Abstract
Lung adenocarcinoma is one of the most common malignant tumors worldwide. Although efforts have been made to clarify its pathology, the underlying molecular mechanisms of lung adenocarcinoma are still not clear. The microarray datasets GSE75037, GSE63459 and GSE32863 were downloaded from the Gene Expression Omnibus (GEO) database to identify biomarkers for effective lung adenocarcinoma diagnosis and therapy. The differentially expressed genes (DEGs) were identified by GEO2R, and function enrichment analyses were conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO). The STRING database and Cytoscape software were used to construct and analyze the protein-protein interaction network (PPI). We identified 376 DEGs, consisting of 83 upregulated genes and 293 downregulated genes. Functional and pathway enrichment showed that the DEGs were mainly focused on regulation of cell proliferation, the transforming growth factor β receptor signaling pathway, cell adhesion, biological adhesion, and responses to hormone stimulus. Sixteen hub genes were identified and biological process analysis showed that these 16 hub genes were mainly involved in the M phase, cell cycle phases, the mitotic cell cycle, and nuclear division. We further confirmed the two genes with the highest node degree, DNA topoisomerase IIα (TOP2A) and aurora kinase A (AURKA), in lung adenocarcinoma cell lines and human samples. Both these genes were upregulated and associated with larger tumor size. Upregulation of AURKA in particular, was associated with lymphatic metastasis. In summary, identification of the DEGs and hub genes in our research enables us to elaborate the molecular mechanisms underlying the genesis and progression of lung adenocarcinoma and identify potential targets for the diagnosis and treatment of lung adenocarcinoma.
Keywords: lung adenocarcinoma, TOP2A, AURKA, biomarker, bioinformatics, human samples
Introduction
Non-small cell lung cancer (NSCLC) is the most common malignant tumor globally and is associated with an extremely high mortality rate (1). The incidence of NSCLC continues to surge globally. As with other tumors, lung adenocarcinoma which accounts for about 35–45% of all lung malignant tumors is a heterogeneous disease characterized by high rates of genetic mutation (2). Despite the emergence of diverse new approaches for the treatment of lung adenocarcinoma, such as targeted and immune therapy, long term survival is still poor (3,4). One of the main reasons for this is that most patients are diagnosed at an advanced stage. Thus, it is necessary to understand the molecular mechanisms behind lung adenocarcinoma genesis, growth and progression, and identify biomarkers that can be detected during the early stages of the disease.
Recently, high-throughput bioinformatic technologies such as microarrays have been widely used to screen for differentially expressed genes (DEGs) and identify the functional pathways involved in the genesis and development of lung adenocarcinoma. However, the reliable results are not easy to obtain due to the false-positive rates that may exist in every independent microarray analysis. Thus, we downloaded three original mRNA data sets (GSE32863, GSE63459 and GSE75037) from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and aimed to identify the DEGs between normal lung and lung adenocarcinoma tissues. Next, the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/) and Gene Ontology (GO; http://www.geneontology.org) databases were used to identify biological processes enriched in DEGs, and integrated protein-protein interaction (PPI) network analysis was used to help us understand the molecular mechanisms underlying lung adenocarcinoma genesis and development. Sixteen hub genes and 376 DEGs were identified, which could be potential target genes and candidate biomarkers for lung adenocarcinoma. To minimize the false-positive rate of the microarray analysis, the results were then confirmed in cell lines and human sample tissues.
Materials and methods
Microarray data
GEO is a public functional genomics data repository supporting MIAME-compliant data submissions. It accepts sequence based and array data. Tools are provided to help users query and download experiments and curated gene expression profiles (5). The GSE32863, GSE63459 and GSE75037 datasets produced by the Illumina HumanWG-6/Ref-8 v3.0 expression beadchip platform (Illumina Inc; http://www.illumina.com) were downloaded for further analysis. The GSE63459 dataset contains data from 33 lung adenocarcinoma tissue samples and 32 adjacent normal tissue samples (6). The GSE32863 dataset contains data from 58 lung adenocarcinoma tissue samples and 58 fresh frozen adjacent non-cancerous samples (7). Moreover, the GSE75037 dataset contains data from 84 lung adenocarcinoma and 84 adjacent non-cancerous lung tissue samples (8).
Identification of DEGs
GEO2R is an online web tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) provided by the GEO for comparing GEO series to identify DEGs across experimental conditions. The cutoff criteria were set to P-value <0.05 and logFC (fold change)>1. We excluded probe sets without exact gene symbols, and genes with two or more probe sets were averaged.
KEGG and GO enrichment analyses of DEGs
The functional annotation tools version 6.7 of the Database for Annotation, Integrated Discovery and Visualization (9) (DAVID; http://david.ncifcrf.gov) were used to extract biological information about our DEGs. KEGG is a public database used for understanding the functions and abilities of biological systems, such as cells, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets acquired by genome sequencing (10). GO was also used to annotate genes and further analyze their biological functions. The DAVID online database was used to analyze the function and biological process of the screened DEGs. P<0.05 was considered to indicate statistical significance.
PPI network construction and analysis
The Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org; version 10.0) was used to construct the PPI network from the DEGs (11). The sources for interactions are text mining, databases, experiments, neighborhood, co-expression, gene co-occurrence and fusion. We set the minimum required interaction score to 0.4. Cytoscape version 3.4.0 software (12) was used to visualize the molecular interaction networks of the DEGs. The APP plug-in, Molecular Complex Detection (MCODE) (13), was used to arrange the network topology to cluster densely connected genes. After the PPI networks was constructed, its key modules were searched by using the MCODE application. The parameter for inclusions are MCODE score >5, degree cutoff=2, node score cutoff=0.2, node density cutoff=0.1, k-score=2 and Max depth=100. Then, DAVID was used to perform the GO and KEGG analyses for these most significant modules.
Hub gene screen and analysis
The criterion for being a hub gene selection was degree ≥10. Further analysis was performed using the cBioPortal online platform (http://www.cbioportal.org) to build the network of the DEGs and co-expressing genes (14). The mutation rates of the hub genes were also measured with the cBioPortal platform (15). Cytoscape's Biological Networks Gene Oncology tool (BiNGO) (version 3.0.3) was used for the biological process analysis and visualization (16). The University of California Santa Cruz (USCS) platform was used to analyze the hierarchical clustering of hub genes (17). Kaplan-Meier curves for overall survival and disease-free survival with these hub genes were obtained from cBioPortal. The expression profiles of DNA topoisomerase IIα (TOP2A) and aurora kinase A (AURKA) in 20 types of malignant tumors were analyzed and displayed using the Oncomine database (http://www.oncomine.com) (18).
Analysis of TOP2A and AURKA expression in cell lines
To confirm our bioinformatics results, reverse transcription and quantitative real-time PCR (RT-qPCR) were conducted on lung adenocarcinoma (HCC827, A549 and H1975) cell lines and a human bronchial epithelial (HBE) cell line. A549, HCC827 and H1975 cells were purchased from the Shanghai Cell Bank (Shanghai, China) and were cultured using Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). The medium was supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.), and 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) under a 5% CO2-containing humidified atmosphere at 37°C. Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The isolated RNA was reverse-transcribed into cDNA using a reverse transcription kit (Takara, Dalian, China). RT-qPCR was performed as described in our previous research, 2 min at 50°C, 10 min at 95°C, 40 cycles at 95°C for 15 sec, and 60°C for 30 sec (19) and the results were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. Primers were as follows: TOP2A (forward, 5-AGGATTCCGCAGTTACGTGG-3 and reverse, 5-CATGTCTGCCGCCCTTAGAA-3) (20) and AURKA (forward, 5-TTGGGTGGTCAGTACATGCTC-3 and reverse, 5-GTGAATTCAACCCGTGAT-3) (21) and GAPDH sense, 5′-CAATGACCCCTTCATTGACC-3′ and reverse, 5′-TGGAAGATGGTGATGGGATT-3′. The statistical analyses were conducted using SPSS version 21 (IBM Corp.). Results are displayed as mean ± SEM and differences between the HBE and cancerous cell lines were analyzed by one-way ANOVA. We further used the Tukey test to determine the significance between each cancer cell line and HBE. P-value <0.05 was considered to indicate statistical significance. Each experiment was repeated three times.
Analysis of TOP2A and AURKA expression in human samples
The Ethics Committee of the First Hospital of the China Medical University (Shenyang, Liaoning, China) approved our research. Written informed consent was received from all participants. Seventy-two lung adenocarcinoma and paired non-cancerous tissues were obtained between February 2013 and June 2014 from 35 women and 37 men, ranging in age from 38 to 75, with a median age of 60. Patients who had received chemotherapy, target therapy and radiotherapy or had a history of malignant tumor were excluded. All of the diagnoses were confirmed by two experienced pathologists. The resected samples were preserved at −80°C until the mRNA of TOP2A and AURKA extraction were needed. Differences between cancerous and non-cancerous tissues were compared using the paired Student's t-test.
Results
Identification of DEGs in lung adenocarcinoma
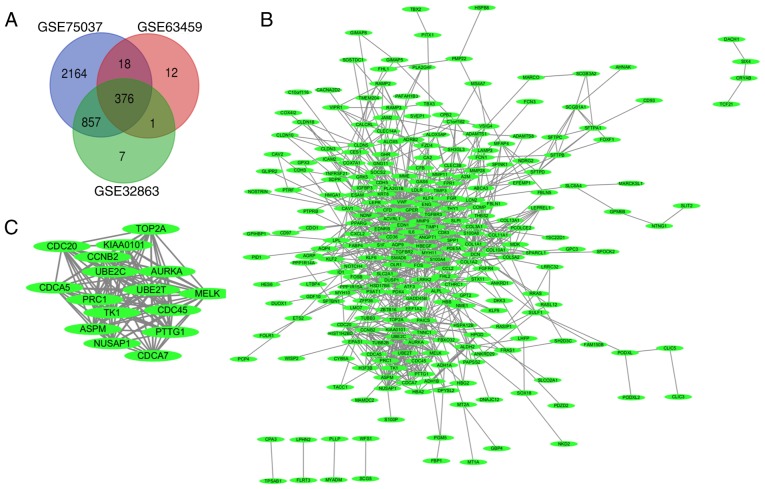
A total of 5,874 genes were found to be differentially expressed in non-cancerous and lung adenocarcinoma tissues (432 in GSE63459, 4,037 in GSE75037 and 1,405 in GSE32863) after standardizing the microarray data. A total of 376 DEGs were found in all three datasets (Venn diagram, Fig. 1A), consisting of 293 downregulated and 83 upregulated genes.
Figure 1.
Venn diagram, protein-protein interaction network and the most significant components of DEGs. (A) DEGs were selected with P-value <0.05 and a fold change >2 among the mRNA expression profiling sets GSE75037, GSE63459 and GSE32863. These three datasets manifested an overlap of 376 genes. (B) The protein-protein interaction network of DEGs was constructed using Cytoscape software. (C) The most significant components of the DEGs were obtained from protein-protein interaction network with 16 nodes. DEGs, differentially expressed genes.
GO and KEGG enrichment analyses of the DEGs
The DAVID online database was used to further analyze the biological classification, as well as functions and pathways enriched in DEGs. GO analysis showed that the biological processes (BP) of the DEGs were mainly involved in regulation of cell proliferation, the transforming growth factor β receptor signaling pathway, cell adhesion, biological adhesion and responses to hormone stimulus (Table I). Examination of their cell component showed that the DEGs were mainly located in the proteinaceous extracellular matrix, cell surface, cell-cell junction, and cell-substrate adherent junction. KEGG pathway analysis showed that the DEGs were mainly over represented in the TGF-β signaling pathway, cell adhesion molecules (CAMs), complement and coagulation cascades and ECM-receptor interaction (Table I).
Table I.
KEGG and GO pathway enrichment analysis of DEGs in the lung adenocarcinoma samples.
| Term | Description | Count in gene set | P-value |
|---|---|---|---|
| GO:0048545 | Response to steroid hormone stimulus | 27 | 9.53E-14 |
| GO:0009725 | Response to hormone stimulus | 33 | 2.60E-11 |
| GO:0009719 | Response to endogenous stimulus | 33 | 3.31E-10 |
| GO:0042127 | Regulation of cell proliferation | 45 | 9.09E-09 |
| GO:0001501 | Skeletal system development | 26 | 3.68E-08 |
| GO:0010033 | Response to organic substance | 41 | 5.71E-08 |
| GO:0007179 | TGF-β receptor signaling pathway | 12 | 6.25E-08 |
| GO:0043627 | Response to estrogen stimulus | 15 | 6.80E-08 |
| GO:0007155 | Cell adhesion | 40 | 7.66E-08 |
| GO:0022610 | Biological adhesion | 40 | 7.78E-08 |
| hsa04512 | ECM-receptor interaction | 11 | 5.33E-05 |
| hsa04670 | Leukocyte transendothelial migration | 10 | 0.003333312 |
| hsa04514 | Cell adhesion molecules (CAMs) | 10 | 0.006973812 |
| hsa04510 | Focal adhesion | 12 | 0.014731626 |
| hsa04350 | TGF- β signaling pathway | 7 | 0.024809421 |
| hsa04610 | Complement and coagulation cascades | 6 | 0.032656397 |
| hsa03320 | PPAR signaling pathway | 6 | 0.032656397 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; DEGs, differentially expressed genes.
PPI network construction and module analysis
Cytoscape was used to build the DEG PPI network (Fig. 1B) and identify the most significant genes of the PPI network (Fig. 1C). Analysis of these genes with the DAVID platform found that they were mainly involved in M phases, cell cycle phase, the mitotic cell cycle and nuclear division (Table II).
Table II.
KEGG and GO pathway enrichment analysis of DEGs in the most significant module.
| Pathway ID | Pathway description | Count in gene set | FDR |
|---|---|---|---|
| GO:0000279 | M phase | 9 | 3.72E-07 |
| GO:0000278 | Mitotic cell cycle | 9 | 9.46E-07 |
| GO:0000280 | Nuclear division | 8 | 1.07E-06 |
| GO:0007067 | Mitosis | 8 | 1.07E-06 |
| GO:0000087 | M phase of mitotic cell cycle | 8 | 1.21E-06 |
| GO:0048285 | Organelle fission | 8 | 1.41E-06 |
| GO:0022403 | Cell cycle phase | 9 | 2.30E-06 |
| GO:0051301 | Cell division | 8 | 8.24E-06 |
| GO:0007049 | Cell cycle | 10 | 1.24E-05 |
| GO:0022402 | Cell cycle process | 9 | 2.65E-05 |
| GO:0007059 | Chromosome segregation | 4 | 0.08986 |
| GO:0030261 | Chromosome condensation | 3 | 0.367997 |
| GO:0000226 | Microtubule cytoskeleton organization | 4 | 0.523613 |
| GO:0007051 | Spindle organization | 3 | 1.195525 |
| GO:0007017 | Microtubule-based process | 4 | 2.500246 |
| hsa04114 | Oocyte meiosis | 4 | 0.096833 |
| hsa04110 | Cell cycle | 4 | 0.141584 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; DEGs, differentially expressed genes; FDR, false discovery rate.
Hub gene selection and analysis
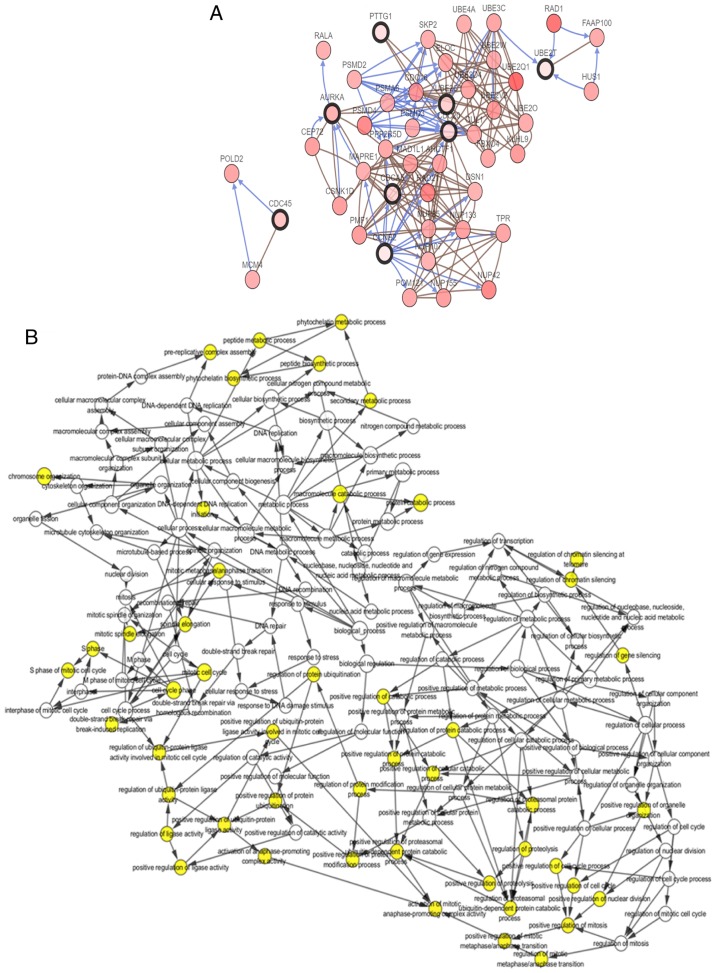
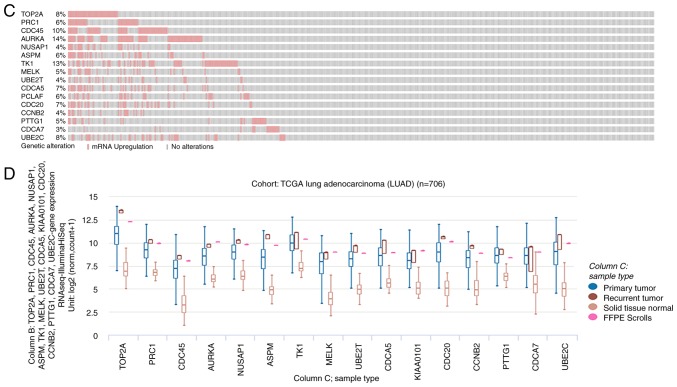
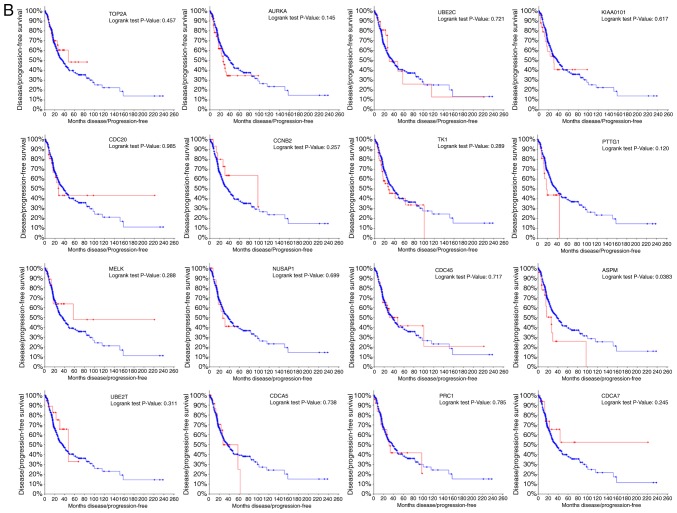
Sixteen genes were identified with a degree ≥10 and were defined as hub genes. The degree of each gene was calculated by the CytoScape software and represented the number of other genes with which it was connected. The hub gene symbol, full name, function and degree are listed in Table III. cBioPortal was then used to construct a network of the 16 hub genes and their co-expressed genes. The results of this and analysis of the BP are shown in Fig. 2A and B, respectively. oncoPrint analysis with the cBioPortal showed that TOP2A and AURKA have the highest genetic mutation rates of the hub genes in lung adenocarcinoma at 8 and 14%, respectively (Fig. 2C). Hierarchical clustering analysis revealed that these 16 hub genes could generally differentiate both primary and recurrent lung adenocarcinoma tissues from their adjacent non-cancerous lung tissues (Fig. 2D).
Table III.
Summary of the hub gene functions.
| No. | Gene symbol | Full name | Function | Degree |
|---|---|---|---|---|
| 1 | TOP2A | DNA topoisomerase II α | TOP2A functions as the target for various anticancer agents and mutations in it are associated with drug resistance | 31 |
| 2 | AURKA | Aurora kinase A | AUEKA plays a role in tumor development and progression | 22 |
| 3 | UBE2C | Ubiquitin conjugating enzyme E2 C | UBE2C is required for the destruction of mitotic cyclins and for cell cycle progression, and is involved in cancer progression | 20 |
| 4 | KIAA0101 (PCLAF) | PCNA clamp associated factor | PCNA-binding protein acts as a regulator of DNA repair during DNA replication | 20 |
| 5 | CDC20 | Cell division cycle 20 | CDC20 is a regulatory protein interacting with several other proteins at multiple points in the cell cycle | 19 |
| 6 | CCNB2 | Cyclin B2 | CCNB2 is one of the essential components of the cell cycle regulatory machinery | 18 |
| 7 | TK1 | Thymidine kinase 1 | High level of TK1 is used as a biomarker for diagnosing and categorizing many types of cancers | 17 |
| 8 | PTTG1 | Pituitary tumor-transforming 1 | PTTG1 product has transforming activity in vitro and tumori genic activity in vivo, and it is highly expressed in various tumors | 17 |
| 9 | MELK | Maternal embryonic leucine zipper kinase | Diseases associated with MELK include uterine corpus endometrial carcinoma. Among its related pathways are Neuroscience | 17 |
| 10 | NUSAP1 | Nucleolar and spindle associ ated protein 1 | NUSAP1 is a nucleolar-spindle-associated protein that plays a role in spindle microtubule organization | 16 |
| 11 | CDC45 | Cell division cycle 45 | The protein encoded by CDC45 is an essential protein required for the initiation of DNA replication | 16 |
| 12 | ASPM | Abnormal spindle microtubule assembly | ASPM is essential for normal mitotic spindle function in embryonic neuroblasts | 16 |
| 13 | UBE2T | Ubiquitin conjugating enzyme E2 T | The protein encoded by UBE2T catalyzes the covalent attachment of ubiquitin to protein substrates. Defects in UBE2T are associated with Fanconi anemia of complementation group T | 15 |
| 14 | CDCA5 | Cell division cycle associated 5 | Among its related pathways are Cell cycle, Mitotic and MicroRNAs in cancer | 15 |
| 15 | PRC1 | Protein regulator of cytokinesis 1 | PRV1 encodes a protein that is involved in cytokinesis which has been shown to be a substrate of several cyclin-dependent kinases | 14 |
| 16 | CDCA7 | Cell division cycle associated 7 | CDCA7 was identified as a c-Myc responsive gene. Overexpression of this gene enhances the transformation of lymphoblastoid cells | 13 |
Figure 2.
Interaction network of the hub genes and the biological process analysis. (A) cBioPortal platform were used to analyze the hub genes and the co-expression genes. The hub genes are marked with a bold outline. Co-expression genes are marked with a thin outline. (B) The plugin of Cytoscape, BiNGO, was adopted to conduct the analysis of biological process. P-value of the ontologies are represented by different color shade. The yellow node indicates higher functional enrichment than white. The numbers of the genes involved in the ontologies are represented by the different size of the node. P<0.05 was considered statistically significant. Interaction network of the hub genes and the biological process analysis. (C) The hub gene alteration rates in lung adenocarcinoma were screened from cBioPortal platform; the red-colored bars represent the upregulation of the gene. (D) The UCSC (University of California Santa Cruz) cancer platform was used to construct the hub gene hierarchical clustering. The primary tumor, recurrent tumor and normal tissue are represented in different colors.
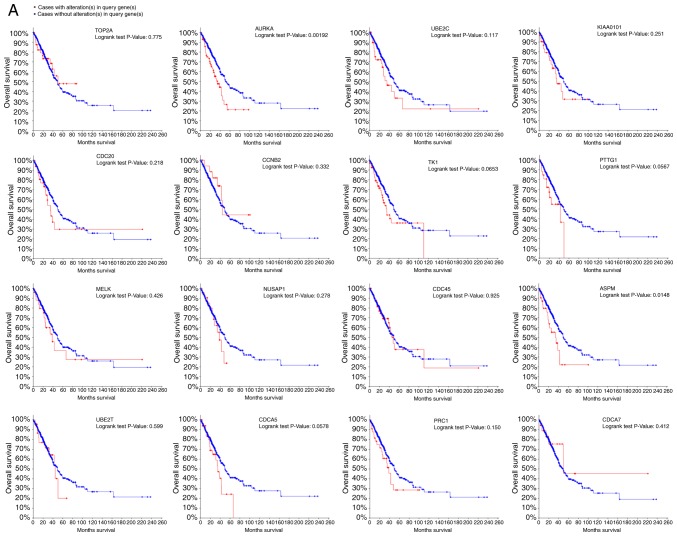
Clinical significance of TOP2A and AURKA
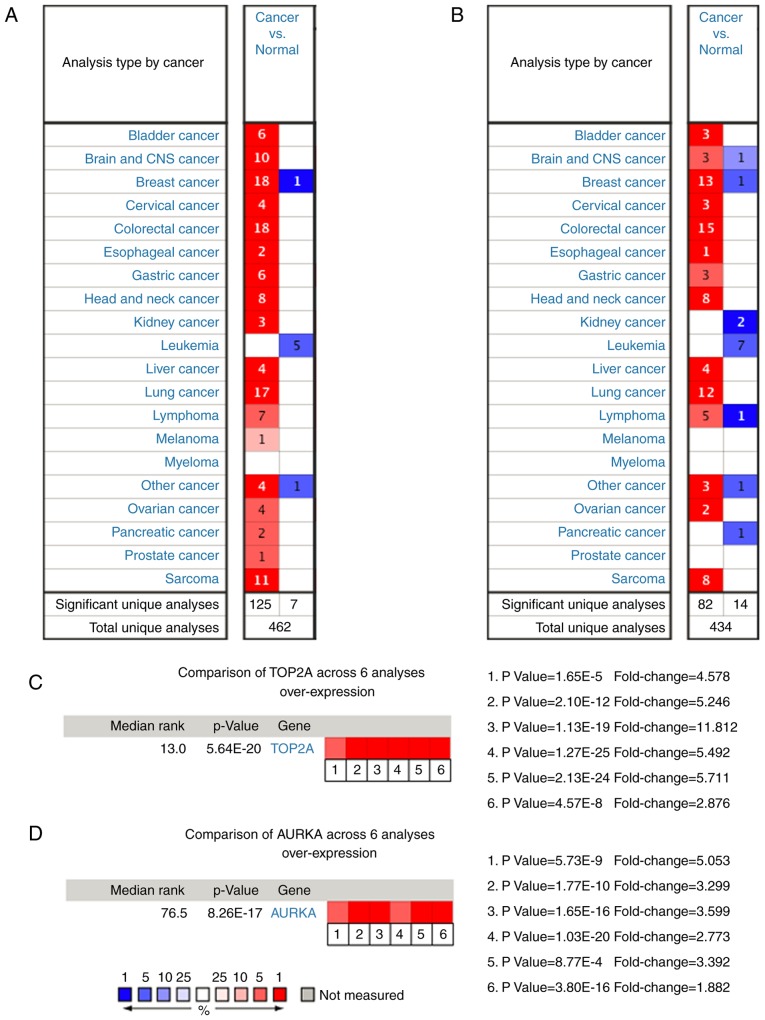
Analysis of the association between these genes and disease-free survival and overall survival analysis was conducted using Kaplan-Meier curves in the cBioPortal platform. Lung adenocarcinoma patients with AURKA mutation had worse overall and disease-free survival and patients with ASPM (abnormal spindle microtubule assembly) mutation had worse disease-free survival (Fig. 3A and B). Moreover, AURKA and TOP2A had the highest node degrees at 22 and 31, respectively, implying that they may play significant roles in the genesis and development of lung adenocarcinoma. When analyzing the data from cBioPortal platform, we discovered that lung adenocarcinoma patients who had an AURKA mutation had reductions in overall survival (P=0.00192). However, this was not true for the TOP2A gene (P=0.775, Fig. 3A and B). The expression profile of AURKA and TOP2A in 20 types of human cancer tissues was displayed using the Oncomine database. TOP2A mRNA levels in bladder, brain, breast, colorectal, esophageal, kidney, gastric and sarcoma cancer tissues were higher than those in matched adjacent normal tissues (Fig. 4A). The AURKA mRNA levels in bladder, brain, breast, cervical, lung and liver cancer tissues were higher than those in adjacent matched normal tissues (Fig. 4B). When we analyzed six different datasets from the Oncomine database, we found that TOP2A and AURKA were significantly overexpressed in lung adenocarcinoma tissues compared with non-cancerous tissues (Fig. 4C and D) (7,22–28).
Figure 3.
The relationship between alteration of the hub genes and overall survival (A) and disease free survival (B) were explored in the cBioPortal platform. P<0.05 was considered statistically significant. To note, the full names of all the gene symbols used in the figures are listed in Table III.
Figure 4.
Expression profiles of (A) TOP2A and (B) AURKA in 20 malignant tumor types are represented using the Oncomine database. The number represent the cases meeting the threshold for TOP2A and AURKA. Heat maps of (C) TOP2A and (D) AURKA gene expression in lung carcinoma samples vs. adjacent lung tissues in the Oncomine database. (C) 1. Lung carcinoma vs. normal lung, Beer et al (2002) (22). 2. Lung carcinoma vs. normal lung, Hou et al (2010) (23). 3. Lung carcinoma vs. normal lung, Landi et al (2008) (24). 4. Lung carcinoma vs. normal lung, Selamat et al (2012) (7). 5. Lung carcinoma vs. normal lung, Su et al (2007) (25). 6. Lung carcinoma vs. normal lung, Yamagata et al (2003) (26). (D) 1. Lung carcinoma vs. normal lung, Bhattacharjee et al (2001) (27). 2. Lung carcinoma vs. normal lung, Garber et al (2001) (28). 3. Lung carcinoma vs. normal lung, Hou et al (2010) (23). 4. Lung carcinoma vs. normal lung, Landi et al (2008) (24). 5. Lung carcinoma vs. normal lung, Selamat et al (2012) (7). 6. Lung carcinoma vs. normal lung, Su et al (2007) (25). The P-value for a gene is its P-value for the median-ranked analysis. The fold change represents the relative expression of the tumor tissue compared with the normal tissue. AURKA, aurora kinase A; TOP2A, DNA topoisomerase II α.
Expression of TOP2A and AURKA in lung adenocarcinoma cell lines
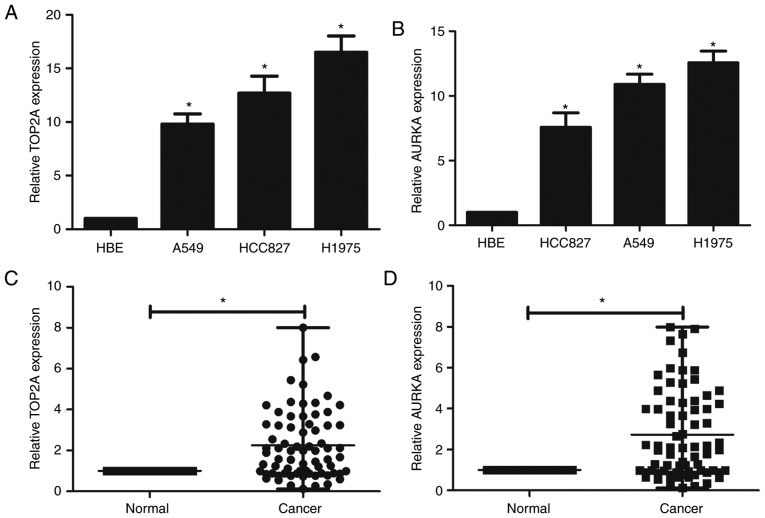
To confirm the bioinformatics results, the expression of these two genes in lung adenocarcinoma cell lines and HBE cells were assessed by RT-qPCR. The mRNA levels of both TOP2A and AURKA were significantly higher in the lung adenocarcinoma cell lines than the HBE cells (Fig. 5A and B).
Figure 5.
RT-qPCR to confirm the upregulation of TOP2A and AURKA in lung adenocarcinoma and HBE cell lines and human samples. (A) TOP2A expression in lung adenocarcinoma cell lines A549, HCC827 and H1975 and a human bronchial epithelial (HBE) cell line. (B) AURKA expression in the lung adenocarcinoma cell lines. (C) TOP2A expression in lung adenocarcinoma tissue and non-cancerous lung tissue samples. (D) AURKA expression in lung adenocarcinoma tissue and non-cancerous lung tissue samples. AURKA, aurora kinase A; TOP2A, DNA topoisomerase II α. *P<0.05.
Expression of TOP2A and AURKA in human lung adenocarcinoma and adjacent normal lung tissues
Table IV shows the clinicopathological characteristics of all the lung adenocarcinoma patients. TOP2A and AURKA were found to have 2.25- and 2.73-times higher expression levels in the cancer tissues than in the normal tissues, respectively (Fig. 5C and D). The 72 samples were then classified into two groups based on the relative expression levels of these two genes: The relatively high TOP2A and AURKA group (n=36, the first 36 cases having relative high TOP2A and AURKA were defined as the high-expression group; the remaining 36 cases were defined as the low-expression group) and the relatively low TOP2A and AURKA group (n=36). χ2 analysis was used to analyze the results. Analysis revealed that increased TOP2A expression was observed in tumors with a larger diameter. Increased AURKA expression was observed in tumors with a larger size and in lymphatic metastatic tumors (Tables IV and V).
Table IV.
Clinicopathologic associations of TOP2A expression in lung adenocarcinoma (N=72).
| Relative TOP2A expression | ||||
|---|---|---|---|---|
| Clinical parameters | No. of cases | Low | High | P-value |
| Age (years) | 0.237 | |||
| >60 | 39 | 22 | 17 | |
| ≤60 | 33 | 14 | 19 | |
| Sex | 0.099 | |||
| Male | 37 | 15 | 22 | |
| Female | 35 | 21 | 14 | |
| Smoking | 1 | |||
| Smoker | 42 | 21 | 21 | |
| Non-smoker | 30 | 15 | 15 | |
| Maximum diameter (cm) | 0.003 | |||
| <3 | 26 | 19 | 7 | |
| ≥3 | 46 | 17 | 29 | |
| Lymphatic metastasis | 0.812 | |||
| Positive | 41 | 21 | 20 | |
| Negative | 31 | 15 | 16 | |
| Metastasis | 0.607 | |||
| M0 | 68 | 33 | 35 | |
| M1 | 4 | 3 | 1 | |
TOP2A, DNA topoisomerase II α.
Table V.
Clinicopathologic associations of AURKA expression in lung adenocarcinoma (N=72).
| Relative AURKA expression | ||||
|---|---|---|---|---|
| Clinical parameters | No. of cases | Low | High | P-value |
| Age (years) | 0.813 | |||
| >60 | 39 | 20 | 19 | |
| ≤60 | 33 | 16 | 17 | |
| Sex | 0.099 | |||
| Male | 37 | 22 | 15 | |
| Female | 35 | 14 | 21 | |
| Smoking | 0.633 | |||
| Smoker | 42 | 20 | 22 | |
| Non-smoker | 20 | 16 | 14 | |
| Maximum diameter (cm) | 0.003 | |||
| <3 | 26 | 19 | 7 | |
| ≥3 | 46 | 17 | 29 | |
| Lymphatic metastasis | 0.002 | |||
| Positive | 41 | 14 | 27 | |
| Negative | 31 | 22 | 9 | |
| Metastasis | 1 | |||
| M0 | 68 | 34 | 34 | |
| M1 | 4 | 2 | 2 | |
AURKA, aurora kinase A.
Discussion
Recent statistics on cancer globally revealed that lung cancer is the most commonly diagnosed malignant tumor and is the leading cause of cancer-related mortality accounting for 11.6% of all malignant tumors and 18.4% of cancer-related deaths. Lung adenocarcinoma is the most common subtype of malignant lung cancer, and its incidence is increasing rapidly (29). Air pollution and smoking are the two main etiological factors for lung adenocarcinoma (30,31). The microarray data of Sekine et al revealed that a human lung mucoepidermoid carcinoma cell line exposed to smoke with a charcoal filter had a total of 1,582 genetic mutations (32). Still, the molecular mechanisms underlying the genesis of lung adenocarcinoma remain unclear. Without an early diagnosis most patients are not candidates for curative therapies leading to the deeply unsatisfactory prognosis for the disease. Therefore, biological markers with satisfactory efficiency for early diagnosis and therapy are desperately needed. With the development of microarray technology, we are able to efficiently screen for changes in gene expression in lung adenocarcinoma, and this approach has been proven to be a very useful method for screening early stage biomarkers in both malignant and benign diseases (33–35).
In the present research, three mRNA microarray datasets were downloaded from the GEO database and subsequently analyzed to acquire differentially expressed genes (DEGs) between lung adenocarcinoma and adjacent non-cancerous lung tissues. In addition, a total of 376 DEGs were identified in the three datasets, consisting of 293 upregulated genes and 83 downregulated genes. The biological roles of the identified DEGs were then studied using KEGG and GO enrichment analyses. The downregulated genes were mainly overrepresented in cell division, nuclear division, DNA replication and second-messenger-mediated signaling, and the upregulated genes were mainly involved in cell cycle process, DNA metabolic process, the transforming growth factor (TGF)-β signaling pathway and angiogenesis. Previous articles have revealed that dysregulation of angiogenesis and activation of the TGF-β signaling pathway are associated with the carcinogenesis and progression of lung adenocarcinoma (36–38). In addition, DNA damage and metabolic process abnormalities often play a significant role in cell cycle regulation dysfunction and are associated with malignant tumors (39). In summary, our results are consistent with these previous reports and theories. KEGG enrichment analysis showed differences mainly in the TGF-β signaling pathway, complement and coagulation cascades, Cell adhesion molecules (CAMs), and ECM-receptor interaction, while changes identified by GO terms were mainly in regulation of cell adhesion, proliferation, the TGF-β receptor signaling pathway, biological adhesion and response to hormone stimulus.
Sixteen genes with degrees ≥10 were defined as hub genes (40). Two of these hub genes, TOP2A and AURKA, had the highest node degrees at 31 and 22, respectively. Gene mutation can promote tumorigenesis, thus the mutation rates of these 16 lung adenocarcinoma hub genes were screened with the cBioPortal platform. The four genes with the highest mutation rates were AURKA, TK1, CDC45 and TOP2A with mutation rates of 14, 13, 10 and 8% respectively, which indicates that these genes may play a significant role in tumorigenesis.
DNA topoisomerase II α (TOP2A) that encodes the DNA topoisomerase, an enzyme that alters and controls the topological state of DNA during transcription, has been shown to be correlated with an increased risk of developing brain metastases, drug resistance and an abnormal cell cycle (41–43). It is regarded as a target for several anticancer agents, such as etoposide and topotecan (44). In our research, the protein-protein interaction network (PPI) network revealed that TOP2A directly interacts with maternal embryonic leucine zipper kinase (MELK), CDC20, CCNB2, UBE2T, KIAA0101 and TK1, indicating that TOP2A plays a key role in lung adenocarcinoma. Two of these genes, CDC20 and MELK, are closely involved in tumorigenesis and the cell cycle. Cell division cycle 20 (CDC20) appears to function as a regulatory protein interacting with other proteins at several important phases in the cell cycle. CDC20 is activated in lung adenocarcinoma and overexpression is correlated with poor prognosis (45). MELK plays a key role in the proliferation and self-renewal of progenitor and tumor stem cells, and is overexpressed in lung adenocarcinoma, contributing to carcinogenesis. MELK is an effective target for kinase drugs (46). Moreover, the expression of TOP2A is upregulated in various tumors, such as colon and ovarian malignant tumors, and may be considered a sensitive biomarker for early detection and therapy of these tumors (47,48).
Aurora kinase A (AURKA) is a putative oncogene, associated with cell cycle-regulated kinase (49). GO annotations associated with this gene include protein tyrosine kinase activity, transferring phosphorus-containing groups and transferase activity. Overexpression of this gene is associated with several common features of malignant tumor cells, such as chromosomal instability, aneuploidy in mammalian cells and centrosomal duplication abnormalities (50,51). It has also been found to be overexpressed in several types of malignant tumors and has been associated with poor prognosis (52,53). The aurora kinase A inhibitor, alisertib, has been approved for therapy for solid tumors especially non-small cell lung cancer (NSCLC) and breast cancer, and has achieved satisfactory results (54). In addition, Chen et al revealed that AURKA antagonists can enhance the cytotoxicity of epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) (55).
The relationships between TOP2A and AURKA expression and survival were further assessed with the cBioPortal platform. We found that changes in TOP2A expression were associated with a decrease in both disease-free and overall survival, although they were not statistically significant. While changes in AURKA expression were associated with significantly worse overall survival rates, but no significant change in disease-free survival rate. This may be explained by our study of 72 paired human samples which found that upregulation of AURKA was associated with lymphatic metastasis and that upregulation of TOP2A was only associated with tumor size. One of the underlying molecular reason may be that the overexpression of TOP2A arises from amplification and mutation, while the survival analysis using the cBioPortal platform was only based on the mutation of TOP2A. Thus, amplification but not mutation may result in overexpression of TOP2A, not related to changes in prognosis, although further study is needed to prove this hypothesis. Oncomine analysis revealed that higher AURKA mRNA levels were observed in colorectal cancer, breast cancer, lung cancer and sarcoma. Additionally, higher TOP2A mRNA levels were observed in colorectal cancer, breast cancer, lung cancer and brain cancer, indicating important roles of these two genes in the carcinogenesis and development of malignant tumors. However, these results also indicate that these two genes can only be used as broad-spectrum tumor markers as they cannot differentiate lung adenocarcinoma from other malignant tumors.
The UCSC (University of California Santa Cruz) cancer platform was used to hierarchically cluster the hub genes. Their expression levels in both the primary and recurrent tumors were upregulated compared with the normal tissues. The expression levels of these hub genes in recurrent tumors were higher than in the primary tumor. TOP2A in particular was found to have the highest expression in the recurrent tumors. We therefore infer that TOP2A and other hub genes may be regarded as early biomarkers for monitoring tumor recurrence.
To confirm our results, we detected the expression of TOP2A and AURKA in lung adenocarcinoma and human bronchial epithelial (HBE) cell lines. Both of these genes were downregulated in the lung adenocarcinoma cell lines. TOP2A and AURKA had their highest expression levels in H1975 cells with approximately 15 times higher expression than that in HBEs cells. Further experiments in human samples found that TOP2A and AURKA were both upregulated in lung adenocarcinoma tissues compared with non-cancerous tissues. The upregulation of TOP2A was found to be associated with larger tumor size, and AURKA was found to be associated with both larger tumor size and positive lymphatic metastasis. Thus, we demonstrated that TOP2A and AURKA are closely involved in lung adenocarcinoma using both bioinformatics and cell experiments. The cell lines we selected have varying levels of EMT. The H1975 cell line (56) has the highest relative level of EMT and also has higher levels of AURKA and TOP2A in our study. We also showed that higher AURKA expression is correlated with poor prognosis. Thus, the higher expression of AURKA may be correlated with higher levels of EMT, which result in metastasis and lead to poor prognosis.
In conclusion, our research aimed to identify DEGs, which may be involved in the genesis and development of lung adenocarcinoma. Two of the 16 hub genes were further studied in cell lines and human samples and may be regarded as biomarkers for the diagnosis of lung adenocarcinoma. Further research is needed to elucidate the mechanisms behind their changes in expression and their biological function in lung adenocarcinoma.
Acknowledgements
Not applicable.
Funding
This research was supported by a grant (no. 81702242) from the National Science Foundation of China and Doctoral Scientific Research Foundation of Liaoning Province (no. 20170520441).
Availability of data and materials
The data used in this study are available from the corresponding author on reasonable request.
Authors' contribution
SD and SX designed the research. WM carried out the data collection and analysis. SY conducted the experiments. SD wrote the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The Ethics Committee of the First Hospital of the China Medical University (Shenyang, Liaoning, China) approved our research. Written informed consent was received from all of the included patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrara R, Mezquita L, Texier M, Lahmar J, Audigier-Valette C, Tessonnier L, Mazieres J, Zalcman G, Brosseau S, Le Moulec S, et al. Hyperprogressive disease in patients with advanced non-small cell lung cancer treated With PD-1/PD-L1 inhibitors or with single-agent chemotherapy. JAMA Oncol. 2018;4:1543–1552. doi: 10.1001/jamaoncol.2018.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park CK, Cho HJ, Choi YD, Oh IJ, Kim YC. A phase II trial of osimertinib in the second-line treatment of non-small cell lung cancer with the EGFR T790M mutation, detected from circulating tumor DNA: LiquidLung-O-Cohort 2. Cancer Res Treat. 2019;51:777–787. doi: 10.4143/crt.2018.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41((Database Issue)):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robles AI, Arai E, Mathé EA, Okayama H, Schetter AJ, Brown D, Petersen D, Bowman ED, Noro R, Welsh JA, et al. An Integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10:1037–1048. doi: 10.1097/JTO.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba II, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res. 2016;22:4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanabe M, Kanehisa M. Using the KEGG database resource. Curr Protoc Bioinformatics Chapter 1. 2012 doi: 10.1002/0471250953.bi0112s38. Unit 1.12. [DOI] [PubMed] [Google Scholar]
- 11.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon S, Kim H, Kim HS. Identification of pharmacologically tractable protein complexes in cancer using the R-based network clustering and visualization program MCODER. BioMed Res Int. 2017;2017:1016305. doi: 10.1155/2017/1016305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demchak B, Hull T, Reich M, Liefeld T, Smoot M, Ideker T, Mesirov JP. Cytoscape: The network visualization tool for GenomeSpace workflows. Version 2. F1000Res. 2014;3:151. doi: 10.12688/f1000research.4492.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 17.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong S, Qu X, Li W, Zhong X, Li P, Yang S, Chen X, Shao M, Zhang L. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol. 2015;8:43. doi: 10.1186/s13045-015-0140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labbé DP, Sweeney CJ, Brown M, Galbo P, Rosario S, Wadosky KM, Ku SY, Sjöström M, Alshalalfa M, Erho N, et al. TOP2A and EZH2 provide early detection of an aggressive prostate cancer subgroup. Clin Cancer Res. 2017;23:7072–7083. doi: 10.1158/1078-0432.CCR-17-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouquier S, Pillaire MJ, Cazaux C, Giorgi D. Expression of the microtubule-associated protein MAP9/ASAP and its partners AURKA and PLK1 in colorectal and breast cancers. Dis Markers. 2014;2014:798170. doi: 10.1155/2014/798170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 23.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagata N, Shyr Y, Yanagisawa K, Edgerton M, Dang TP, Gonzalez A, Nadaf S, Larsen P, Roberts JR, Nesbitt JC, et al. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clin Cancer Res. 2003;9:4695–4704. [PubMed] [Google Scholar]
- 27.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Juhasz AL, Ma LQ, Cui X. Inhalation bioaccessibility of PAHs in PM2.5: Implications for risk assessment and toxicity prediction. Sci Total Environ. 2019;650:56–64. doi: 10.1016/j.scitotenv.2018.08.246. [DOI] [PubMed] [Google Scholar]
- 31.Papadopoulos A, Guida F, Cénée S, Cyr D, Schmaus A, Radoï L, Paget-Bailly S, Carton M, Tarnaud C, Menvielle G, et al. Cigarette smoking and lung cancer in women: Results of the French ICARE case-control study. Lung Cancer. 2011;74:369–377. doi: 10.1016/j.lungcan.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Sekine T, Sakaguchi C, Fukano Y. Investigation by microarray analysis of effects of cigarette design characteristics on gene expression in human lung mucoepidermoid cancer cells NCI-H292 exposed to cigarette smoke. Exp Toxicol Pathol. 2015;67:143–151. doi: 10.1016/j.etp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Huang Z, Su R, Qing C, Peng Y, Luo Q, Li J. Plasma circular RNAs hsa_circ_0001953 and hsa_circ_0009024 as diagnostic biomarkers for active tuberculosis. Front Microbiol. 2018;9:2010. doi: 10.3389/fmicb.2018.02010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan TZ, Rouanne M, Tan KT, Huang RY, Thiery JP. Molecular subtypes of urothelial bladder cancer: Results from a meta-cohort analysis of 2411 tumors. Eur Urol. 2019;75:423–432. doi: 10.1016/j.eururo.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Li SY, Wu HC, Mai HF, Zhen JX, Li GS, Chen SJ. Microarray-based analysis of whole-genome DNA methylation profiling in early detection of breast cancer. J Cell Biochem. 2019;120:658–670. doi: 10.1002/jcb.27423. [DOI] [PubMed] [Google Scholar]
- 36.Miao S, Qiu T, Zhao Y, Wang H, Sun X, Wang Y, Xuan Y, Qin Y, Jiao W. Overexpression of S100A13 protein is associated with tumor angiogenesis and poor survival in patients with early-stage non-small cell lung cancer. Thorac Cancer. 2018;9:1136–1144. doi: 10.1111/1759-7714.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eser PO, Jänne PA. TGFβ pathway inhibition in the treatment of non-small cell lung cancer. Pharmacol Ther. 2018;184:112–130. doi: 10.1016/j.pharmthera.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Basu AK. DNA damage, mutagenesis and cancer. Int J Mol Sci. 2018;19:E970. doi: 10.3390/ijms19061767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park CW, Bak Y, Kim MJ, Srinivasrao G, Hwang J, Sung NK, Kim BY, Yu JH, Hong JT, Yoon DY. The novel small molecule STK899704 promotes senescence of the human A549 NSCLC cells by inducing DNA damage responses and cell cycle arrest. Front Pharmacol. 2018;9:163. doi: 10.3389/fphar.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol Rep. 2017;38:2607–2618. doi: 10.3892/or.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knez L, Sodja E, Kern I, Košnik M, Cufer T. Predictive value of multidrug resistance proteins, topoisomerases II and ERCC1 in small cell lung cancer: A systematic review. Lung Cancer. 2011;72:271–279. doi: 10.1016/j.lungcan.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Liu J, Meng Q, Niu G. Multidrug resistance protein and topoisomerase 2 alpha expression in non-small cell lung cancer are related with brain metastasis postoperatively. Int J Clin Exp Pathol. 2015;8:11537–11542. [PMC free article] [PubMed] [Google Scholar]
- 43.Sudan S, Rupasinghe HP. Quercetin-3-O-glucoside induces human DNA topoisomerase II inhibition, cell cycle arrest and apoptosis in hepatocellular carcinoma cells. Anticancer Res. 2014;34:1691–1699. [PubMed] [Google Scholar]
- 44.Kaur G, Reinhart RA, Monks A, Evans D, Morris J, Polley E, Teicher BA. Bromodomain and hedgehog pathway targets in small cell lung cancer. Cancer Lett. 2016;371:225–239. doi: 10.1016/j.canlet.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi R, Sun Q, Sun J, Wang X, Xia W, Dong G, Wang A, Jiang F, Xu L. Cell division cycle 20 overexpression predicts poor prognosis for patients with lung adenocarcinoma. Tumour Biol. 2017;39:1010428317692233. doi: 10.1177/1010428317692233. [DOI] [PubMed] [Google Scholar]
- 46.Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, Reinecke M, Ruprecht B, Petzoldt S, Meng C, et al. The target landscape of clinical kinase drugs. Science. 2017;358:eaan4368. doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lazaris AC, Kavantzas NG, Zorzos HS, Tsavaris NV, Davaris PS. Markers of drug resistance in relapsing colon cancer. J Cancer Res Clin Oncol. 2002;128:114–118. doi: 10.1007/s00432-001-0310-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa MJ, Hansen CL, Holden JA, Guinee D., Jr Topoisomerase II alpha: Prognostic predictor and cell cycle marker in surface epithelial neoplasms of the ovary and peritoneum. Int J Gynecol Pathol. 2000;19:248–257. doi: 10.1097/00004347-200007000-00009. [DOI] [PubMed] [Google Scholar]
- 49.Taylor NJ, Bensen JT, Poole C, Troester MA, Gammon MD, Luo J, Millikan RC, Olshan AF. Genetic variation in cell cycle regulatory gene AURKA and association with intrinsic breast cancer subtype. Mol Carcinog. 2015;54:1668–1677. doi: 10.1002/mc.22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoque A, Carter J, Xia W, Hung MC, Sahin AA, Sen S, Lippman SM. Loss of aurora A/STK15/BTAK overexpression correlates with transition of in situ to invasive ductal carcinoma of the breast. Cancer Epidemiol Biomarkers Prev. 2003;12:1518–1522. [PubMed] [Google Scholar]
- 51.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 52.Staff S, Isola J, Jumppanen M, Tanner M. Aurora-A gene is frequently amplified in basal-like breast cancer. Oncol Rep. 2010;23:307–312. [PubMed] [Google Scholar]
- 53.Lukasiewicz KB, Lingle WL. Aurora A, centrosome structure, and the centrosome cycle. Environ Mol Mutagen. 2009;50:602–619. doi: 10.1002/em.20533. [DOI] [PubMed] [Google Scholar]
- 54.Melichar B, Adenis A, Lockhart AC, Bennouna J, Dees EC, Kayaleh O, Obermannova R, DeMichele A, Zatloukal P, Zhang B, et al. Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: A five-arm phase 2 study. Lancet Oncol. 2015;16:395–405. doi: 10.1016/S1470-2045(15)70051-3. [DOI] [PubMed] [Google Scholar]
- 55.Chen J, Lu H, Zhou W, Yin H, Zhu L, Liu C, Zhang P, Hu H, Yang Y, Han H. AURKA upregulation plays a role in fibroblast-reduced gefitinib sensitivity in the NSCLC cell line HCC827. Oncol Rep. 2015;33:1860–1866. doi: 10.3892/or.2015.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duan L, Ye L, Zhuang L, Zou X, Liu S, Zhang Y, Zhang L, Jin C, Huang Y. VEGFC/VEGFR3 axis mediates TGFβ1-induced epithelial-to-mesenchymal transition in non-small cell lung cancer cells. PLoS One. 2018;13:e0200452. doi: 10.1371/journal.pone.0200452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in this study are available from the corresponding author on reasonable request.