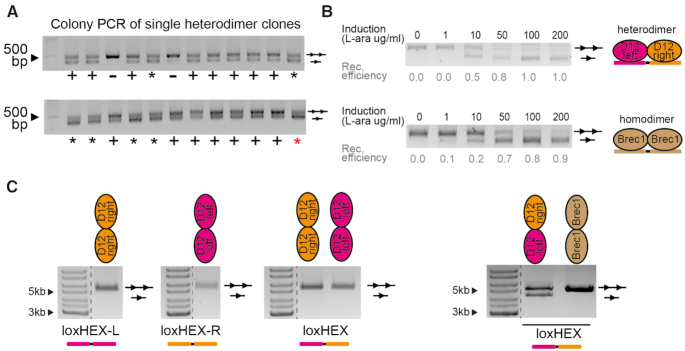
After evolving a library of heterodimers that recombines the loxHEX target sites, clones of recombinase heterodimers were tested for their activity on the final human genomic target site in bacteria. Twenty-four heterodimer clones were grown and recombinase expression was induced with l-arabinose for 16 h. Analysis of the samples showed that seven heterodimers had high recombination activity, fifteen heterodimers moderate activity, and only two heterodimers were inactive (Figure 4A).
Figure 4.
Molecular characterization of recombinase heterodimer clones. (A) Recombination efficiency of 24 heterodimer clones on the final loxHEX target sites analyzed with a PCR-based assay. Heterodimer clones marked with a plus (+) showed activity (two bands), whereas inactive heterodimer clones (single band) are marked with a minus (–). Heterodimer clones marked with an asterisk show recombination activity of at least 50%. The clone with the highest activity is marked with a red asterisk (hereinafter referred to as D12). (B) l-arabinose concentration dependent activity of the D12 heterodimer on loxHEX analyzed by a restriction digest-based assay. As a comparison the recombination efficiency of Brec1 on loxBTR (brown) is presented. The displayed numbers show relative recombination efficiencies calculated as a ratio of the recombined (lower) to the nonrecombined (upper) band intensities. (C) Specificity tests of D12. Recombination characteristics of monomers evolved on either loxHEX-L (D12-left) or loxHEX-R (D12-right) are shown for different target-sites. Note that the individual recombinases neither recombine the target site of the other recombinase, nor do they recombine the final loxHEX site alone. Only the heterodimers recombine loxHEX (right panel). Brec1 was used as a negative control.
The heterodimer clone with the highest recombination activity (hereafter referred to as D12—row D clone 12) was further analyzed for its activity on the final target site, using different recombinase expression levels. D12 recombined loxHEX in a dose dependent manner with virtually full recombination activity at l-arabinose concentrations of 100 μg/ml or above (Figure 4B). To be able to benchmark the recombination activity of this clone, we compared it to the previously evolved Brec1 recombinase (31). D12 showed comparable activity in this assay, justifying its further investigation. Moreover, we could show that the individual monomers of D12 did not show cross-reactivity on the other respective target sites, nor did they recombine the final loxHEX site (Figure 4C).
Sequencing of the two monomers of D12 (D12-left and D12-right) and deep sequencing of the final libraries revealed the amino acid changes that were acquired during the evolution process on the loxHEX-L or loxHEX-R target sites (Supplementary Figures S6 and S7).
We defined three categories of changes in the amino acid sequences, based on where they occurred. The first group consists of common changes that are found in recombinases evolved on loxHEX-R or loxHEX-L (V7L, P15L, Y77H, S108G, A175S, E262Q, N317T and I320S). These changes are potentially important to stabilize the protein, independent of the nucleotide sequence of the target-site. Indeed, mutations V7L, P15L and Y77H have been suggested to play an important role in protein stability (36).
The second set of changes is exclusively found in the recombinases evolved on loxHEX-R (K43R, M44V, A84T, K86N, G93A, K244R, E266A, A285T, P307A and N319E). The last set is defined as changes that were solely found in the recombinases evolved on loxHEX-L (R243S, N245Y, G263R and T268A). Hence, mutations of the last two categories might play a role in target site selectivity.
Interestingly, some of the changes that only occurred in recombinases for loxHEX-R have previously been found in Brec1 (M44V, K86N, P307A or N319E) (31). Moreover, some of the changes that have been identified in Tre were also found in recombinases evolved for loxHEX-L (N245Y and G263R) (30). These common changes might have evolved due to the partial similarity of loxHEX-R compared to loxBTR and loxHEX-L compared to loxLTR. Therefore, these mutations may provide an entry point for a more rational understanding of how DNA recombination specificity is achieved for designer-recombinases.
Assessment of off-target recombination of D12
Genome editing tools should ideally modify their target sequence without altering the genome at any other position. To identify possible off-target sequences in the human genome, we performed a bioinformatic analysis to classify sequences with the highest degree of similarity to the loxHEX sequences (see Materials and Methods) and experimentally tested the four most similar sequences.
The first off-target site (loxHEX-Off1) is composed of loxHEX on chromosome 7 and another sequence on chromosome 8 with the same spacer, but with 8 asymmetric positions in total (Supplementary Figure S8A). The other off-target sites (loxHEX-Off2, loxHEX-Off3 and loxHEX-Off4) are either similar to the full on-target site or similar to the palindromic loxHEX-L or loxHEX-R sites (Supplementary Figure S8A). We cloned these target sites into the pEVO backbone and expressed the D12 heterodimer in E. coli, followed by extraction and analyses of the plasmid DNA.
We did not detect activity of the D12 heterodimer on loxHEX-Off2 and loxHEX-Off3, and only very weak activity on loxHEX-Off4 (<10%, Supplementary Figure S8B). However, loxHEX-Off1 was visibly recombined, although less efficient than the loxHEX on-target site. Thus, the heterospecific recombinases can recombine other loxHEX-like sequences present in the human genome under the artificial conditions of a plasmid-based assay.
To investigate whether the loxHEX-Off1 sequence is also a potential off-target in the context of the human genome, we developed a PCR-based assay that would reveal recombination events between the loxHEX target sites on chromosome 7 and the loxHEX-Off1 site on chromosome 8. Notably, we could not detect any recombination on genomic DNA after expressing the recombinase heterodimer (Supplementary Figure S8C), indicating that recombination of these two target sites does not take place in human cells at frequencies measurable by PCR.
Dual designer-recombinase induced genomic deletion (dDRiGD) in human cells
For initial testing of D12 in human cells, we transiently co-transfected two recombinase expression plasmids (each expressing one recombinase of D12) together with a recombination-reporter plasmid into HeLa cells. The reporter plasmid constitutively expresses red fluorescent protein (mCherry) after recombination, whereas enhanced green fluorescent protein (EGFP) is co-expressed together with the recombinases (Figure 5A). Cells transfected with the empty expression plasmid and the reporter plasmid did not reveal any mCherry positive cells 48 h after transfection (Figure 5B). In contrast, co-transfection of D12 with the reporter plasmid resulted in double positive cells for GFP and mCherry (Figure 5B). Quantification by flow cytometry revealed that ∼60% of the transfected cells had recombined the reporter plasmid after 48 h (Supplementary Figure S9). This result demonstrates that two recombinases can form active heterodimers in human cells, and that this complex recombines its target in a transient co-transfection experiment.
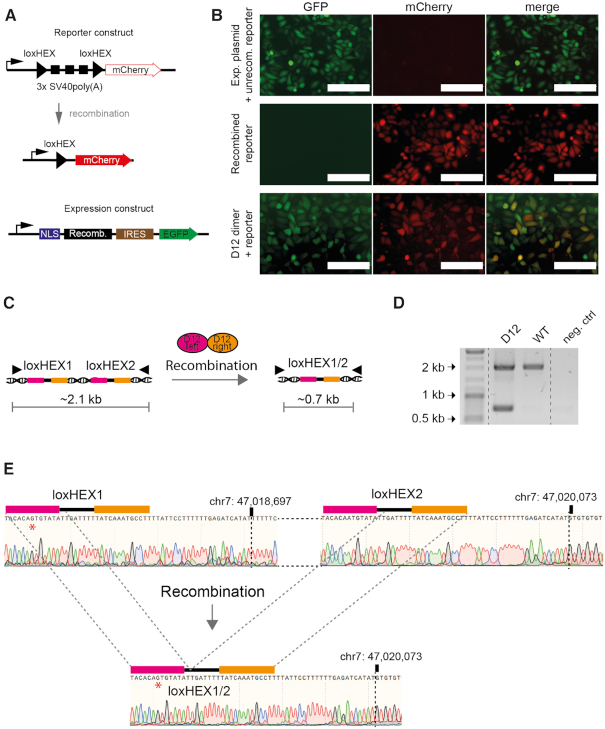
Figure 5.
Recombinase heterodimers are active in mammalian cells. (A) Graphic presentation of the mammalian reporter and expression plasmids. Upon recombination of the reporter construct the 3xSV40poly(A) will be excised allowing for the expression of mCherry (red). Important elements in the reporter and recombinase expression vectors are indicated. Black triangles represent loxHEX target-sites. NLS, nuclear localization signal; Recomb., recombinase coding sequence; IRES, internal ribosome entry site. (B) Fluorescence microscopy images of HeLa cells transfected with the empty expression plasmid and nonrecombined reporter plasmid (top panel), the recombined reporter control (middle panel) or co-transfection of the reporter with the D12 recombinase expression plasmids (lower panel) are shown. Scale bars represent 200 μm. (C) Graphical scheme of the recombination event on genomic DNA. Filled triangles in the illustration represent primer binding sites. After PCR on genomic DNA the nonrecombined locus will be amplified as a ∼2.1 kb fragment whereas the recombined locus will result in a ∼0.7 kb fragment. (D) The D12 heterodimer recombines the endogenous loxHEX locus in human cells. PCR analysis on genomic DNA isolated from cells transfected with D12 and nontransfected control cells are shown. Neg. ctrl. = water control. (E) Sanger-sequencing reads of the WT and recombined PCR fragment on the genomic DNA obtained from WT HeLa cells or transfected HeLa cells with the D12 heterodimer are shown. A repeated region of 23 bp after loxHEX1 and loxHEX2 is indicated. The exact positions on chromosome 7 where the WT differs from the recombined locus is marked with a dotted line. The red asterisk indicates a single nucleotide difference in the loxHEX1 site compared to the loxHEX2 site. A deletion of 1376 bp is observed and demonstrates the precise excision between the two loxHEX target sites.
In order to investigate if the D12 heterodimer can also recombine the endogenous loxHEX sites located on human chromosome 7 (Figure 5C), the plasmids containing D12 were co-transfected into HeLa cells. Forty-eight hours post transfection, genomic DNA was extracted and used in a PCR reaction with primers flanking the two loxHEX sites. As expected, the PCR reaction amplified a 2.1 kb fragment in the non-treated control. In contrast, a 2.1 kb and a 0.7 kb fragment were amplified in cells that had been transfected with D12, indicating that the recombinases had excised a ∼1.4 kb fragment from the genome (Figure 5D). A semi-quantitative PCR assay revealed that ∼30% of the transfected cells had excised the 1.4 kb fragment (Supplementary Figure S10) from the HeLa genome, demonstrating robust recombination efficiency of D12. Moreover, DNA sequencing of the ∼0.7 kb PCR fragment indeed confirmed the exact genomic modification between the loxHEX sites (Figure 5E). To test whether D12 is active in another human cell type, using a different delivery method, we transfected D12 mRNAs into human induced pluripotent stem (iPS) cells. Using the same PCR assay, we observed the characteristic ∼2.1 kb and ∼0.7 kb DNA fragments as in HeLa cells, demonstrating that the recombinases are active in primary stem cells and that mRNA transfection is a productive method to induce the genomic deletion in these cells (Supplementary Figure S10A). Overall, these results demonstrate that co-expression of two recombinases result in the desired and precise excision of the genomic DNA fragment, providing proof-of-concept that dual designer-recombinase induced genomic deletion (dDRiGD) is a feasible approach to delete a defined locus from the human genome.
Bioinformatic analysis of loxP-like sites in human protein-coding genes
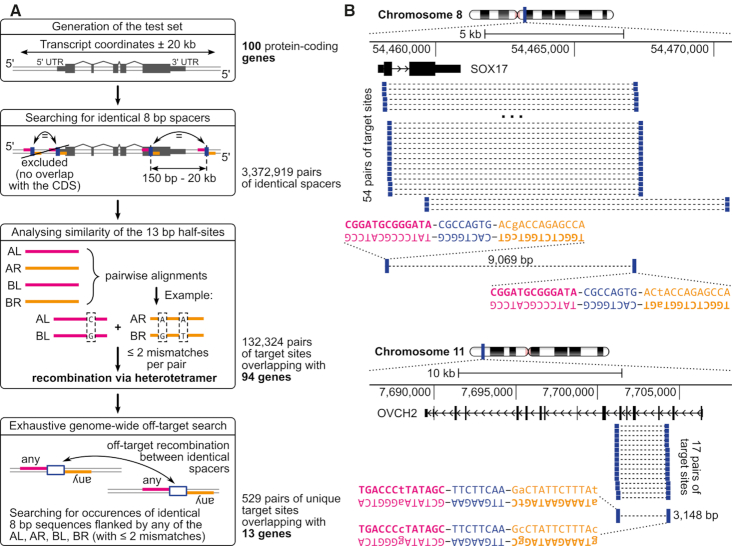
Based on the proof-of-concept that dDRiGD is a possible way to delete native genomic fragments from the human genome, we wanted to know how many protein-coding genes might be targetable by this approach. Considering the computational cost of the off-target analysis, we decided to focus on a representative set of 100 protein-coding genes to predict the percentage of genes that could be targeted by dDRiGD. To generate the test set, we took all protein-coding genes annotated by the HAVANA team (http://www.sanger.ac.uk/HGP/havana/) that are also members of the Consensus Coding Sequence (CCDS) set (52). For each gene, we extracted the main functional isoform by using transcript annotations from the APPRIS database (53). This analysis created a gene-set of 16 864 transcripts. We sorted the list by the coding sequence (CDS) length and selected every 168th gene. As a result, we obtained a test set composed of 100 transcripts, representing the size-differences of human protein-coding genes. We applied our computational pipeline to genomic sequences enclosing the selected genes and identified over 3 million spacer pairs intersecting the coding sequences (Figure 6). 132 324 putative target sites for dual recombinases were classified that differ at no more than two positions per half-site. After testing all pairs of target sites for potential off-targets (see Materials and Methods), we obtained a final set of 529 target-pairs overlapping with 13 protein-coding genes (Figure 6 and Supplementary Table S5). Hence, these genes could likely be targeted by dDRiGD. Extrapolating to the full gene set, this result suggests that with the current parameters around 13% of all protein-coding genes could be conditionally inactivated by the generation of two designer recombinases.
Figure 6.
Evaluation of 100 protein-coding genes in respect to a possibility of performing gene knockouts via dDRiGD. (A) The workflow starts with extracting genomic sequences covering transcript coordinates extended by 20 kb in each direction. In the next step, each sequence is scanned for repeats of identical 8 bp sequences (blue bars) and their 13 bp-flanking sequences (in orange and magenta, AL, AR, BL, BR). Pairs that do not flank coding sequences are discarded. By performing pairwise sequence similarity analysis, half-site sequences are checked to ensure that the target sites can be recombined with two SSRs (no more than two mismatches allowed). In the final step, off-target analyses are performed. Count statistics, on the right of the workflow, indicate abundance of target site pairs generated at each step and counts of genes covered by the target sites. (B) Two example protein coding genes amenable for dDRiGD with their nominated target sites are presented. Each example shows a zoom-in on a chromosomal location, with numbers indicating genomic coordinates, accompanied by a scale bar. Black rectangles connected by arrowed lines represent exons and introns, with arrows indicating direction of transcription. All pairs of potential target sites overlapping with coding sequences (thicker sections of the rectangles) are shown as blue bars connected with dashed lines. For each example, one pair is shown in detail with sequences of target sites and the distance in base pairs (bp) between them. Spacer sequences are highlighted in blue, and half-sites to be bound by the same recombinase are shown in either magenta or orange.
DISCUSSION
The interest in precision genome engineering has considerably increased in the last two decades (54,55). The majority of the work in this area has focused on nuclease-based genome manipulation where specific DNA breaks are introduced at the genomic position of interest. These DNA strand breaks are subsequently corrected by cellular DNA repair pathways, many of which are error-prone (56,57). While useful for numerous genome engineering exercises, various applications would benefit from genetic alterations that work precisely at the nucleotide level and are independent of the cellular repair machinery. Cre-like recombinases with altered DNA-specificity can be utilized as powerful tools for genome editing that fulfill these requirements (16). However, their generation is currently difficult, labor-intensive and time-consuming (31,58). Moreover, suitable target sites to evolve single recombinases occur very rarely in eukaryotic genomes, making this approach ineffective.
With this study we expand the potential of evolved recombinases to remove natural sequences found in the human genome by developing dual designer-recombinase induced genomic deletion (dDRiGD), an approach that operates with two different recombinases. An important development from our work is that Cre-like recombinases, evolved separately to recombine symmetric target sites, can be combined into an active heterodimer to specifically recombine a genomic asymmetric target site. The previous finding that Cre WT together with a Cre mutant is able to recombine an asymmetric loxP-like site indicated that the wild-type enzyme can function as a heterodimer on an artificial target site (51,59,60). Our work demonstrates that two separately evolved Cre-based enzymes can also form active heterodimers and that this approach does not only allow recombination of an artificial loxP-like target-site, but that it is able to excise a natural human target sequence in a genomic context. Moreover, we demonstrate that each single recombinase from the heterodimer is specific to its target site and only recombines the final target in conjunction with the partner recombinase.
High efficiency and low off-target effects are desirable features for any genome editing technology. Without further optimization, the evolved D12 heterodimer was able to delete the desired genomic DNA in ∼30% of cells within 48 h, demonstrating robust recombination efficacy. In terms of specificity we found that D12 can recombine other sequences from the human genome in a plasmid-based assay in bacteria. Hence, the D12 heterodimer is not absolutely specific to its target sequence. Off-target recombination at these sites was not detectable in a genomic context when the recombinases were expressed in cultured human cells, likely because recombinases did not form a functional tetrameric complex on the two target sites for recombination to occur. The presence of the off-target (loxHEX-Off1) sequence on a different chromosome possibly prevented recombination at measurable frequencies. Nevertheless, if therapeutic use of the designer recombinases is envisioned, the highest possible specificity is needed to avoid unwanted alterations of the genome. We did not optimize the described recombinases further, because they do not target a sequence that is therapeutically relevant. However, it is known that high levels of Cre WT expression can result in undesired DNA alterations and have anti-proliferative effects in mammalian cells (61). In order to reduce theses genotoxic effects several methods to improve specificity of SSRs have been described, including counter-selection, generation of obligate heterotetramers and introduction of mutations that improve recombination accuracy (31,60,62,63). Therefore, off-target activity can be addressed with different computational and experimental strategies. Ultimately, off-target activity should be assessed in an advanced and humanized in vivo model to minimize potential side effects.
Being able to use two designer recombinases as heterodimers offers the opportunity to identify more potential target sites in genomes compared to single SSRs. Calculations for single designer recombinases revealed that only 14 suitable target sites could be identified in the human genome, of which merely four target sites would be amenable for the inactivation of a protein-coding gene. In contrast, we predict that 13% (2,192) of human protein-coding genes could be conditionally inactivated with two designer-recombinases applying the same parameters. Relaxing our parameters to identify more potential target sites would substantially increase the number of targetable genes. This could be achieved by allowing more than two asymmetries per half-site, expanding the number of individual recombinase monomers or increasing the search distance. However, relaxing the parameters could go at the cost of efficacy and/or specificity of the recombinases. Further experiments are necessary to define the best parameters that reveal the maximal number of target sites in the human genome without compromising activity and/or specificity of the designer recombinases.
In the long run, we envision that it will be possible to create novel Cre-like recombinases for genomic target sites in a faster time frame. In this study we developed improved protocols for the evolution process that reduced the time per evolution cycle by 50%. Additionally, we have presented a strategy to identify critical residues in evolved recombinases that are important for target site recognition. We showed that it is possible to mutate a library of recombinases simultaneously at 12 of these critical positions resulting in immediate activity on the desired target site. Taken together, the new evolution protocols and strategies to hypermutate specific amino acid positions will make designer recombinases more competitive compared to other genome engineering methods. Nonetheless, engineering the genome with designer recombinases will likely remain more laborious than genome editing with CRISPR/Cas systems. However, the precision during the genome editing process and the versatility of possible recombination reactions, such as excision, inversion and cassette exchange, represent important advantages over non-recombinase-based approaches (64–67). Most certainly, adding dual designer-recombinase induced genomic deletions (dDRiGD) to the spectrum of the Cre/loxP system will substantially broaden its utility. Moreover, if one could demonstrate that dDRiGD or other dual recombinase-based approaches are safe and efficient in whole organisms, they would qualify as potent approaches for future gene and cell therapies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Buchholz laboratory for fruitful discussions. We thank Jenna Hoersten for her careful and critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union [ERC 742133, H2020 UPGRADE 825825]; German Research Foundation [DFG BU 1400/7-1]; BMBF GO-Bio [031B0633]. Funding for open access charge: BMBF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gaj T., Gersbach C.A., Barbas C.F.. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013; 31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Komor A.C., Badran A.H., Liu D.R.. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017; 168:20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cho S.W., Kim S., Kim J.M., Kim J.-S.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013; 31:230–232. [DOI] [PubMed] [Google Scholar]
- 4. Sfeir A., Symington L.S.. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jasin M., Rothstein R.. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013; 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. et al.. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018; 24:939–946. [DOI] [PubMed] [Google Scholar]
- 10. Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J.. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018; 24:927–930. [DOI] [PubMed] [Google Scholar]
- 11. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amabile A., Migliara A., Capasso P., Biffi M., Cittaro D., Naldini L., Lombardo A.. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell. 2016; 167:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A. et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; doi:10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meinke G., Bohm A., Hauber J., Pisabarro M.T., Buchholz F.. Cre recombinase and other tyrosine recombinases. Chem. Rev. 2016; 116:12785–12820. [DOI] [PubMed] [Google Scholar]
- 17. Saleh-Gohari N., Helleday T.. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004; 32:3683–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yeh W.-H., Chiang H., Rees H.A., Edge A.S.B., Liu D.R.. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 2018; 9:2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sternberg N., Hamilton D.. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 1981; 150:467–486. [DOI] [PubMed] [Google Scholar]
- 21. Hoess R.H., Ziese M., Sternberg N.. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:3398–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo F., Gopaul D.N., Van Duyne G.D.. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997; 389:40–46. [DOI] [PubMed] [Google Scholar]
- 23. Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1987; 7:2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sauer B., Henderson N.. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990; 2:441–449. [PubMed] [Google Scholar]
- 25. Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998; 14:381–392. [DOI] [PubMed] [Google Scholar]
- 26. Thummel R., Burket C.T., Brewer J.L., Sarras M.P., Li L., Perry M., McDermott J.P., Sauer B., Hyde D.R., Godwin A.R.. Cre-mediated site-specific recombination in zebrafish embryos. Dev. Dyn. 2005; 233:1366–1377. [DOI] [PubMed] [Google Scholar]
- 27. Gilbertson L. Cre-lox recombination: Cre-ative tools for plant biotechnology. Trends Biotechnol. 2003; 21:550–555. [DOI] [PubMed] [Google Scholar]
- 28. Buchholz F., Stewart A.F.. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001; 19:1047–1052. [DOI] [PubMed] [Google Scholar]
- 29. Santoro S.W., Schultz P.G.. Directed evolution of the site specificity of Cre recombinase. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:4185–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarkar I., Hauber I., Hauber J., Buchholz F.. HIV-1 Proviral DNA excision using an evolved recombinase. Science. 2007; 316:1912–1915. [DOI] [PubMed] [Google Scholar]
- 31. Karpinski J., Hauber I., Chemnitz J., Schäfer C., Paszkowski-Rogacz M., Chakraborty D., Beschorner N., Hofmann-Sieber H., Lange U.C., Grundhoff A. et al.. Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat. Biotechnol. 2016; 34:401–409. [DOI] [PubMed] [Google Scholar]
- 32. Hauber I., Hofmann-Sieber H., Chemnitz J., Dubrau D., Chusainow J., Stucka R., Hartjen P., Schambach A., Ziegler P., Hackmann K. et al.. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS Pathog. 2013; 9:e1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buchholz F., Hauber J.. In vitro evolution and analysis of HIV-1 LTR-specific recombinases. Methods. 2011; 53:102–109. [DOI] [PubMed] [Google Scholar]
- 34. Hermann M., Stillhard P., Wildner H., Seruggia D., Kapp V., Sánchez-Iranzo H., Mercader N., Montoliu L., Zeilhofer H.U., Pelczar P.. Binary recombinase systems for high-resolution conditional mutagenesis. Nucleic Acids Res. 2014; 42:3894–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baldwin E.P., Martin S.S., Abel J., Gelato K.A., Kim H., Schultz P.G., Santoro S.W.. A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water. Chem. Biol. 2003; 10:1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abi-Ghanem J., Chusainow J., Karimova M., Spiegel C., Hofmann-Sieber H., Hauber J., Buchholz F., Pisabarro M.T.. Engineering of a target site-specific recombinase by a combined evolution- and structure-guided approach. Nucleic Acids Res. 2013; 41:2394–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meinke G., Karpinski J., Buchholz F., Bohm A.. Crystal structure of an engineered, HIV-specific recombinase for removal of integrated proviral DNA. Nucleic Acids Res. 2017; 45:9726–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herman A., Tawfik D.S.. Incorporating synthetic oligonucleotides via gene reassembly (ISOR): a versatile tool for generating targeted libraries. Protein Eng. Des. Sel. 2007; 20:219–226. [DOI] [PubMed] [Google Scholar]
- 39. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Slater G.S.C., Birney E.. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005; 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quinlan A.R. BEDTools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014; 47:doi:10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N. et al.. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019; 47:D853–D858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klöckner A., Pinto N., Lee Y., Catanzaro B., Ivanov P., Fasih A.. PyCUDA and PyOpenCL: a scripting-based approach to GPU run-time code generation. Parallel Comput. 2012; 38:157–174. [Google Scholar]
- 45. van der Walt S., Colbert S.C., Varoquaux G.. The NumPy Array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011; 13:22–30. [Google Scholar]
- 46. Payseur B.A., Jing P., Haasl R.J.. A genomic portrait of human microsatellite variation. Mol. Biol. Evol. 2011; 28:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vergnaud G., Denoeud F.. Minisatellites: mutability and genome architecture. Genome Res. 2000; 10:899–907. [DOI] [PubMed] [Google Scholar]
- 48. de Koning A.P.J., Gu W., Castoe T.A., Batzer M.A., Pollock D.D.. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011; 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ringrose L., Chabanis S., Angrand P.O., Woodroofe C., Stewart A.F.. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 1999; 18:6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uemura M., Niwa Y., Kakazu N., Adachi N., Kinoshita K.. Chromosomal manipulation by site-specific recombinases and fluorescent protein-based vectors. PLoS One. 2010; 5:e9846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saraf-Levy T., Santoro S.W., Volpin H., Kushnirsky T., Eyal Y., Schultz P.G., Gidoni D., Carmi N.. Site-specific recombination of asymmetric lox sites mediated by a heterotetrameric Cre recombinase complex. Bioorg. Med. Chem. 2006; 14:3081–3089. [DOI] [PubMed] [Google Scholar]
- 52. Farrell C.M., O’Leary N.A., Harte R.A., Loveland J.E., Wilming L.G., Wallin C., Diekhans M., Barrell D., Searle S.M.J., Aken B. et al.. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 2014; 42:D865–D872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodriguez J.M., Maietta P., Ezkurdia I., Pietrelli A., Wesselink J.-J., Lopez G., Valencia A., Tress M.L.. APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013; 41:D110–D117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018; 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carroll D. Genome editing: past, present, and future. Yale J. Biol. Med. 2017; 90:653–659. [PMC free article] [PubMed] [Google Scholar]
- 56. Bibikova M., Golic M., Golic K.G., Carroll D.. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002; 161:1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bogdanove A.J., Bohm A., Miller J.C., Morgan R.D., Stoddard B.L.. Engineering altered protein-DNA recognition specificity. Nucleic Acids Res. 2018; 46:4845–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gelato K.A., Martin S.S., Liu P.H., Saunders A.A., Baldwin E.P.. Spatially directed assembly of a heterotetrameric cre-lox synapse restricts recombination specificity. J. Mol. Biol. 2008; 378:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang C., Myers C.A., Qi Z., Mitra R.D., Corbo J.C., Havranek J.J.. Redesign of the monomer-monomer interface of Cre recombinase yields an obligate heterotetrameric complex. Nucleic Acids Res. 2015; 43:9076–9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Loonstra A., Vooijs M., Beverloo H.B., Allak B.A., van Drunen E., Kanaar R., Berns A., Jonkers J.. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carlson J.C., Badran A.H., Guggiana-Nilo D.A., Liu D.R.. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 2014; 10:216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eroshenko N., Church G.M.. Mutants of Cre recombinase with improved accuracy. Nat. Commun. 2013; 4:2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schlake T., Bode J.. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994; 33:12746–12751. [DOI] [PubMed] [Google Scholar]
- 65. Turan S., Zehe C., Kuehle J., Qiao J., Bode J.. Recombinase-mediated cassette exchange (RMCE) — a rapidly-expanding toolbox for targeted genomic modifications. Gene. 2013; 515:1–27. [DOI] [PubMed] [Google Scholar]
- 66. Voziyanova E., Malchin N., Anderson R.P., Yagil E., Kolot M., Voziyanov Y.. Efficient Flp-Int HK022 dual RMCE in mammalian cells. Nucleic Acids Res. 2013; 41:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Voziyanova E., Anderson R.P., Voziyanov Y.. Dual recombinase-mediated cassette exchange by tyrosine site-specific recombinases. Methods Mol. Biol. 2017; 1642:53–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.