Abstract
Background. This study was performed to evaluate the associations of newly recognized viruses, namely, human metapneumovirus (hMPV), human coronavirus (HCoV)–NL63, and human bocavirus (HBoV) with lower respiratory tract infections (LRTIs) in previously healthy children.
Methods. To determine the prevalences of 11 viruses—respiratory syncytial virus (RSV), adenovirus, rhinovirus, parainfluenza viruses (PIVs) 1 and 3, influenza viruses A and B, hMPV, HCoV, HCoV-NL63, and HBoV—among infants or children with LRTIs, in association with their epidemiologic characteristics, we performed multiplex reverse-transcriptase polymerase chain reaction on nasopharyngeal aspirates obtained from 515 children 5 years old with LRTIs during the period 2000–2005.
Results. Viruses were identified in 312 (60.6%) of the 515 patients. RSV was detected in 122 (23.7%), HBoV in 58 (11.3%), adenovirus in 35 (6.8%), PIV-3 in 32 (6.2%), rhinovirus in 30 (5.8%), hMPV in 24 (4.7%), influenza A in 24 (4.7%), PIV-1 in 9 (1.7%), influenza B in 9 (1.7%), and HCoV-NL63 in 8 (1.6%). Coinfections with 2 viruses were observed in 36 patients (11.5%). Twenty-two patients (37.9%) infected with HBoV had a coinfection. Bronchiolitis was frequently diagnosed in patients who tested positive for RSV, PIV-3, or rhinovirus, whereas influenza A, PIV-1, and HCoV-NL63 were commonly found in patients with croup. The age distributions of patients with viral infections differed; notably, RSV was responsible for 77% of LRTIs that occurred in infants 3 months old. The number of hMPV infections peaked between February and April, whereas the number of HCoV-NL63 infections peaked between April and May.
Conclusions. This study describes the features of LRTIs associated with newly identified viruses in children, compared with those associated with known viruses. Additional investigations are required to define the role of HBoV in LRTI.
Acute viral respiratory illnesses are major health problems in infants and children. Traditionally, respiratory syncytial virus (RSV), parainfluenza virus (PIV), influenza virus, and adenovirus have been viewed as being the leading causes of acute viral lower respiratory tract infections (LRTIs) [1–4].
However, in addition to previously known viruses, many respiratory viruses have been recently identified as causative agents of lower respiratory illnesses in children. Rhinovirus, which generally has been considered a cause of mild upper respiratory illnesses in children and adults, is now considered to be a major cause of acute LRTIs and asthma exacerbations [5–7]. However, the extent to which rhinoviruses contribute to LRTIs in otherwise healthy children is unclear. More recently, new viruses, such as human metapneumovirus (hMPV) [8] and human coronavirus (HCoV)–NL63 [9, 10], have been suggested to cause LRTIs in children. Moreover, human bocavirus (HBoV) has been identified in the respiratory tracts by molecular screening [11], but its role as a causative agent of LRTI remains to be proven.
To determine the relative contributions made by these newly recognized respiratory viruses to LRTIs in childhood and to characterize their epidemiologic and clinical features, over a period of 5 years (2000–2005) we performed virological studies of Korean infants and children with acute LRTIs. Specifically, we evaluated the relative prevalences and the epidemiologic characteristics of rhinovirus, hMPV, conventional HCoV, HCoV-NL63, and HBoV and compared these findings with those of common respiratory viruses, such as RSV, PIV, adenovirus, and influenza virus.
Patients, Materials, and Methods
Patients and respiratory specimens. The study population consisted of children 5 years old with acute LRTIs (i.e., bronchiolitis, pneumonia, or croup). All illnesses were diagnosed at the Seoul National University Children's Hospital (Korea) or the Seoul National University Bundang Hospital (Korea) between September 2000 and August 2005. Samples obtained from children with major risk factors other than recurrent episodes of wheezing were excluded, as were those obtained from children with hospital-related infections. Diagnostic definitions were as follows [12, 13]: pneumonia required rales on auscultation or demonstration of an infiltrate by chest X-ray; bronchiolitis was characterized by a cough, tachypnea, retraction, and expiratory wheezes, often accompanied by rales; and croup required a barking cough with stridor. The principal investigator determined the clinical diagnosis on the basis of a review of medical records conducted by 5 of the investigators. The predominant clinical diagnosis was determined when >1 diagnosis was present.
Nasopharyngeal aspirates were prospectively collected from all subjects. Specimens were obtained either at the time of visiting an emergency department or immediately following hospital admission. Viral RNA was detected in nasopharyngeal aspirates using RT-PCR.
Viral diagnosis. Samples of nasopharyngeal aspirates were kept frozen at -70°C. Viral RNA in nasopharyngeal aspirates was extracted using a QIAamp Viral RNA Mini kit (Qiagen), in accordance with the manufacturer's instructions. cDNA was synthesized using random hexamers and Superscript RT (Invitrogen). Multiplex RT-PCR assays were developed to detect 11 viruses, namely, RSV and PIV-3 (panel 1), hMPV and rhinovirus (panel 2), influenza viruses A and B and PIV-1 (panel 3), coronaviruses OC43 and 229E and HCoV-NL63 (panel 4), and HBoV (panel 5). Adenovirus was detected by culture in HEp-2 monolayers, because this cell culture–based assay has a diagnostic sensitivity similar to that of adenovirus-specific PCR [14].
In brief, 17.5 µL of total RNA was mixed with 1 µL of random hexamers at a concentration of 15 µM and then incubated at 65°C and chilled on ice. A reaction mix of 11.5 µL containing 6 µL of 5x first strand buffer, 3 µL of 100 mM dithiothreitol, 1 µL of deoxyribonucleotide triphosphate mix at a concentration of 10 mM, 1 µL of RNase inhibitor, and 100 units of SuperScript II reverse transcriptase (Invitrogen) was then added. The reaction was incubated at 25°C for 10 min and at 42°C for 60 min, and after a denaturation step at 94°C for 3 min, the cDNA was used as a template for subsequent PCR. The 20-µL reaction mixtures consisted of 1x GeneAmp PCR buffer Gold (Applied Biosystems), 2.0 mM MgCl2, each deoxyribonucleotide triphosphate at a concentration of 0.2 mM, 20 pmol of primers, and 2.5 units of AmpliTaq Gold DNA polymerase (Applied Biosystems). After incubation for 5 min at 95°C, amplication was performed at 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min. The primers used were designed in this study or modified from the published methods [15–19] to amplify the virus-specific genomes (table 1). Amplified products were then separated on agarose gels, and virus-specific PCR products were identified.
Table 1.
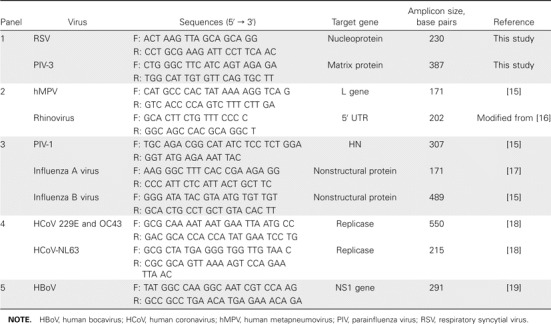
Primer sequences for multiplex RT-PCR.
The sensitivity of the multiplex RT-PCR for 5 viruses (RSV, PIV-1, PIV-3, hMPV, and influenza viruses A and B) was determined for each of the targets using virus suspensions in cell culture media that were quantified in terms of TCID50 using standard virological techniques [20].
All specimens were tested by the multiplex RT-PCR for the 11 respiratory viruses and by viral culture and immunofluorescent antigen detection for 6 viruses (RSV, PIV-1, PIV-3, adenovirus, and influenza viruses A and B), as described elsewhere [3]. Samples were considered to be positive if the 2 PCR results using separately extracted copies of viral RNA were positive or if a single positive PCR result was confirmed by viral culture or immunofluorescent antigen detection methods. Each PCR included distilled water as a negative control, as well as positive controls for the corresponding copies of viral cDNA in each panel.
Clinical database. Respiratory symptoms and signs were recorded on a standardized form during the emergency department visit or while the patient was hospitalized. Medical records were reviewed to determine the clinical manifestations and any underlying conditions of patients. Clinical data were entered into a database by a person who did not have knowledge of virus identities.
Statistical analysis. The γ2 test or Fisher's exact test was used to compare samples with respect to percentage of each sex, symptoms, and the clinical diagnoses, and the Mann-Whitney U test was used to compare the mean age at onset of illness between the various groups. Analyses were performed using GraphPad InStat software, version 3.06 (GraphPad Software).
Results
Patient characteristics. A total of 2198 nasopharyngeal aspirates samples associated with a diagnosis of LRTI in the previously healthy children (5 years old) were collected during the study period. Tested specimens were selected by a random number assignment from the list of archived samples. A total of 515 nonconsecutive specimens (23.4%) were chosen for RT-PCR after excluding unavailable samples and repeated samples collected from the same patient. The monthly proportion of selected samples ranged from 20% to 28% of the total number of archived specimens (figure 1). Of the study population, 306 (59%) were male. The mean age among children was 15.4 months; 20% were infants <4 months old, 28% were 4–12 months old, 25% were 13–24 months old, and 27% were >24 months old. The remaining 1683 samples that were not selected for this study did not differ significantly from selected samples with respect to mean age at disease onset, sex, seasonal distribution, or clinical diagnosis.
Figure 1.
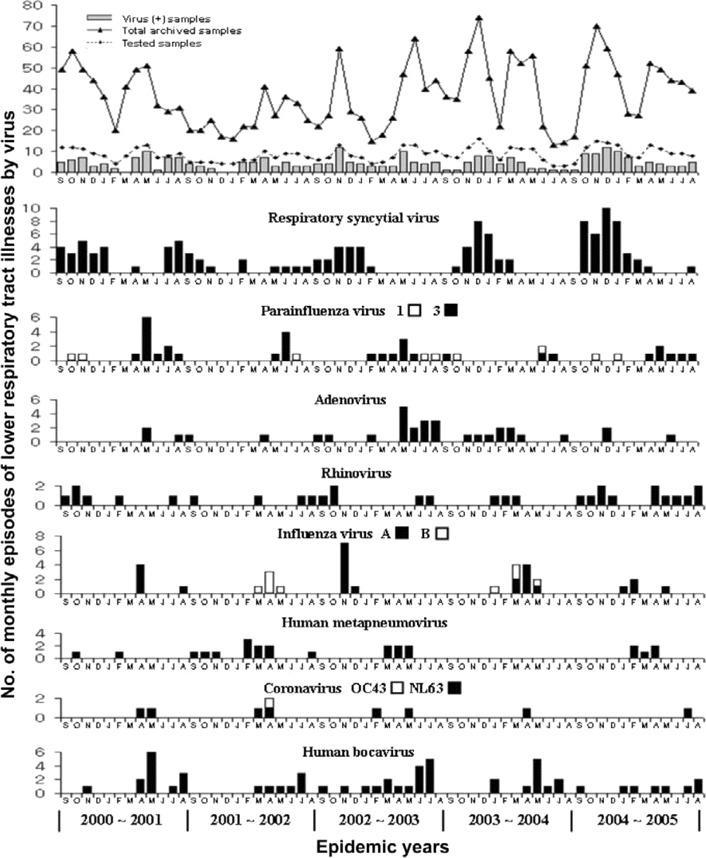
Monthly occurrence of acute lower respiratory tract infections associated with 11 respiratory viruses isolated from children over the 5-year period 2000–2005.
Prevalence of respiratory viruses among children with an LRTI. The detection sensitivity of the assay for 5 viruses (RSV, PIV-1, PIV-3, hMPV, and influenza viruses A and B) was greatest for influenza A, with a value of 10-4 TCID50. Similarly, the assay detected 5 × 10-3 TCID50 of RSV and PIV-3 and 10-3 TCID50 of PIV-1, hMPV, and influenza B. Detectable viral RNA concentrations for rhinovirus, coronaviruses OC43 and 229E, HCoV-NL63, and HBoV were 5–10 ng of the total RNA extracted from the cell culture supernatants or clinical specimens. Of the 515 samples tested, 312 (60.6%) were positive for any of the 11 respiratory viruses (table 2). The detection rate of multiplex RT-PCR was increased because of additional viruses that can be detected by this method and because of greater sensitivity, compared with viral culture or antigen detection methods. Of these 312 virus-positive samples, 36 were also positive for additional viruses, resulting in a coinfection rate of 11.5%. Virus frequencies were as follows: RSV was detected in 122 children (23.7%), HBoV in 58 (11.3%), adenovirus in 35 (6.8%), PIV-3 in 32 (6.2%), rhinovirus in 30 (5.8%), hMPV in 24 (4.7%), influenza A in 24 (4.7%), PIV-1 in 9 (1.7%), influenza B in 9 (1.7%), HCoV-NL63 in 8 (1.6%), and conventional HCoV in 1 (replicase 1a gene sequencing analysis revealed this to be HCoV-OC43). Rhinoviruses were found to be associated with acute LRTI at a relatively high frequency. The prevalence of hMPV was similar to that of influenza virus A. In contrast to these, HCoV-NL63 was present at a low prevalence in the study group.
Table 2.
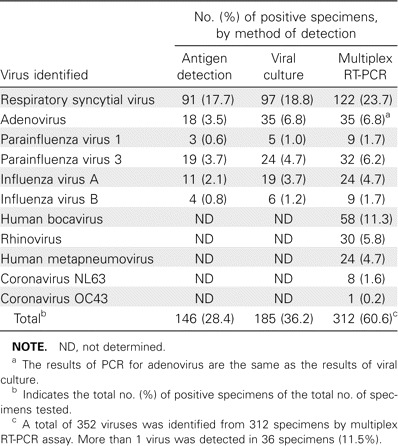
Viruses identified in 515 nasopharyngeal aspirates obtained from previously healthy children with acute lower respiratory tract infections.
Seasonal distribution. The monthly distributions of detected viruses over the 5-year study period are shown in figure 1. Monthly variations in percentage contributions to the diagnosed viral LRTIs ranged from 0% to 88%. The number of RSV infections increased during late fall and peaked between November and January. PIV-3 was prevalent from April to June, and rhinovirus was detected year-round, with a peak occurring during late summer and fall. The prevalence of hMPV increased during late winter and peaked between February and April (68% of total isolates). hMPV was not detected during the 2003–2004 epidemic period, although comparable numbers of samples were tested. HCoV-NL63 was identified, with peaks occurring between April and May (44% of total isolates).
Clinical characteristics. The clinical features of children who were tested for these viruses are summarized in table 3. The age distributions of infants and children associated with each virus differed. LRTIs caused by RSV were predominant among younger infants (mean age, 9 months), compared with LRTIs associated with adenovirus, HBoV, hMPV, or influenza A (P < .01 for each comparison). In particular, a greater proportion of RSV infections occurred in infants 3 months old, compared with other virus-associated LRTIs (P < .04 for all comparisons between RSV and 6 viruses [adenovirus, HBoV, PIV-3, influenza virus A, hMPV, and HCoV-NL63]) (figure 2). In addition, rhinovirus accounted for a larger proportion of LRTIs in young infants 3 months than adenovirus or HBoV (P < .03). In contrast, adenovirus, hMPV, and influenza A were more frequently detected in children >24 months old.
Table 3.
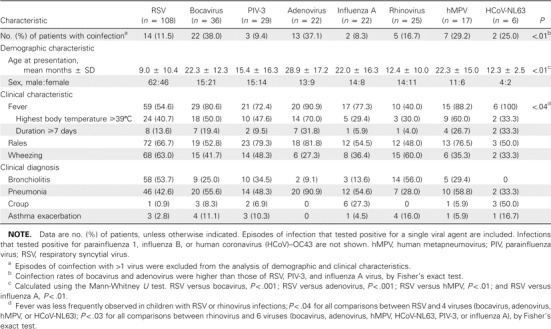
Comparison of the characteristics of the 265 children with acute lower respiratory tract infections with respect to the 8 major respiratory viruses.
Figure 2.
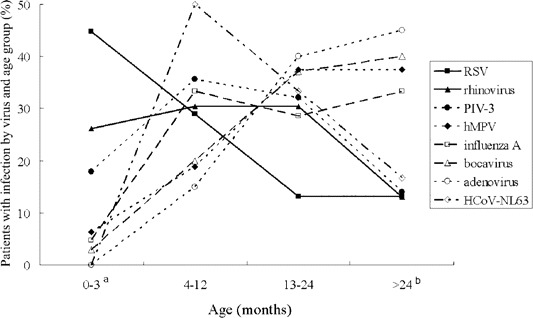
Age distribution of children with lower respiratory tract infections associated with viral agents. The percentage of patients infected with individual viruses is shown for each age group. The sum of the proportion of persons infected with each virus in each of 4 age groups is 100%. aThe proportion of infants 3 months old; P < .04 for all comparisons between respiratory syncytial virus (RSV) and 6 viruses (adenovirus, bocavirus, parainfluenza virus [PIV]–3, influenza virus A, human metapneumovirus [hMPV], or human coronavirus [HCoV]–NL63; P < .03 for rhinovirus versus adenovirus or bocavirus, by Fisher's exact test. bComparison of the proportion of children >24 months old; P < .05 for RSV versus adenovirus, influenza A virus, bocavirus, or hMPV; P < .04 for rhinovirus versus adenovirus or bocavirus; and P < .03 for PIV-3 versus adenovirus or bocavirus, by Fisher's exact test.
The clinical diagnoses of the 265 patients with LRTIs associated with 8 common viruses are summarized in table 3. Bronchiolitis or pneumonia was frequently observed in patients with positive results for RSV, HBoV, PIV-3, hMPV, or rhinovirus. Of these, RSV and rhinovirus were more likely to cause bronchiolitis, whereas by our definition, hMPV showed a tendency to cause pneumonia. Croup was frequently associated with influenza A, PIV-1, or HCoV-NL63. Fever was more frequently observed in hMPV-infected children (88.2%) or HCoV-NL63–infected children (100%) than in those infected with RSV (54%; P < .04 for both comparisons) or rhinovirus (40%; P < .03 for both comparisons). The frequency of rales was similar for all viruses, whereas wheezing occurred more frequently in patients infected with RSV than in patients with an LRTI associated with adenovirus or hMPV. No differences were observed between children with coinfections and single infections with respect to disease severity.
Characteristics of HBoV infection. As shown in table 2, HBoV was detected in 58 children (11.3%). Of these, 22 (37.9%) were found to be coinfected with other viruses (table 2). HBoV was identified in 22 (61%) of 36 patients coinfected with >1 virus. In view of the observation that adenovirus was more frequently found in patients with mixed viral infections (13 [37.1%] of 35 adenovirus-positive samples) than were the other viruses, the above findings indicate that HBoV was associated with high rates of mixed infections. Viruses that were frequently found to be coinfecting with HBoV were as follows, in descending order: adenovirus (7 cases), hMPV (5 cases), RSV (5 cases), and PIV-3 (3 cases). HBoV was identified throughout each year during the 5-year study period, with a peak (57% of total isolates) occurring from May to July, and the mean age of HBoV-infected children was 22.3 months. The clinical diagnoses of 36 HBoV-associated LRTIs were bronchiolitis for 9 patients (25%), pneumonia for 20 patients (55.6%), croup for 3 patients (8.3%), and asthma exacerbation for 4 patients (11.1%).
Discussion
In this study, we evaluated the overall prevalences of 11 respiratory viruses identified by multiplex RT-PCR in a cohort of previously healthy children with acute LRTIs. In addition, we compared the relative contributions and clinical features between the acute LRTIs associated with the recently identified viruses (rhinovirus, hMPV, HCoV-NL63, and HBoV) and those caused by the previously known viruses (RSV, parainfluenza viruses, adenovirus, and influenza viruses) during 5 consecutive years, 2000–2005.
Since both hMPV and HCoV-NL63 were recognized as being the etiologic agents of LRTIs [8, 9, 21–23], comparative studies on the relative prevalences and clinical characteristics of respiratory tract infections caused by newly identified viruses in children have been hampered, primarily because of the different sensitivities of the diagnostic tests employed [24–26]. In addition, the majority of previous studies have used molecular methods to investigate the roles of hMPV and HCoV-NL63 in acute respiratory diseases by using subjects that differ with respect to the number of respiratory specimens tested or results obtained by viral culture or by antigen detection methods [22, 27]. Moreover, simultaneous infections by other viruses are often documented; thus, contributions made by individual viruses to respiratory diseases may be over- or underestimated.
In this study, we used multiplex RT-PCR, which has been widely used to identify recently recognized viruses, such as hMPV, HCoV-NL63, and HBoV, to determine the prevalences of these viruses and to reassess the prevalences of older viruses, such as coronavirus and rhinovirus, among children with LRTIs. Eleven tested viruses were found in 312 (60.6%) of the 515 patients.
The clinical characteristics of patients infected with different viruses were relatively distinct. RSV, rhinovirus, and PIV-3 were frequently observed in the patients with bronchiolitis. In contrast, influenza virus, PIV-1, and HCoV-NL63 were found to be major viral agents of croup. Although previous studies have shown that hMPV is more likely to be associated with bronchiolitis or wheezy bronchitis than pneumonia or croup [27, 28], we found that hMPV was frequently associated with viral pneumonia in this cohort. This observation may reflect differences in diagnostic definitions or study populations between reports; nonetheless, the present study indicates that hMPV is a common virus detected in bronchiolitis patients, because it was found in an additional 3 patients among 4 who were coinfected with other viruses.
Previous studies of acute respiratory tract infections in children have demonstrated detection rates of 0.7%–8.8% for HCoV-NL63 [18, 24, 29–32]. Moreover, although HCoV-NL63 may be associated with mild upper respiratory tract infections, it has also been suggested to contribute to the development of bronchiolitis or croup [30, 33]. The present study indicates that HCoV-NL63 is a less common respiratory pathogen in cases of acute LRTIs in Korean children. In fact, HCoV-NL63 only accounted for 1.6% of cases, and HCoV-OC 43 was rarely found. However, despite the small number of children identified with HCoV-NL63 infection, when it was found, it was frequently associated with croup (3 of 6 cases). Moreover, the proportion of HCoV-NL63–positive croup cases was found to be similar to the proportion of cases caused by influenza virus or PIV-1, which are the 2 most common causes of croup in the fall and winter seasons [3, 34]. These findings suggest that HCoV-NL63 is an important etiologic agent of croup during the spring in Korea.
Our study also has several limitations. It was based on an analysis of samples selected from a list of cases of acute LRTI in a hospital database. Therefore, we could not determine the population-based prevalence. Viruses associated with low prevalence and mild symptoms may not be fully described, either. However, comparable numbers of the archived samples were selected in each study year; thus, we believe that the data obtained in the present study may represent the overall activities of viruses responsible for acute LRTIs and their epidemiologic characteristics. During the 5-year observation period, hMPV infection appeared to be rare in the 1-year period of 2003–2004. This finding suggests that hMPV-associated LRTIs are subject to annual variations, which contrasts with the relatively stable annual incidence of RSV infections [35, 36]. These results may be the consequence of spurious observations affected by the selection bias of this study. However, annual variability has also been demonstrated in other studies of periods of only 2 or 3 years and of archived samples during a longer period [26, 37, 38]. Additional prospective studies are needed to characterize the epidemiologic features of hMPV infection.
HBoV, a potential causative agent of LRTI, demonstrated seasonal periodicity during each study year, with a peak prevalence occurring from May to July. The number of HBoV infections peaked slightly later than the number of PIV-3 infections, and during its peak months, the total prevalence of the other 10 viral agents excluding HBoV was 34.7% (50 cases among 144 nasopharyngeal aspirates), which is lower than the overall detection rate. The detection rate of 11 viruses during the same period increased from 34.7% to 59%, similar to that of the total detection rate of 60.6%. These findings suggest that HBoV is a major viral agent of respiratory episodes during late spring to early summer. However, the issue of a causal relationship remains unresolved. It is noteworthy that the detection of HBoV DNA was associated with a higher rate of coinfection than other viral agents. As demonstrated above, the prevalence of coinfection of HBoV was similar to that of adenovirus, which is frequently observed as a copathogen because of its long shedding period. In 2 previous studies, HBoV has also been found with other agents with prevalences of 17.5% and 55.6% [11, 19]. Thus, HBoV infection might occur incidentally to respiratory infections caused by other viral agents. At the present time, despite the periodicity shown by HBoV, its role as a true pathogen remains uncertain. Additional studies are required to determine its asymptomatic prevalence in the population, pathogenicity, and viral shedding characteristics.
Acknowledgments
Financial support. Korean Ministry of Health and Welfare (grant 0412-BM01-716-0001).
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Henderson FW, Clyde WA, Jr, Collier AM, et al. The etiologic and epidemiologic spectrum of bronchiolitis in pediatric practice. J Pediatr. 1979;95:183–90. doi: 10.1016/s0022-3476(79)80647-2. [DOI] [PubMed] [Google Scholar]
- 2.Hall CB, Walsh EE, Schnabel KC, et al. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162:1283–90. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- 3.Yun BY, Kim MR, Park JY, Choi EH, Lee HJ, Yun CK. Viral etiology and epidemiology of acute lower respiratory tract infections in Korean children. Pediatr Infect Dis J. 1995;14:1054–9. doi: 10.1097/00006454-199512000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Hong JY, Lee HJ, Piedra PA, et al. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin Infect Dis. 2001;32:1423–9. doi: 10.1086/320146. [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos NG, Moustaki M, Tsolia M, et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002;165:1285–9. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- 6.Friedlander SL, Busse WW. The role of rhinovirus in asthma exacerbations. J Allergy Clin Immunol. 2005;116:267–73. doi: 10.1016/j.jaci.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Xatzipsalti M, Kyrana S, Tsolia M, et al. Rhinovirus viremia in children with respiratory infections. Am J Respir Crit Care Med. 2005;172:1037–40. doi: 10.1164/rccm.200502-315OC. [DOI] [PubMed] [Google Scholar]
- 8.van den Hoogen BG, de Jong JC, Groen J, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier RA, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102:12891–6. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIntosh K, Chao RK, Krause HE, Wasil R, Mocega HE, Mufson MA. Coronavirus infection in acute lower respiratory tract disease of infants. J Infect Dis. 1974;130:502–7. doi: 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny FW, Clyde WA., Jr Acute lower respiratory tract infections in nonhospitalized children. J Pediatr. 1986;108:635–46. doi: 10.1016/s0022-3476(86)81034-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee JA, Kim NH, Kim SJ, Choi EH, Lee HJ. Rapid identification of human adenovirus types 3 and 7 from respiratory specimens via multiplex type-specific PCR. J Clin Microbiol. 2005;43:5509–14. doi: 10.1128/JCM.43.11.5509-5514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruteke P, Glas AS, Dierdorp M, Vreede WB, Pilon JW, Bruisten SM. Practical implementation of a multiplex PCR for acute respiratory tract infections in children. J Clin Microbiol. 2004;42:5596–603. doi: 10.1128/JCM.42.12.5596-5603.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulos NG, Hunter J, Sanderson G, Meyer J, Johnston SL. Rhinovirus identification by BglI digestion of picornavirus RT-PCR amplicons. J Virol Methods. 1999;80:179–85. doi: 10.1016/S0166-0934(99)00045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Class EC, Sprenger MJ, Kleter GE, van Beek R, Quint WG, Masurel N. Type-specific identification of influenza viruses A, B, and C by the polymerase chain reaction. J Virol Methods. 1992;39:1–13. doi: 10.1016/0166-0934(92)90120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis. 2005;191:492–8. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karber G. 50% Endpoint calculation. Arch Exp Pathol Pharmak. 1931;162:480–3. [Google Scholar]
- 21.Boivin G, De Serres G, Cote S, et al. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis. 2003;9:634–40. doi: 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAdam AJ, Hasenbein ME, Feldman HA, et al. Human metapneumovirus in children tested at a tertiary-care hospital. J Infect Dis. 2004;190:20–6. doi: 10.1086/421120. [DOI] [PubMed] [Google Scholar]
- 23.Arden KE, Nissen MD, Sloots TP, Mackay IM. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005;75:455–62. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu SS, Chan KH, Chu KW, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–9. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson JL, Lee BE, Bastien N, Li Y. Seasonality and clinical features of human metapneumovirus infection in children in Northern Alberta. J Med Virol. 2005;76:98–105. doi: 10.1002/jmv.20329. [DOI] [PubMed] [Google Scholar]
- 26.Kim YK, Lee HJ. Human metapneumovirus-associated lower respiratory tract infections in Korean infants and young children. Pediatr Infect Dis J. 2005;24:1111–2. doi: 10.1097/01.inf.0000190042.65120.23. [DOI] [PubMed] [Google Scholar]
- 27.Williams JV, Harris PA, Tollefson SJ, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350:443–50. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebihara T, Endo R, Kikuta H, et al. Human metapneumovirus infection in Japanese children. J Clin Microbiol. 2004;42:126–32. doi: 10.1128/JCM.42.1.126-132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebihara T, Endo R, Ma X, Ishiguro N, Kikuta H. Detection of human coronavirus NL63 in young children with bronchiolitis. J Med Virol. 2005;75:463–5. doi: 10.1002/jmv.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastien N, Robinson JL, Tse A, Lee BE, Hart L, Li Y. Human coronavirus NL-63 infections in children: a 1-year study. J Clin Microbiol. 2005;43:4567–73. doi: 10.1128/JCM.43.9.4567-4573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki A, Okamoto M, Ohmi A, Watanabe O, Miyabayashi S, Nishimura H. Detection of human coronavirus-NL63 in children in Japan. Pediatr Infect Dis J. 2005;24:645–6. doi: 10.1097/01.inf.0000168846.71517.ee. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser L, Regamey N, Roiha H, Deffernez C, Frey U. Human coronavirus NL63 associated with lower respiratory tract symptoms in early life. Pediatr Infect Dis J. 2005;24:1015–7. doi: 10.1097/01.inf.0000183773.80217.12. [DOI] [PubMed] [Google Scholar]
- 33.van der Hoek L, Sure K, Ihorst G, et al. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:e240. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny FW, Murphy TF, Clyde WA, Jr, Collier AM, Henderson FW. Croup: an 11-year study in a pediatric practice. Pediatrics. 1983;71:871–6. [PubMed] [Google Scholar]
- 35.Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001;344:1917–28. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 36.Choi EH, Lee HJ. Genetic diversity and molecular epidemiology of the G protein of subgroups A and B of respiratory syncytial viruses isolated over 9 consecutive epidemics in Korea. J Infect Dis. 2000;181:1547–56. doi: 10.1086/315468. [DOI] [PubMed] [Google Scholar]
- 37.Williams JV, Wang CK, Yang CF, et al. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J Infect Dis. 2006;193:387–95. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galiano M, Videla C, Puch SS, Martinez A, Echavarria M, Carballal G. Evidence of human metapneumovirus in children in Argentina. J Med Virol. 2004;72:299–303. doi: 10.1002/jmv.10536. [DOI] [PubMed] [Google Scholar]


