In a study of viral load, shedding, and immune response in 37 cases of Middle East respiratory syndrome coronavirus infection, virus was not eliminated upon development of neutralizing serum antibodies. Vaccination strategies should not be restricted to eliciting neutralizing antibodies.
Keywords: MERS, viral load, antibodies, shedding, clearance
Abstract
Background. The Middle East respiratory syndrome (MERS) coronavirus causes isolated cases and outbreaks of severe respiratory disease. Essential features of the natural history of disease are poorly understood.
Methods. We studied 37 adult patients infected with MERS coronavirus for viral load in the lower and upper respiratory tracts (LRT and URT, respectively), blood, stool, and urine. Antibodies and serum neutralizing activities were determined over the course of disease.
Results. One hundred ninety-nine LRT samples collected during the 3 weeks following diagnosis yielded virus RNA in 93% of tests. Average (maximum) viral loads were 5 × 106 (6 × 1010) copies/mL. Viral loads (positive detection frequencies) in 84 URT samples were 1.9 × 104 copies/mL (47.6%). Thirty-three percent of all 108 serum samples tested yielded viral RNA. Only 14.6% of stool and 2.4% of urine samples yielded viral RNA. All seroconversions occurred during the first 2 weeks after diagnosis, which corresponds to the second and third week after symptom onset. Immunoglobulin M detection provided no advantage in sensitivity over immunoglobulin G (IgG) detection. All surviving patients, but only slightly more than half of all fatal cases, produced IgG and neutralizing antibodies. The levels of IgG and neutralizing antibodies were weakly and inversely correlated with LRT viral loads. Presence of antibodies did not lead to the elimination of virus from LRT.
Conclusions. The timing and intensity of respiratory viral shedding in patients with MERS closely matches that of those with severe acute respiratory syndrome. Blood viral RNA does not seem to be infectious. Extrapulmonary loci of virus replication seem possible. Neutralizing antibodies do not suffice to clear the infection.
The Middle East respiratory syndrome coronavirus (MERS-CoV) was first isolated in 2012 in Saudi Arabia [1]. Since 2012, at least 1595 laboratory-confirmed cases of MERS-CoV infection, mostly with respiratory tract illness, have been reported; 571 of these were fatal [2]. Known cases and outbreaks have been linked to countries in the Arabian peninsula [3]. Large nosocomial outbreaks, such as in Jeddah, Kingdom of Saudi Arabia in 2014 and the Republic of Korea in 2015, have demonstrated the potential of the virus to spread in healthcare settings [4–6].
Due to the sporadic nature of the disease, with cases and small outbreaks distributed over a wide geographic area, investigation of the natural history of infection has been limited. Except for individual case descriptions, chronological data summarizing the main viral diagnostic parameters, such as viral load or antibodies, are lacking. Better knowledge of the kinetics of viral shedding from different body regions could help prevent nosocomial transmission and inform clinical management. Knowledge of serological features, such as the kinetics of antibody production, could guide decisions regarding diagnostic protocols and provide essential information regarding immunity and virus elimination. Quantitative data, such as viral loads and antibody titers, could enable comparisons with related diseases, in particular, severe acute respiratory syndrome (SARS), for which studies of natural history were conducted in the aftermath of the 2002–2003 epidemic [7].
MATERIALS AND METHODS
Patients and Samples
Patients were seen during a hospital-associated outbreak between 5 March and 1 May 2014. There was no prospective planning of statistical power. Patients were selected for MERS-CoV testing by reverse transcription polymerase chain reaction (RT-PCR) based on general clinical condition, oxygen saturation, and their need for invasive or noninvasive ventilation. Samples of patients who tested positive were taken at least daily, starting from 0–7 days after initial submission of samples for MERS-CoV diagnosis. The day of the first sample testing positive by RT-PCR was defined as the day of diagnosis. The mean delay between first positive sampling and return of laboratory results was 3 days. Stored samples, for retrospective analysis, were not available.
Specimens were taken from tracheal secretions via suction catheters, from the throat and the eyes using sterile swabs, and from urine and stool via sterile containers. Baseline information on enrolled patients is provided in Supplementary Table 1. Institutional review board approval was obtained from the Research Ethics Committee at Prince Sultan Military Medical City.
MERS-CoV RT-PCR Testing
Real-time RT-PCR was performed on RNA extracts using the upE and ORF1A target genes as described in [8, 9]. Raw RNA concentrations were transformed to absolute viral loads by conversion factors, according to sample type (Supplementary Table 2).
MERS-CoV Isolation in Cell Culture
Virus isolation, with increased sensitivity via the use of CaCo2 human colon carcinoma cells, was performed as described in [10].
MERS CoV Serology
Recombinant Enzyme-Linked Immunosorbent Assay
A recombinant enzyme-linked immunosorbent assay (ELISA; Anti-MERS-CoV ELISA IgG, Euroimmun, Lübeck, Germany) was based on soluble MERS-CoV spike protein S1 domain expressed in HEK-293T cells [11]. The test was conducted as described previously [12, 13]. Sera were tested twice and the arithmetic mean of the 2 measurements was used.
IgM Immunofluorescence Assay
Detection of immunoglobulin M (IgM) antibodies was done using immunofluorescence slides carrying Vero cells infected with full MERS-CoV, as described in Corman et al [9]. These were converted into a homogenous reagent format by an in vitro diagnostics manufacturer (Anti-MERS-CoV-IIFT; Euroimmun). All sera were depleted of immunoglobulin G (IgG) antibodies using Eurosorb (Euroimmun) reagent according to manufacturer instructions.
Serum Neutralization Assay
A MERS-CoV microneutralization test (NT) was performed as described in [13–15]. Predilution before setting up log2-dilution series was 1:10, defining 1:20 as the lowest possible significant titer for categorizing a sample as positive.
Statistical Analyses
Statistical analyses were done using SPSS software (version 22). In all cases, correlation analyses and preliminary multiple regression analyses were conducted to exclude confounding due to patient age or disease duration.
RESULTS
Patient Characteristics
To determine kinetic virological parameters in MERS-CoV infection, we followed 37 hospitalized patients. Mean age was 63 years (range, 24–90 years), and 73% of patients were male. MERS-CoV infection had been established in all cases by RT-PCR. Sixty-five percent of all patients died during the course of study.
Sequencing of full or partial genomes from 35 of the study patients revealed the existence of at least 6 closely related virus lineages (Supplementary Figure 1 and Table 1). Some sequences had already been seen in an earlier study [5]. Patients belonged to at least 3 nosocomial transmission clusters. Three cases could not be associated with clusters.
At time of positive diagnosis, patients had spent 11 days in hospital on average, with a maximum of 108 days. Only 20 of the 37 patients had been hospitalized for less than a week. Because of the unresolved timing of transmission events in nosocomial clusters and the existence of comorbidities in most patients, it was impossible to determine the day of onset of symptoms in the majority of patients. Unambiguous knowledge of the day of onset of symptoms was available for only 9 patients. Mean and median duration between symptom onset and admission was 3 days (range, 0–8 days). In these 9 cases, mean and median duration between onset and diagnosis was 8 days (range, 1–16 days). The mean age of the 9 cases was not significantly different from the mean age of all patients under study. To provide a common point of reference in the clinical course of all patients, the day of diagnosis (day of first RT-PCR–positive sample) was defined as day 0 in the subsequent analyses.
Eight hundred twenty-three specimens from the 37 patients were tested, including 661 tests for viral load in 6 different sample categories (Supplementary Table 2). Because of the variable latency between diagnosis and enrollment, clinical samples were not evenly distributed over patients' courses of disease (Supplementary Figure 2).
Cross-sectional Virus RNA Detection and Courses of Viral Load
Absolute viral RNA concentrations and positive proportion of samples were determined in 661 samples. Data are illustrated in Figure 1 and Supplementary Table 2. Lower respiratory tract (LRT) samples had the highest viral loads, up to 6.3 × 1010 copies/mL (mean, 5.01 × 106 copies/mL). Average viral loads in all other sample types were significantly lower (2-tailed t test, P < .0001 for all comparisons). Virus isolation trials using the 6 stool samples with the highest RNA concentration had negative outcomes.
Figure 1.
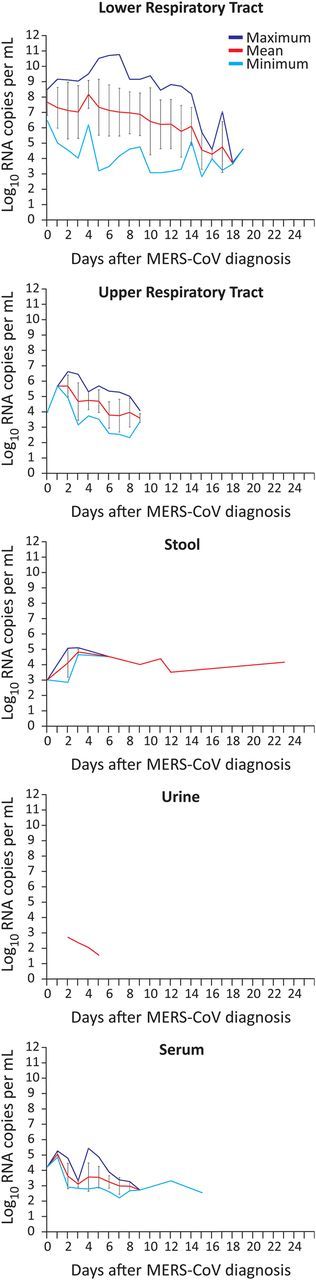
Viral loads in patients with Middle East respiratory syndrome coronavirus (MERS-CoV). Mean viral loads in positive-testing samples per day and specimen type. Maximum and minimum viral loads are shown as purple and cyan lines, respectively. Error bars represent standard deviation. Sample numbers and proportion of positive samples are summarized in Supplementary Figure 2.
Almost half of all sera showed detectable viral loads during the first week after diagnosis (25 of 51 sera tested). Virus isolation from 20 viremic serum samples (10 with and 10 without neutralizing antibodies) failed, despite a highly optimized protocol [10]. There was an inverse correlation between in vitro serum neutralization activity and viremia in 45 sera (Pearson R = −0.31, P < .03). However, viral RNA and neutralizing antibodies were codetected in several cases, suggesting that the detected viral RNA may only in part represent infectious virions (Figure 2A). Concentrations of RNA in serum did not correlate significantly with those in LRT samples collected on the same day (n = 31 pairs of serum and LRT samples; Spearman correlation, P = .08; Figure 2B).
Figure 2.
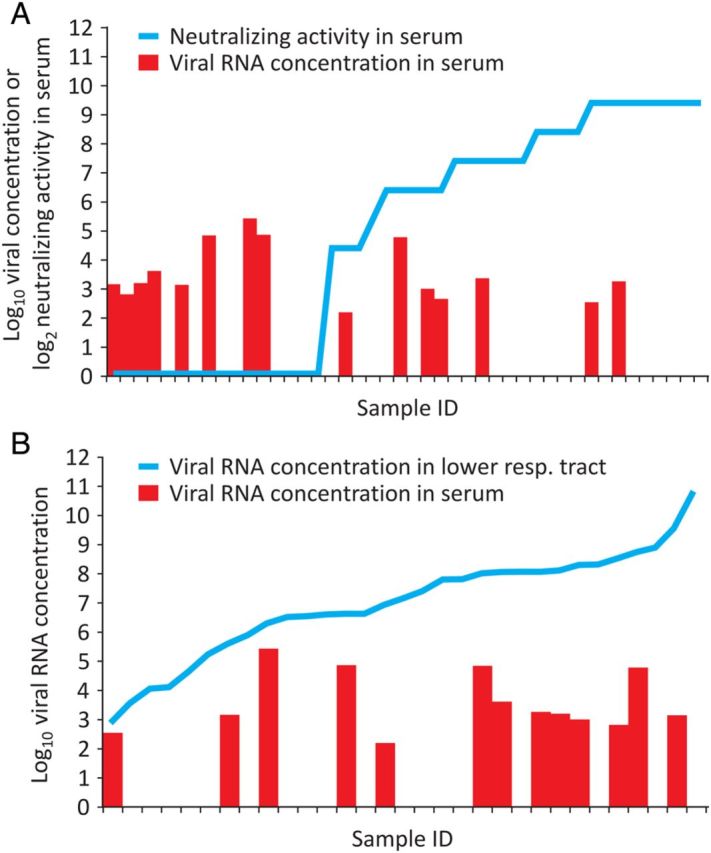
Correlation of serum viral RNA detection with neutralizing antibodies and viral RNA concentration in respiratory samples. A, Neutralizing antibodies; B, Viral RNA concentration in lower respiratory tract samples. Columns in both panels show serum viral load. Empty spaces represent serum samples that tested negative for viral RNA.
Distributions of average LRT viral load per patient were summarized in 3 subsequent time windows (Figure 3). In particular, during the first 5 days after diagnosis, average viral loads were not normally distributed but had a skewed distribution with a preponderance of patients with high viral load. Of note, the 17 patients in the 2 highest viral load categories (right-most columns in the top panel in Figure 3) did not show a significantly increased proportion of fatal outcomes (χ2 test, P = .12).
Figure 3.
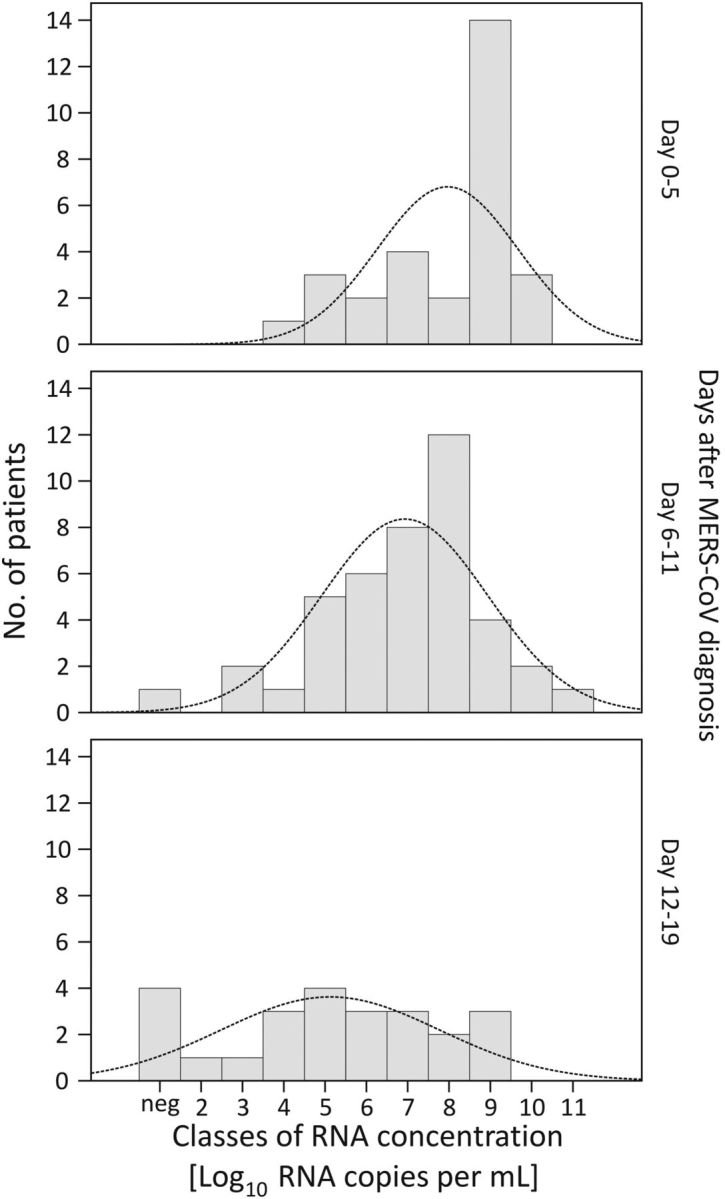
Distribution of RNA viral loads in lower respiratory tract Middle East respiratory syndrome coronavirus (MERS-CoV) samples in 3 time windows. Columns show viral loads for each patient averaged over the time windows indicated to the right of each panel. Curves represent ideal normal distributions based on sample means and variance.
The average viral load during the first week after diagnosis was 5 × 107 copies/mL in fatal cases and 3.9 × 106 copies/mL in survivors (2-tailed t test, P < .007). Divergence of viral loads between survivors and fatal cases was more pronounced in the second week (1.6 × 105 and 7.8 × 106 copies/mL, respectively; P < .0006).
Time Course of Antibody Production
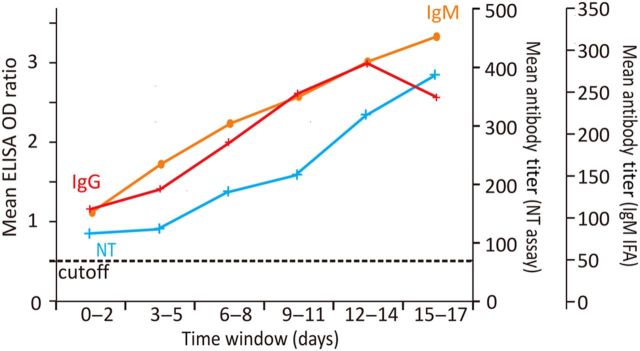
Serological courses could be followed for 35 patients. Almost half of these (n = 17) were already reactive (via ELISA) on the day of diagnosis. Among 27 patients with complete serological follow-up during the first week after diagnosis, 89% (n = 24) had antibodies by end of the week in both ELISA and neutralization tests. Eighteen of these patients tested positive for IgM by immunofluorescence assay (IFA) (titers >1:10) by end of the week. Only 1 of the IgM-positive patients did not have a concomitant positive ELISA result by end of the week. All of the 12 patients with 2 weeks of serological follow-up seroconverted (ELISA and neutralization tests). Eleven of these 12 patients developed IgM detectable by IFA. Antibody kinetics averaged over all samples and tests are summarized in Figure 4.
Figure 4.
Kinetics of antibody production. The red line shows mean immunoglobulin G (IgG) titer, represented as optical density (OD) ratios obtained from S1 enzyme-linked immunosorbent assay (ELISA). The orange line shows mean immunoglobulin M (IgM) titer from an immunofluorescence assay (IFA). The cyan line shows virus microneutralization titer (NT). Titers from each patient are averaged over successive 3-day time intervals.
Information on outcome was available for 34 patients with serological follow-up. All 12 patients who survived their infection showed seroconversion (ELISA and NT) during the first week. All developed IgM antibodies concomitantly. Among 22 fatal cases, 14 showed seroconversion by ELISA prior to death, the latest seroconversion occurring by day 11 postdiagnosis. Twelve of 22 fatal cases developed neutralizing antibodies, and 11 developed detectable IgM. Antibody levels (ELISA optical density [OD] and log2 NT titers) during the first week after diagnosis were not significantly different between surviving and fatal cases (2-tailed t test, P = .8). During the second week after diagnosis, the average ELISA OD values in survivors were significantly higher than in fatal cases (2.9 vs. 2.1, 2-tailed t test, P < .02). Also, average NT titers were higher in survivors during week 2, but with less significant discrimination (27.5 vs 25.4 in survivors and fatal cases, respectively; 2-tailed t test, P < .06).
Correlation of ELISA Antibodies, NT Titers, and Viral Loads
ELISA OD values and log2 NT titers were compared against log10 viral loads in LRT samples. From 30 patients, ELISA and viral load data were available based on matched serum and respiratory tract samples taken on the same days (198 matched data pairs, covering days 0–17 postdiagnosis). Because of workload and biosafety, the number of NT assays had to be restricted. However, combined ELISA, NT, and viral load data were available from 26 patients, with 91 matched datasets that covered days 1–17 after diagnosis. Supplementary Figure 3 summarizes the distribution of matched samples over time. Pearson test identified significant linear correlation between antibodies and log10 viral loads (ELISA, R = −0.6; NT, R = −0.51; P < .001 in both analyses). However, plots of ELISA and NT antibody results in matched sample pairs yielded no evidence of mutually exclusive occurrence of virus and antibodies (Figure 5A and 5B).
Figure 5.
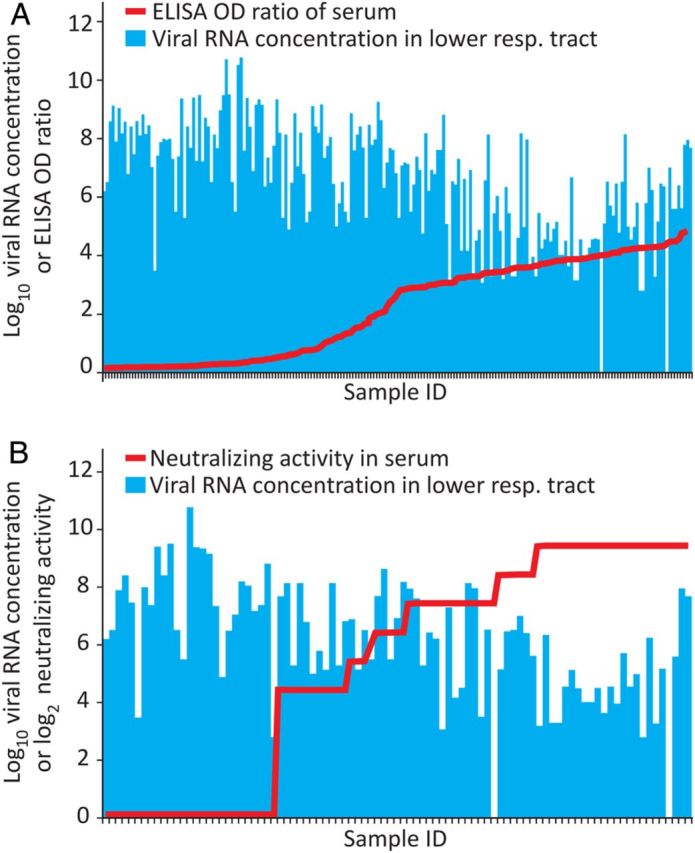
Effect of serum antibodies on lower respiratory tract (LRT) viral loads. This analysis is based on paired serum and LRT samples taken from the same patient on the same day. Antibodies are shown as line graphs. Viral loads in the corresponding LRT samples are shown as columns. The panels show samples sorted according to increasing levels of enzyme-linked immunosorbent assay (ELISA) optical density (OD) ratios (A) or neutralizing antibody titers (B). Sample numbers for this analysis are summarized in Supplementary Figure 3.
DISCUSSION
We studied quantitative viral excretion and serum antibody kinetics of a substantial group of hospitalized patients infected with MERS-CoV. The time of diagnosis chosen as a chronological reference point represents the time when an infection is suspected in hospitalized patients, or when outpatients report to hospitals due to worsening symptoms. Even though the day of onset was unknown in many of the studied cases, the presented virological courses represent the typical situation encountered during MERS treatment in hospital settings.
By providing absolute quantitative measures of virus excretion, we can for the first time compare results between MERS and SARS, a disease that is now thought to have involved higher pandemic potential than MERS [16]. In our patients and elsewhere, the LRT was found to be the main source of MERS-CoV excretion [17]. Unfortunately, there are few data on LRT virus excretion for SARS-CoV, because endotracheal sampling was widely discouraged during SARS outbreaks to avoid nosocomial risks. We have shown, in one of the few available studies, that SARS-CoV was excreted from the LRT at mean concentrations of 1.2–2.8 × 106 copies/mL, reaching a maximum of 1010 copies/mL [18]. That study was conducted in a similar clinical context (a treatment center in Singapore) with similar timing of samples and clinical courses. Determination of viral load was performed by the same laboratory, using equivalent calibrators and conversion factors. From this comparison, we can conclude that average and peak LRT viral loads in MERS are equal to those in SARS [18]. More data are available for upper respiratory tract (URT) viral loads in SARS patients. Peak URT RNA concentrations can reach up to 5 × 105 copies/sample between days 7 and 10 after onset [18, 19]. The corresponding number for MERS (peaking at 4.1 × 106 copies/sample), is equivalent or higher. Also, the 47.6% proportion positive for MERS in URT exceeded the 38% proportion positive in 98 URT samples for SARS patients in Hong Kong [19].
The timing of excretion is more difficult to compare, as many patients in studies on SARS were outpatients who entered hospital because of their SARS infection, whereas our MERS patients were mostly inpatients [20]. In SARS cases that occurred before Hong Kong authorities started active community contact tracing, the average time from symptom onset to admission was 4–7 days [20]. Because case definitions and diagnostics were well-established during SARS in Hong Kong, diagnostic samples would have been taken immediately upon admission. This can be aligned with the timing in our study based on a subcohort of patients for whom an onset of symptoms could be reliably determined. In these patients, the time from onset of symptoms to the initiation of laboratory diagnostics (8 days) was similar to that of early Hong Kong SARS cases. The shedding peak in SARS patients occurred after approximately 10–12 days from symptom onset, which is very similar to the shedding maximum observed in our study, under the assumption that the day of first diagnosis in our MERS cases plausibly falls around the eighth day of symptoms (ie, directly after admission) [21, 22]. In summary, neither the virus concentration nor the timing of respiratory shedding provides an explanation for differences in transmissibility between MERS-CoV and SARS-CoV. Experimental data suggest a higher sensitivity of MERS-CoV to respiratory epithelium–associated type I interferon, which might provide a plausible explanation for its lower transmissibility in comparison with SARS-CoV [23]. Many other explanations are possible, however.
The detection of MERS-CoV in serum is another similarity with SARS. Up to 79% of serum samples were found to contain SARS-CoV RNA during the first week of illness, and around 50% during the second week [24–26]. These numbers match our observations for MERS. Free viremia seems unlikely as a cause of nosocomial transmission of MERS, as no infectious virus was isolated from serum. There is evidence of SARS-CoV replicating in peripheral blood mononuclear cells, macrophages, and dendritic cells, albeit at low levels [27–30]. In the present study, the absence of correlation of serum viral load with LRT viral load points to potential extrapulmonary replication. Viremia despite the presence of neutralizing antibodies indicates a body region that is not accessible to neutralizing antibodies but which releases virus into the blood. SARS-CoV has been shown to replicate in several extrapulmonary organs without evidence of tissue damage [27]. One organ implicated in MERS is the kidney. Kidney failure has been reported in many cases, and MERS-CoV was originally isolated in kidney cells that express DPP4, the MERS-CoV entry receptor [11]. However, earlier healthcare-associated outbreaks have been centered near dialysis centers and nephrology departments, and have affected metabolically compromised patients who are predisposed to kidney failure when suffering from systemic disease affecting blood pressure and circulation. Although patients with SARS-CoV showed viral RNA detection rates up to 50% in urine [7, 22], we rarely found urine-associated MERS-CoV RNA in this study. As in SARS, kidney failure in MERS patients might well be explained by severe inflammatory reaction combined with the administration of potentially nephrotoxic drugs during intensive care [31]. Nevertheless, it will be highly important to conduct postmortem examinations, particularly of the kidney, of patients who die during acute MERS-CoV infection.
A clear difference from SARS was the detection of viral RNA in stool. In SARS, the RNA prevalence in stool samples was so high that testing of stool was proposed as a reliable and sensitive way to routinely diagnose the disease [7, 22]. Active replication in the gut with live virus isolation has been demonstrated [32]. For MERS, we found stool-associated RNA in only 14.6% of samples, with rather low RNA concentration, and had no success in isolating infectious virus. Based on these data, fecal excretion may not have played a relevant role for nosocomial spread of MERS-CoV among the patients under study.
As in SARS, MERS-CoV nosocomial transmission was repeatedly ascribed to the potential of some patients to act as super-shedders or super-spreaders [6, 20]. Our analysis of viral loads, particularly in the early acute phase of disease, supports the existence of a limited number of patients with extraordinarily high viral loads. As these patients were not more likely to die of the infection, they might not have had more severe symptoms, and thus might have been able to engage in social contact despite their disease.
The course of MERS antibody development resembles that of SARS. Patients infected with SARS seroconverted during weeks 2 and 3 after onset [7]. Most of the patients studied here had already seroconverted during the first week after diagnosis, which putatively represents the second week after onset. As in SARS, IgM was not detected earlier than IgG, which limits its diagnostic utility, in particular when considering that IgM against more prevalent human coronaviruses may cross-react with MERS-CoV [12, 33]. With current methodology, IgM testing should be restricted to cases that require proof of recent and overcome MERS-CoV infection.
Information on the prognostic value of antibody response in SARS is less clear. In the present study on MERS, 36% (ELISA) and 45% (NT) of fatal cases failed to mount an antibody response prior to death. However, differences became apparent only in the second week after diagnosis, pointing to only a weak protective effect against lung disease. The development of antibodies in serum was not followed by a rapid elimination of viral RNA from the lung. Neutralizing antibodies normally include immunoglobulin A (IgA) secreted in respiratory fluids and saliva. We have recently shown that anti–MERS-CoV IgA is indeed secreted in respiratory fluids [10], suggesting that the development of IgA comes too late to confer timely reduction of viral replication in infected mucosa. Based on these data, vaccines against MERS-CoV should be designed so as to include and enhance cellular immune responses.
Supplementary Material
Notes
Acknowledgments. We thank Artem Siemens, Heike Kirberg, Monika Eschbach-Bludau, Sebastian Brünink, Stephan Kallies, and Tobias Bleicker (Institute of Virology, Bonn, Germany) for excellent technical assistance. We also thank Dr Terry C. Jones, Department of Zoology, University of Cambridge, United Kingdom, for editing the final manuscript.
Financial support. This work was supported by European Commission grant PREPARE (contract number 602525), as well as the Deutsche Forschungsgemeinschaft (DFG DR772/7-1).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–20. [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization. Disease outbreak news: Middle East respiratory syndrome coronavirus (MERS-CoV). Available at: http://www.who.int/csr/don/12-october-2015-mers-saudi-arabia/en/ Accessed 12 October 2015.
- 3. World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV): summary of current situation, literature update and risk assessment. WHO/MERS/RA/15.1. Available at: http://www.who.int/iris/bitstream/10665/179184/http://apps.who.int//iris/bitstream/10665/179184/2/WHO_MERS_RA_15.1_eng.pdf Accessed 7 July 2015.
- 4. Oboho IK, Tomczyk SM, Al-Asmari AM et al. . 2014 MERS-CoV outbreak in Jeddah—a link to health care facilities. N Engl J Med 2015; 372:846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drosten C, Muth D, Corman VM et al. . An observational, laboratory-based study of outbreaks of Middle East respiratory syndrome coronavirus in Jeddah and Riyadh, Kingdom of Saudi Arabia, 2014. Clin Infect Dis 2015; 60:369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, Wu JT. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Euro Surveill 2015; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poon LL, Guan Y, Nicholls JM, Yuen KY, Peiris JS. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis 2004; 4:663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corman VM, Eckerle I, Bleicker T et al. . Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill 2012; 17. [DOI] [PubMed] [Google Scholar]
- 9. Corman VM, Muller MA, Costabel U et al. . Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill 2012; 17. [DOI] [PubMed] [Google Scholar]
- 10. Muth D, Corman VM, Meyer B et al. . Infectious Middle East respiratory syndrome coronavirus excretion and serotype variability based on live virus isolates from patients in Saudi Arabia. J Clin Microbiol 2015; 53:2951–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raj VS, Mou H, Smits SL et al. . Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013; 495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drosten C, Meyer B, Muller MA et al. . Transmission of MERS-coronavirus in household contacts. N Engl J Med 2014; 371:828–35. [DOI] [PubMed] [Google Scholar]
- 13. Muller MA, Meyer B, Corman VM et al. . Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect Dis 2015; 15:629. [DOI] [PubMed] [Google Scholar]
- 14. Meyer B, Muller MA, Corman VM et al. . Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis 2014; 20:552–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muller MA, Corman VM, Jores J et al. . MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg Infect Dis 2014; 20:2093–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015; 386:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Memish ZA, Al-Tawfiq JA, Makhdoom HQ et al. . Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome. J Infect Dis 2014; 210:1590–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drosten C, Chiu LL, Panning M et al. . Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol 2004; 42:2043–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poon LL, Chan KH, Wong OK et al. . Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin Chem 2004; 50:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leung GM, Hedley AJ, Ho LM et al. . The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: an analysis of all 1755 patients. Ann Intern Med 2004; 141:662–73. [DOI] [PubMed] [Google Scholar]
- 21. Chan KH, Poon LL, Cheng VC et al. . Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis 2004; 10:294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peiris JS, Chu CM, Cheng VC et al. . Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003; 361:1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kindler E, Jonsdottir HR, Muth D et al. . Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 2013; 4:e00611–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grant PR, Garson JA, Tedder RS, Chan PK, Tam JS, Sung JJ. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med 2003; 349:2468–9. [DOI] [PubMed] [Google Scholar]
- 25. Ng EK, Hui DS, Chan KC et al. . Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin Chem 2003; 49:1976–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang WK, Fang CT, Chen HL et al. . Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J Clin Microbiol 2005; 43:962–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gu J, Gong E, Zhang B et al. . Multiple organ infection and the pathogenesis of SARS. J Exp Med 2005; 202:415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Law HK, Cheung CY, Ng HY et al. . Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 2005; 106:2366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L, Wo J, Shao J et al. . SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J Clin Virol 2003; 28:239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yilla M, Harcourt BH, Hickman CJ et al. . SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res 2005; 107:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chu KH, Tsang WK, Tang CS et al. . Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int 2005; 67:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leung WK, To KF, Chan PK et al. . Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 2003; 125:1011–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buchholz U, Muller MA, Nitsche A et al. . Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October-November 2012. Euro Surveill 2013; 18. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.