Abstract
Post-transcriptional mechanisms are an important approach in the treatment of cancer, and may also be hijacked by tumor cells to help adapt to the local microenvironment. Filamin B (FLNB), an actin-binding protein that provides crucial scaffolds for cell motility and signaling, has also been identified as an RNA-binding protein. Recent studies demonstrated that FLNB might play an important role, not only in skeletal development, but also in regulating tumorigenesis; however, the effects of dysregulated expression of FLNB at the molecular level are not clear. In the present study, RNA-sequencing was performed to analyze changes in overall transcriptional and alternative splicing between the knocked-down FLNB and the control in HeLa cells. Decreased FLNB levels resulted in significantly lower apoptosis compared with control cells. FLNB knockdown extensively regulated the expression of genes in cell apoptosis, tumorigenesis, metastases, transmembrane transport and cartilage development. Moreover, FLNB regulated alternative splicing of a large number of genes involved in ‘cell death’ and the ‘apoptotic process’. Some genes and alternative splicing related to skeletal development were enriched and regulated by FLNB. Reverse transcription-quantitative-PCR identified FLNB-regulated transcription and alternative splicing of genes, such as NLR family apoptosis inhibitory protein, interleukin 23 subunit α, metastasis associated lung adenocarcinoma transcript 1, phosphofurin acidic cluster sorting protein 2, bone morphogenetic protein 7, matrix metallopeptidase 13, collagen type II α 1 chain, fibroblast growth factor receptor 2 and vitamin D receptor. The present study is the first study, to the best of the authors’ knowledge, to provide transcriptome-wide analysis of differential gene expression and alternative splicing upon FLNB silencing. The present results suggested that FLNB may play an important regulatory role in cervical cancer cell apoptosis via regulation of transcription and alternative splicing, which provide insight for the current understanding of the mechanisms of FLNB-mediated gene regulation.
Keywords: filamin B, FLNB, HeLa cell, apoptosis, alternative splicing, RNA binding, RNA-sequencing
Introduction
Filamin B (FLNB) is a member of the filamin family, which is a group of highly conserved actin-binding proteins that is responsible for anchoring the actin network to cellular membranes and transmembrane receptors. The FLNB gene was first isolated using a yeast two-hybrid system by Takafuta et al (1). The FLNB gene is located at chromosome 3p14.3 and encodes a protein of 2,602 amino acids (2). FLNB, as a cytoskeletal protein, is broadly distributed across the entire cell (more in the cytoplasm than in the nucleus) and expressed in different tissues, such as blood vessels, colon, breast, prostate and skeletal muscle (3,4). FLNB consists of an N-terminal actin-binding domain, followed by immunoglobulin-like repeat domains that form a receptor-binding region at the C-terminus (5). The FLNB structure facilitates execution of dual functions in two ways: Assisting to form a three-dimensional network of actin through the actin-binding domain; and acting as scaffolding proteins for receptor activation and signal transduction, then directing various cell functions, including membrane stability, ion channel transport, adhesion, proliferation, protrusion and motility (6). FLNB has been identified to play vital roles in skeletal disorders. It was previously identified that FLNB mutations or deficiencies cause multiple skeletal malformations, including scoliosis, spondylocarpotarsal synostosis, Larsen syndrome, atelosteogenesis, boomerang dysplasia, clubfoot, joint dislocation and other unique skeletal abnormalities (7,8).
It has been recently demonstrated that FLNB plays an important role in cancer. Several previous studies demonstrated that FLNB expression was highly correlated with tumor proliferation, metastasis and invasiveness. For example, Bandaru et al (9) identified that a FLNB gene deficiency in mouse embryonic fibroblasts increased the expression and proteolytic activity of matrix metalloproteinase-9 (MMP-9), as well as cell invasion mediated by the RAS/ERK pathway. Another previous in vitro study demonstrated that FLNB is highly expressed in several cancer cells, such as A549 (adenocarcinomic human alveolar basal epithelial cells) and HT1080 (fibrosarcoma cell line) cells, which exhibit high invasiveness (10). In particular, an alternative splicing (AS) switch in FLNB promotes the epithelial-to-mesenchymal transition (EMT) in human breast cancer (11).
Notably, Baltz et al (12) identified that FLNB functions as an RNA-binding protein (RBP) using a photoreactive nucleotide-enhanced UV cross-linking and oligo(dT) purification method. RBPs are involved in virtually all steps of this post-transcriptional regulation, including RNA splicing, polyadenylation, stability, localization, translation and degradation, and defects in RBPs have been linked to many human disorders, including cancer, immunologic disorders and neurodegenerative diseases (13–15). Therefore, it was hypothesized that FLNB has important regulatory functions in regulation of AS and transcription in cancer, which has not been previously reported, to the best of the authors’ knowledge.
In the present study, FLNB was knocked down using short hairpin RNA (shRNA) in HeLa cells derived from human cervical cancer cells. It was demonstrated that cell apoptosis was significantly inhibited. Then, high-throughput RNA-sequencing (RNA-seq) was performed to comprehensively analyze the transcriptome changes of the knocked-down FLNB compared with controls. The present results identified that FLNB regulated the transcription and AS of subsets of genes, especially those involved in apoptosis, proliferation and chondrocyte development signaling pathways. The present results may provide insight for the current understanding of FLNB in regulating gene transcription and AS in cancer.
Materials and methods
Sample preparation and FLNB knockdown
Human HeLa cells (CCTCC@GDC0009) were obtained from The China Center for Type Culture Collection. HeLa cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 100 µg/ml streptomycin, 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), and 100 U/ml penicillin at 37°C in 5% CO2. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform plasmid transfection of HeLa, according to the manufacturer's protocol. In total, 2.5 µg plasmid was used to transfect ~5×105 Hela cells in each well of a 6-well plate. The shRNA sequence for silencing FLNB was 5′-CCTTCAGGAATCGGGATTAA-3′. The primer was synthesized by Sangon Biotech Co., Ltd. HeLa cells which were transfected with empty vector were used as controls.
FLNB knockdown efficiency was assessed by reverse transcription-quantitative (RT-qPCR) by comparing shFLNB groups and control groups with Student's t-test, and GAPDH was used as a control after 48 h of transfection. RNA was extracted using TRIzol® kit (Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA using M-MLV Reverse Transcriptase (Vazyme Biotech Co., Ltd.). The RT procedure included incubation at 37°C for 15 min and then 85°C for 5 sec. Real-time PCR was performed with the StepOne RealTime PCR system (Thermo Fisher Scientific, Inc.) using PrimeScript™ RT reagent kit (Takara Bio, Inc). The PCR conditions consisted of denaturing at 95°C for 10 min, 40 cycles of denaturing at 95°C for 15 sec, annealing and extension at 60°C for 30 sec. PCR amplifications were performed in triplicate for each sample that normalized with the reference gene GAPDH, and the data was assayed using the comparative Cq (2−∆∆Cq) method (16). Primers of FLNB and GAPDH for qPCR analysis are presented in Table SI.
Cell proliferation and apoptosis assay
Cell proliferation was assessed using an MTT assay. In total, 5×103 cell/well HeLa cells were cultured in 96-well plates. Then, cells were transfected with the vector using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After incubation at 37°C for 48 h, MTT solution (5 mg/ml; 0.025 ml) was added to each well. The cells were incubated for another 4 h and the supernatant was removed from each well. The colored formazan crystals produced by MTT were dissolved in DMSO (0.15 ml) and the optical density was measured at 490 nm.
For the flow cytometric analysis of cell apoptosis, the transfected cells were incubated at 37°C for 48 h, and the live cells were harvested and washed twice with ice-cold 1X PBS. Viable cells were double-stained with 7-amino actinomycin D and FITC-conjugated Annexin V (Beijing 4A Biotech Co., Ltd.). The percentage of cell apoptosis was calculated as the sum of the right lower and upper quadrants. The number of stained cells was quantified using a flow cytometer (CytoFLEX; Beckman Coulter, Inc.). Cell cycle distribution was quantified using multi-cycle software (FlowJo 10.5.3; FlowJo, LLC).
Statistical analysis
Student's t-test was used to assess differences between the FLNB knockdown and control groups. The results of each experiment are presented as the mean ± SD of three experiments. The data were analyzed using SPSS 19.0 software (IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
RNA-seq library preparation and sequencing
Total RNA was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and then was purified with two phenol-chloroform treatments. The RNA-seq library was prepared with 1 µg total RNA for each sample using the VAHTS Stranded mRNA-seq Library Prep kit (Vazyme Biotech Co., Ltd.). The libraries were then applied to the Illumina HiSeq X Ten system (Illumina, Inc.) for 150 nt paired-end sequencing.
RNA-Seq and differentially expressed gene (DEG) analysis
Adaptors were removed from raw reads using cutadapt and low-quality bases were trimmed using FASTX-Toolkit (version 0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/index.html). Reads >16 nt were aligned to the human GRch38 genome using Tophat2 (17). Uniquely mapped reads were used for counting gene reads. Fragments per kilobase of transcript per million fragments mapped (FPKM) (18) was calculated for the gene expression level. edgeR (19) and DESeq2 (20) was utilized to screen out the DEGs. A false discovery rate (FDR)<0.05 and log2 fold change ≥1 or ≤-1 were set as the cut-off criteria for identifying DEGs. Pearson's correlation analysis was performed to assess sample distance.
AS analysis
The AS analysis was performed using the ABLas pipeline, as previously described (21,22). ABLas detects 10 types of alternative splicing events (ASEs) based on the splice junction reads and uniquely mapped reads. These ASEs include exon skipping (ES), alternative 3′splice site (A3SS), intron retention, alternative 5′splice site (A5SS), mutually exclusive exons (MXEs), mutually exclusive 3′untranslated regions (UTRs), mutually exclusive 5′UTRs, cassette exon, A3SS&ES and A5SS&ES.
To assess RBP-regulated ASEs, ABLas used Student's t-test to evaluate the significance of the ratio alteration of ASEs. Those events that were significant at the cut-off of FDR≤5% were considered RBP-regulated ASEs.
Validation of DEGs and ASEs using RT-qPCR and western blotting
RT-qPCR reactions were performed for validation of transcription levels and AS changes. The primer information is presented in Table SI. Total RNA remaining from the RNA-seq library preparation was used for RT-qPCR as previously described. The level of RNA expression for all genes was normalized against GAPDH. For ASEs validation, the primers for detecting ASEs are presented in Table SI. To detect alternative isoforms, a boundary-spanning primer of constitutive and alternative exons was used, as well as an opposing primer in one constitutive exon. The boundary-spanning primer of the alternative exon was designed according to a ‘model exon’ to detect model splicing, or an ‘altered exon’ to detect altered splicing.
Western blot analysis was performed for validation of protein expression. Radioimmunoprecipitation assay lysis buffer with protease inhibitors (Sigma-Aldrich; Merck KGaA) was used to lyse treated cells on ice for 30 min, according to the manufacturer's protocol. The protein concentration was determined by the bicinchoninic acid protein assay kit (Sangon Biotech Co., Ltd.). Equal amounts (30 µg/load) of protein samples were resolved by SDS-PAGE on on 10% gels (Bio-Rad Laboratories, Inc.) and transferred to polyvinylidene fluoride difluoride membranes (Merck KGaA). Subsequently, 5% bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc.) was selected to block the membranes for 1 h at room temperature, which were then incubated with primary antibodies against FLNB (ABclonal Biotechnology Co., Ltd.; 1:500, cat. no. A2481), NLR family apoptosis inhibitory protein (NAIP; BIOSS; 1:1,000; cat. no. bs-5804R), mitogen-activated protein kinase kinase 7 (MAP2K7; ABclonal Biotechnology Co., Ltd.; 1:1,000; cat. no. A2186) and GAPDH (ABclonal Biotechnology Co., Ltd.; 1:50,000; cat. no. AC036) at 4°C overnight with gentle rocking. The membranes were then washed three times, and then incubated with a secondary antibody [peroxidase-conjugated AffiniPure goat Anti-Rabbit IgG (H+L); 1:5,000; cat. no. ZB-2301; Origene Technologies, Inc.] at room temperature for 2 h, and immunoreactivity was detected with ECL substrate (Thermo Fisher Scientific, Inc.). GAPDH was used as an internal control.
Functional enrichment analysis
To determine functional categories of DEGs, Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG Database 2017/6/5; http://www.genome.jp/kegg/) pathways enrichment analysis was performed using the KOBAS 2.0 server (23). The most enriched 10 terms were selected and plotted.
Results
FLNB knockdown significantly inhibits apoptosis of HeLa cells
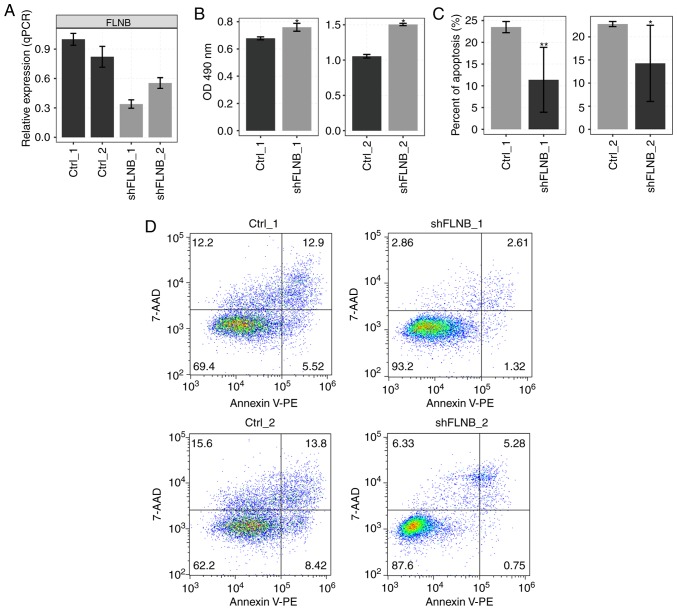
Previous studies demonstrated that FLNB regulates proliferation of chondrocytes (24) and FLNB plays a causal role in the regulation of EMT in human breast cancer (11). Therefore, it was hypothesized that FLNB might regulate proliferation or apoptosis in cancer cells. A cell model was constructed by knocking down FLNB using shRNA in HeLa cells. The expression of FLNB was examined in HeLa cells, which were transfected with empty vector or shRNA with three experimental repeats by RT-qPCR. The mRNA level of FLNB was decreased in shRNA-transfected cells (Fig. 1A). Cell proliferation was significantly increased by knockdown of FLNB (shFLNB; P=0.025; Fig. 1B); however, shFLNB significantly reduced cell apoptosis (P<0.05; Fig. 1C and D). These results suggested that shFLNB significantly inhibited apoptosis in HeLa cells.
Figure 1.
Knockdown of FLNB promotes proliferation and inhibits apoptosis of HeLa cells. (A) Relative mRNA expression of FLNB in HeLa cells after it was knocked down using FLNB-specific shRNA was determined by reverse transcription-qPCR. shFLNB_1 and shFLNB_2 indicate two biological repeats of HeLa cells transfected with FLNB-specific shRNA. Ctrl_1 and Ctrl_2 indicate two biological repeats of HeLa cells transfected with empty vector as controls. (B) Cell proliferation of shFLNB was measured by an MTT assay in HeLa cells. Cell apoptosis of cells transfected with shFLNB was measured by (C) flow cytometry and (D) subsequent analysis by a 7-ADD and Annexin V assay. *P<0.05 and **P<0.01 vs. respective Ctrl. FLNB, filamin B; sh, short hairpin; qPCR, quantitative PCR; Ctrl, control; 7-AAD, 7-amino actinomycin D; OD, optical density; PE, phycoerythrin.
RNA-seq and DEG analysis
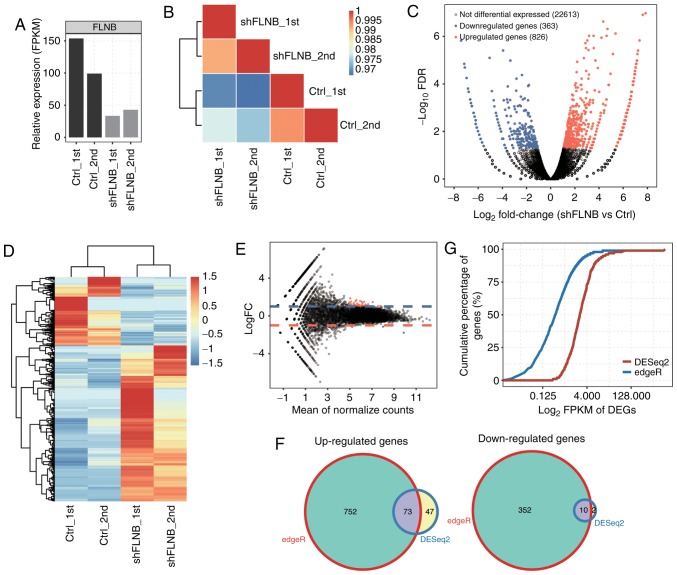
To investigate FLNB-mediated transcriptional regulation, cDNA libraries on shFLNB and control cells were constructed for RNA-seq. Utilizing the Illumina HiSeq X Ten platform, two biological replicates were used and yielded a total of 81.7±8.6 M 150 nucleotide paired-end raw reads per sample. After removing adaptors and low-quality reads, around 78.3±8.4 M high-quality reads were generated, 84.15–89.69% of the reads from each sample could be uniquely aligned to the human GRCH38 genome using TopHat2 (Table SII). Uniquely mapped reads were used to estimate the normalized transcription level as FPKM. In total, 23,763 expressed genes were detected from RNA-seq (Table SIII). The effective knockdown of FLNB was further confirmed with RNA-seq data (Fig. 2A), which was consistent with the RT-qPCR results (Fig. 1A). FPKM values for all 23,763 genes were used to generate a Pearson's distance correlation matrix to compare the transcriptomes from each sample. The heatmap demonstrated that shFLNB and the control were distinguished, and two biological replicates were highly correlated (Fig. 2B).
Figure 2.
RNA-seq and DEG analysis. (A) Comparison of FLNB expression levels (FPKM) between shFLNB and control using RNA-seq data. (B) Pearson's correlation analysis of the gene expression between all biological RNA samples. (C) Volcano plot presents DEGs by shFLNB compared with control cells from two samples in each group as experimental replicates. Red indicates upregulated genes (log2 FC≥1 and FDR<0.05) and blue indicates downregulated genes (log2 FC≤-1 and FDR<0.05). (D) Expression heatmap of all 1,189 significant DEGs between shFLNB and control cells. (E) MA-plot for differential expression analysis using DESeq2. The x-axis indicates the mean of normalized counts and the y-axis indicates the log2 fold change. (F) Venn diagram showing the overlapped upregulated (left) and downregulated (right) genes detected by edgeR and DESeq2. (G) Cumulative profiles of genes expression level of DEGs from DESeq2 and edgeR. RNA-seq, RNA-sequencing; DEG, differentially expressed gene; FLNB, filamin B; FPKM, fragments per kilobase of transcript per million fragments mapped; sh, short hairpin; FC, fold change; FDR, false discovery rate; Ctrl, control.
The DEGs between the shFLNB and control cells were investigated using the edgeR package (19). In total, 826 upregulated and 363 downregulated DEGs (Table SIV) were identified (log2 fold change≥1 and FDR <5%). The DEGs associated with shFLNB are presented in a volcano plot (Fig. 2C). The expression heatmap of DEGs indicated that more genes were upregulated upon shFLNB transfection (Fig. 2D). It was additionally identified that the mRNA expression of MMP-9 was downregulated after knockdown of FLNB in HeLa cells (Table SIII), which is not consistent with that reported in mouse embryonic fibroblasts (9).
DESeq2 (20) was used to detect DEGs, to compare the results with edgeR. A total of 120 upregulated and 12 downregulated DEGs were identified (FDR <5%; Fig. 2E). This suggested that DESeq2 was more stringent than edgeR in recovering DEGs. The Venn diagram demonstrated that 63% of DEGs based on DESeq2 were also identified by edgeR (Fig. 2F). DESeq2 is more inclined to identify DEGs with higher expression compared with edgeR (Fig. 2G).
Knockdown of FLNB in HeLa cells preferentially regulates transcription level variation of tumorigenesis and cartiliage development-related genes
To further examine the potential biological roles of these DEGs regulated by FLNB, GO and KEGG enrichment analysis was performed. The top 10 biological process terms of the GO analysis demonstrated that shFLNB-upregulated genes were strongly enriched in ‘membrane depolarization involved in regulation of action potential’, ‘transmembrane transport’, ‘negative regulation of neuron apoptotic process’, ‘cartilage development’ and ‘extracellular matrix organization’ (Fig. 3A). The downregulated genes were enriched in ‘signal transduction’, ‘cell adhesion’, ‘angiogenesis’, ‘inflammatory response’, ‘innate immune response’ and ‘cell differentiation’ (Fig. 3B). Based on the KEGG analysis (Fig. 3C), the pathways of upregulated gene sets were mainly associated with the ‘GABAergic synapse’, ‘ABC transporters’ and ‘ECM-receptor interaction’ signaling pathway. ‘Leukocyte transendothelial migration’, ‘cell adhesion molecules (CAMs)’ and ‘transcriptional misregulation in cancer’ pathways were significantly enriched in downregulated gene sets (Fig. 3D; Table SV).
Figure 3.
GO and KEGG analysis of differentially expressed genes between short hairpin FLNB-transfected and control HeLa cells. Top 10 most enriched GO terms (biological process) of (A) upregulated and (B) downregulated genes upon FLNB knockdown. Rectangles around GO terms indicate notable cancer-related and cartilage development terms. Top 10 most enriched KEGG pathways of (C) upregulated and (D) downregulated genes upon FLNB knockdown. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FLNB, filamin B.
FLNB regulates AS in HeLa cells
Since RNA-seq technology provides an opportunity to study AS, transcriptome data were used to examine ASEs that might be regulated by FLNB in HeLa cells. Spliced reads account for 48.46–53.99% of all uniquely mapped reads obtained from shFLNB and control cells (Table SII). Of the 367,321 annotated exons from the reference genome, 224,199 were detected. Using Tophat2, 151,708 annotated splice junctions and 108,767 novel splice junctions were detected, accounting for 40% of the total. A total of 16,783 known ASEs (defined in the reference genome) and 46,959 new splicing events (not including intron retention) were identified using the previously published ABLas software tool (Table SVI). These results suggested that splicing in human is more complex than previously hypothesized.
For each ASE, the differential splicing patterns usage between shFLNB and control cells was calculated. A total of 402 significant differential ASEs in 363 genes was identified (AS ratio difference ≥0.2 and P-value ≤0.05; Table SVII). In detail, these ASEs include 48 ES, 49 cassette exon, 75 A5SS, 65 A3SS and 119 intron retention (Fig. 4A). Compared with the control cells, an enrichment of ES and a depletion of cassette exon in shFLNB were observed, suggesting that exon inclusion is suppressed by FLNB silencing (Fig. 4A). To examine the changes in ASEs attributed to transcription regulation, genes with differential expression level and ASEs were overlapped. The present results identified that there were only four DEGs with significantly altered splicing. Notably, metastasis associated lung adenocarcinoma transcript 1 (MALAT1), as a well-known long non-coding RNA associated with cancer metastasis, demonstrated changes in expression and AS after FLNB knockdown (Fig. 4B).
Figure 4.
Alternative splicing analysis by shFLNB. (A) Classification of significant changes in alternative splicing after FLNB knockdown. (B) Overlap of DEGs and genes with differential alternative splicing (RASG). Top 10 (C) GO biological process terms and (D) KEGG pathways of genes with differential alternative splicing events between shFLNB and the control. Rectangles around GO terms /KEGG pathways indicate notable apototic-related and cartilage development terms. sh, short hairpin; FLNB, filamin B; DEG, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; 3pMXE, mutually exclusive 3′untranslated regions; 5pMXE, mutually exclusive 5′untranslated regions; A3SS, alternative 3′splice site; A5SS, alternative 5′splice site; ES, exon skipping; IntronR, intron retention; RASE, regulated alternative splicing events; RASG, genes with differential alternative splicing; KIF9, kinesin family member 9; SLC25A36, solute carrier family 25 member 36; MALAT1, metastasis associated lung adenocarcinoma transcript 1.
GO functional clustering analysis demonstrated that the alternative spliced genes were enriched in ‘organ morphogenesis’, ‘apoptotic process’, ‘cell death’ and ‘signal transduction’ (Fig. 4C). KEGG pathways were most enriched in those involved in ‘Adherens junctions’, ‘GnRH signaling pathway’, ‘Axon guidance’, ‘Bacterial invasion of epithelial cells’ and ‘Mineral absorption’ pathways (Fig. 4D; Table SVIII). These results suggested that FLNB knockdown markedly regulated AS of apoptosis-related genes and genes involved in skeletal development.
RT-qPCR validation of DEGs and ASEs by FLNB knockdown in HeLa cells
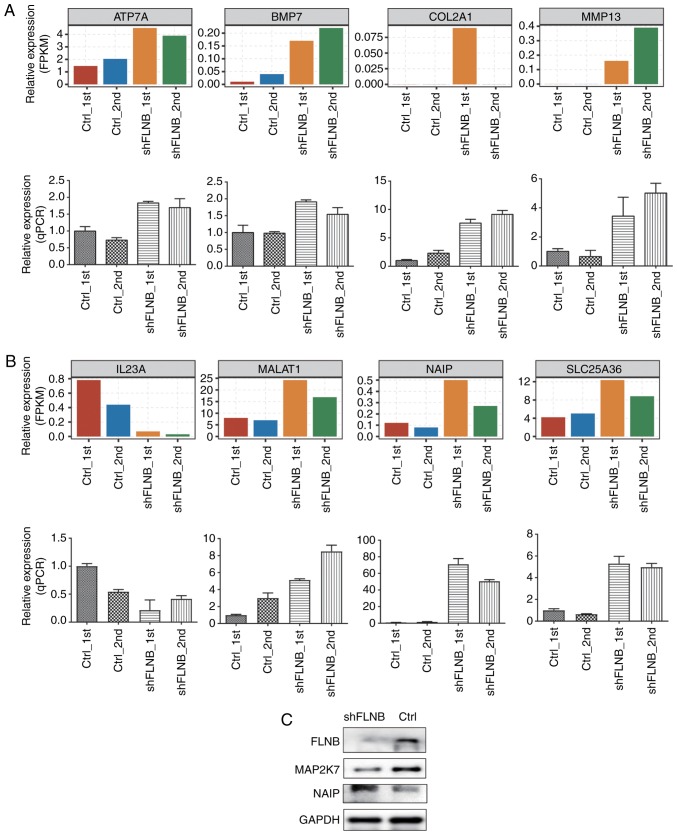
A number of important tumorigenesis and skeletal development-related DEGs and ASEs in HeLa cells were further validated by RT-qPCR. A total of nine DEGs, including ATPase copper transporting α (ATP7A), bone morphogenetic protein 7 (BMP7), collagen type II α 1 chain (COL2A1), matrix metallopeptidase 13 (MMP13), interleukin 23 subunit α (IL23A), MALAT1, NAIP, solute carrier family 25 member 36 and ghrelin and obestatin prepropeptide (GHRL) were evaluated, although expression of GHRL did not yield sufficient PCR signals. The results of all other eight genes upon FLNB silencing matched well with the RNA-seq data, suggesting that FLNB-regulated DEGs identified from RNA-seq data are acceptable (Fig. 5A and B). A total of two apoptotic-related proteins were detected by western blotting in the shFLNB and control cells; NAIP (negative regulator of apoptosis) was upregulated and MAP2K7 (positive regulator of apoptosis) was downregulated in the shFLNB cells compared with the control cells (Fig. 5C). A total of four FLNB-regulated splicing events located in vitamin D receptor (VDR), phosphofurin acidic cluster sorting protein 2 (PACS2), MAP2K7 and MALAT1 were validated by designing PCR primer pairs (Table SI), which amplify both long and short splicing isoforms in the same reaction (Fig. 6). These four ASEs validated by qPCR were in agreement with the RNA-seq results. The ratio of two splicing patterns in each ASE were significantly altered after FLNB-knockdown. These experimental results further suggested that FLNB plays an important regulatory role in gene expression and AS in HeLa cell.
Figure 5.
Validation of FLNB-regulated genes (DEGs). (A) Relative expression level (FPKM, up) and RT-qPCR measurement (down) of cartilage development-related DEGs. (B) Related expression level (FPKM, up) and RT-qPCR measurement (down) of apoptotic-related DEGs. (C) Western blot analysis of two apoptotic-related proteins in shFLNB and Ctrl HeLa cells. FLNB, filamin B; DEGs, differentially expressed genes; FPKM, fragments per kilobase of transcript per million fragments mapped; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; Ctrl, control; ATP7A, ATPase copper transporting α; BMP7, bone morphogenetic protein 7; COL2A1, collagen type II α 1 chain; MMP13, matrix metallopeptidase 13; IL23A, interleukin 23 subunit α; MALAT1, metastasis associated lung adenocarcinoma transcript 1; NAIP, NLR family apoptosis inhibitory protein; SLC25A36, solute carrier family 25 member 36; MAP2K7, mitogen-activated protein kinase kinase 7.
Figure 6.
Validation of FLNB-affected ASEs. Genome visualization (left panel) shows FLNB-regulated ASEs in shFLNB and control. (A) Validation of an ASE of VDR in HeLa cells. (B) Validation of an ASE of PACS2 in HeLa cells. (C) Validation of an ASE of MAP2K7 in HeLa cells. (D) Validation of an ASE of MALAT1 in HeLa cells. The number of junction reads were marked on the line representing splice junction composing ASE. The structures of ASEs are depicted in the top-right panel. The altered ratio of ASEs in RNA-sequencing and in reverse transcription-quantitative PCR were calculated and plotted (right panel, bottom). FLNB, filamin B; ASEs, alternative splicing events; sh/SH, short hairpin; NC, negative control; VDR, vitamin D receptor; PACS2, phosphofurin acidic cluster sorting protein 2; MAP2K7, mitogen-activated protein kinase kinase 7; MALAT1, metastasis associated lung adenocarcinoma transcript 1. *P<0.05 and ***P<0.001 vs. respectively NC.
Discussion
The present study is the first study, to the best of the authors’ knowledge, to perform entire transcriptome analysis with the knockdown of FLNB in a cancer cell line (HeLa cells). The widespread effects of inactivation of FLNB on multiple biological functions were identified, especially tumorigenesis and skeletal development. After FLNB knockdown in HeLa cells, compared with the control, it was identified that the level of apoptosis significantly decreased, consistent with the suppressive role in tumor progression of FLNB as reported in mouse embryonic fibroblasts (9). Furthermore, the transcriptome analysis demonstrated that FLNB knockdown led to upregulation of genes that can negatively regulate the apoptotic process, including genes such as NAIP, while significantly downregulating inflammatory and immune-related genes. Additionally, FLNB knockdown resulted in the deregulation of the AS of hundreds of genes; many were enriched in the top three GO functional terms associated with apoptosis. Notably, it was observed that FLNB-regulated the expression of genes in skeletal development-related pathways, such as ‘cartilage development’ and ‘mineral absorption’ at both the AS and mRNA levels. These findings together suggested that FLNB can extensively regulate pre-mRNA AS in cancer cells, which may be exerted via its RNA binding capability. The FLNB-regulated mRNA expression level could be resulted from either FLNB-regulated mRNA stability or transcription directly or indirectly. The present data supported a model that FLNB-knockdown can inhibit cancer cell apoptosis via regulation of AS and mRNA levels of the apoptotic genes. The present results demonstrated that FLNB regulates the AS and mRNA levels of functional related genes, which may provide insight for the current understanding for FLNB biology and regulatory mechanisms, as well as for FLNB-targeted anti-cancer therapies.
A number of previous studies demonstrated that FLNB mutations or deficiencies result in multiple skeletal hypogenesis; the clinical symptoms usually manifest as a reduction of bone mineral density (BMD), ossification of intervertebral disc, disturbance of proliferation, differentiation and apoptosis in chondrocytes, and impairment of angiogenesis (24–26). The present DEG analysis identified that FLNB knockdown increased the expression of genes relevant to cartilage development, including MMP13, COL2A1, BMP7 and ATP7A in HeLa cells, which need further verification in cartilage cells (27). Additionally, previous studies (28,29) demonstrated that FLNB mutations are associated with a reduction in BMD; however, the effect of FLNB mutations on bone remodeling is largely unclarified based on functional studies. In the present study, it was observed that knockdown of FLNB altered the splicing pattern of VDR. VDR is a member of the nuclear hormone receptor superfamily, and is a ligand-inducible transcription factor (30). A recent study suggested that alternatively spliced transcript variants encoding different isoforms in VDR genes are important in regulating the metabolic processes that affect BMD (31). Indeed, the VDR gene FokI and BsmI polymorphisms are significantly associated with low BMD (32). These results provide insight into the molecular mechanism of BMD caused by FLNB dysfunction, and cartilage cells should be used in future studies to verify the regulatory role of FLNB.
Recently, FLNB has been considered one of the biomarkers for cancer. Splicing variants of FLNB expression have been reported to play key roles in the proliferation and differentiation of tumor cells (33). A recent study demonstrated that in HMLE cells (immortalized human mammary epithelial cells), FLNB interacts with forkhead box C1 (FOXC1) to form a complex (11). FLNB exon skipping leads to a decrease in FLNB protein located in the nucleus, thus the reduced interaction between FLNB, FOXC1 and PBX homeobox 1 leads to the release of FOXC1 to promote EMT (11). Th{Li, 2018 #58}is finding suggested that FLNB plays an important regulatory role in tumorigenesis. In the present study, it was demonstrated that FLNB also had transcriptional and post-transcriptional regulatory functions in HeLa cells.
DEGs between shFLNB and control HeLa cells were obtained. GO and KEGG enrichment analyses demonstrated that upregulated genes were enriched in the' negative regulation of the neuron apoptotic process' pathway, and downregulated genes were enriched in ‘inflammatory response’ and ‘innate immune response’, which may contribute to the survival of cancer cells. Notably, it was verified that FLNB knockdown increased the expression of NAIP and MALAT1.
IL23A associates with interleukin 12B to form interleukin (IL)-23, a heterodimeric cytokine which induces autoimmune inflammation, and is related to tumor incidence and growth (34). Low concentrations of IL-23 promote the proliferation of lung cancer cells, whereas high concentrations of IL-23 inhibit proliferation (35). This is consistent with the fact that FLNB knockdown promotes cell proliferation. NAIP is one of eight human inhibitors of apoptosis proteins that have been identified (36). NAIP has been suggested to have an important role in protecting cells against apoptosis (37). Although NAIP is generally thought to be expressed in nerve cells, NAIP is weakly linked to unfavorable prognosis in multiple cancer types, which indicates the precise role of NAIP in the dysregulation of apoptosis in cancer (38). The present results suggested that FLNB may control the expression of NAIP, and thus affect apoptosis of cancer cells. MALAT1, a long non-coding RNA, is a transcriptional regulator for numerous genes, including some genes related with cancer metastasis and cell migration, and MALAT1 is involved in cell cycle regulation (39). Upregulation of MALAT1 in multiple cancerous tissues was associated with the proliferation and metastasis of tumor cells (39). In conclusion, FLNB knockdown results in abnormal expression of many important genes associated with tumorigenesis, demonstrating the important regulatory role of FLNB in the process of cancer.
Furthermore, it was identified that AS genes regulated by FLNB were significantly enriched in the ‘apoptotic process’ and ‘cell death’ pathways. The FLNB-regulated AS genes in these pathways included PACS2, MALAT1 and MAP2K7, which were validated by RT-qPCR. In particular, PACS2 has been shown to be a multi-functional sorting protein, which regulates endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis (40). PACS2 also mediates tumor necrosis factor-related apoptosis-inducing ligand lysosomes, interacting with Bim and Bax to permeabilize lysosomes and induce apoptosis (41). The present findings regarding FLNB-mediated AS of apoptotic-regulating genes suggested a more complex network among the apoptotic regulation process in cancer, which supports a previous study demonstrating that FLNB knockdown in mouse embryonic fibroblasts leads to increased proteolytic activity of MMP-9 (9). In the present study, it was identified that the mRNA expression of MMP-9 was downregulated after knockdown of FLNB in HeLa cells. Further research is required to determine whether or not FLNB regulates transcription and AS by binding to RNA as an RBP.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Yi Zhang (ABLife BioBigData Institute) for her helpful discussions.
Funding
The present study was supported by The National Natural Science Foundation of China (grant no. 81860746) and The Joint Fund of the National Natural Science Foundation of China (grant no. U1503221).
Availability of data and materials
RNA-sequencing data in the present study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO (https://www.ncbi.nlm.nih.gov/geo) series (accession no. GSE134769).
Authors' contributions
HRM and LC contributed to the study design. FW, CC, RJ, HKZ, SW and ZQ conducted the experiments and/or performed data analysis. HRM and CC prepared the manuscript. ZX made the part analysis of bioinformation data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Takafuta T, Wu G, Murphy GF, Shapiro SS. Human beta-filamin is a new protein that interacts with the cytoplasmic tail of glycoprotein Ibalpha. J Biol Chem. 1998;273:17531–17538. doi: 10.1074/jbc.273.28.17531. [DOI] [PubMed] [Google Scholar]
- 2.Brocker F, Bardenheuer W, Vieten L, Jülicher K, Werner N, Marquitan G, Michael D, Opalka B, Schütte J. Assignment of human filamin gene FLNB to human chromosome band 3p14.3 and identification of YACs containing the complete FLNB transcribed region. Cytogenet Cell Genet. 1999;85:267–268. doi: 10.1159/000015309. [DOI] [PubMed] [Google Scholar]
- 3.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Walsh CA. The many faces of filamin: A versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 5.Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): A molecular leaf spring. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldassarre M, Razinia Z, Burande CF, Lamsoul I, Lutz PG, Calderwood DA. Filamins regulate cell spreading and initiation of cell migration. PLoS One. 2009;4:e7830. doi: 10.1371/journal.pone.0007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang CF, Wang CH, Siong H'ng W, Chang CP, Lin WD, Chen YT, Wu JY, Tsai FJ. Filamin B loss-of-function mutation in dimerization domain causes autosomal-recessive spondylocarpotarsal synostosis syndrome with Rib anomalies. Hum Mutat. 2017;38:540–547. doi: 10.1002/humu.23186. [DOI] [PubMed] [Google Scholar]
- 8.Bicknell LS, Morgan T, Bonafe L, Wessels MW, Bialer MG, Willems PJ, Cohn DH, Krakow D, Robertson SP. Mutations in FLNB cause boomerang dysplasia. J Med Genet. 2005;42:e43. doi: 10.1136/jmg.2004.029967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandaru S, Zhou AX, Rouhi P, Zhang Y, Bergo MO, Cao Y, Akyürek LM. Targeting filamin B induces tumor growth and metastasis via enhanced activity of matrix metalloproteinase-9 and secretion of VEGF-A. Oncogenesis. 2014;3:e119. doi: 10.1038/oncsis.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iguchi Y, Ishihara S, Uchida Y, Tajima K, Mizutani T, Kawabata K, Haga H. Filamin B Enhances the invasiveness of cancer cells into 3D collagen matrices. Cell Struct Funct. 2015;40:61–67. doi: 10.1247/csf.15001. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Choi PS, Chaffer CL, Labella K, Hwang JH, Giacomelli AO, Kim JW, Ilic N, Doench JG, Ly SH, et al. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife. 2018;7:e37184. doi: 10.7554/eLife.37184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol. 2014;15:492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Mangelsdorf M, Liu J, Zhu L, Wu JY. RNA-binding proteins in neurological diseases. Sci China Life Sci. 2014;57:432–444. doi: 10.1007/s11427-014-4647-9. [DOI] [PubMed] [Google Scholar]
- 15.Wurth L, Gebauer F. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta. 2015;1849:881–886. doi: 10.1016/j.bbagrm.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y, Zhang L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech. 2017;1860:911–921. doi: 10.1016/j.bbagrm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Li G, Yu D, Huang W, Cheng C, Liao S, Wu Q, Zhang Y. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus Verticillium dahliae. BMC Genomics. 2017;18:130. doi: 10.1186/s12864-017-3507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39((Web Server Issue)):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Lu J, Lian G, Ferland RJ, Dettenhofer M, Sheen VL. Formin 1 and Filamin B physically interact to coordinate chondrocyte proliferation and differentiation in the growth plate. Hum Mol Genet. 2014;23:4663–4673. doi: 10.1093/hmg/ddu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson SG, Jones MR, Mullin BH, Dick IM, Richards JB, Pastinen TM, Grundberg E, Ljunggren O, Surdulescu GL, Dudbridge F, et al. Common sequence variation in FLNB regulates bone structure in women in the general population and FLNB mRNA expression in osteoblasts in vitro. J Bone Miner Res. 2009;24:1989–1997. doi: 10.1359/jbmr.090530. [DOI] [PubMed] [Google Scholar]
- 26.Zieba J, Forlenza KN, Khatra JS, Sarukhanov A, Duran I, Rigueur D, Lyons KM, Cohn DH, Merrill AE, Krakow D. TGFβ and BMP dependent cell fate changes due to loss of filamin B produces disc degeneration and progressive vertebral fusions. PLoS Genet. 2016;12:e1005936. doi: 10.1371/journal.pgen.1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Lian G, Lenkinski R, De Grand A, Vaid RR, Bryce T, Stasenko M, Boskey A, Walsh C, Sheen V. Filamin B mutations cause chondrocyte defects in skeletal development. Hum Mol Genet. 2007;16:1661–1675. doi: 10.1093/hmg/ddm114. [DOI] [PubMed] [Google Scholar]
- 28.Mullin BH, Mamotte C, Prince RL, Spector TD, Dudbridge F, Wilson SG. Conditional testing of multiple variants associated with bone mineral density in the FLNB gene region suggests that they represent a single association signal. BMC Genet. 2013;14:107. doi: 10.1186/1471-2156-14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Q, Wu N, Cui L, Wu Z, Qiu G. Filamin B: The next hotspot in skeletal research? J Genet Genomics. 2017;44:335–342. doi: 10.1016/j.jgg.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Campbell MJ, Adorini L. The vitamin D receptor as a therapeutic target. Expert Opin Ther Targets. 2006;10:735–748. doi: 10.1517/14728222.10.5.735. [DOI] [PubMed] [Google Scholar]
- 31.Nakamichi Y, Udagawa N, Suda T, Takahashi N. Mechanisms involved in bone resorption regulated by vitamin D. J Steroid Biochem Mol Biol. 2018;177:70–76. doi: 10.1016/j.jsbmb.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Bao L, Chen M, Lei Y, Zhou Z, Shen H, Le F. Association between vitamin D receptor BsmI polymorphism and bone mineral density in pediatric patients: A meta-analysis and systematic review of observational studies. Medicine (Baltimore) 2017;96:e6718. doi: 10.1097/MD.0000000000006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsui JC, Lau CP, Cheung AC, Wong KC, Huang L, Tsui SK, Kumta SM. Differential expression of filamin B splice variants in giant cell tumor cells. Oncol Rep. 2016;36:3181–3187. doi: 10.3892/or.2016.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Zhang L, Zhang J, Wei Y, Li K, Huang L, Zhang S, Gao B, Wang X, Lin P. Interleukin 23 regulates proliferation of lung cancer cells in a concentration-dependent way in association with the interleukin-23 receptor. Carcinogenesis. 2013;34:658–666. doi: 10.1093/carcin/bgs384. [DOI] [PubMed] [Google Scholar]
- 36.Davoodi J, Ghahremani MH, Es-Haghi A, Mohammad-Gholi A, Mackenzie A. Neuronal apoptosis inhibitory protein, NAIP, is an inhibitor of procaspase-9. Int J Biochem Cell Biol. 2010;42:958–964. doi: 10.1016/j.biocel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, Nalps and Naip: Intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 2003;5:581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- 38.Finlay D, Teriete P, Vamos M, Cosford NDP, Vuori K. Inducing death in tumor cells: Roles of the inhibitor of apoptosis proteins. F1000Res. 2017;6:587. doi: 10.12688/f1000research.10625.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: A potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–6768. doi: 10.2147/CMAR.S169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Carmine Simmen K, Cooper TJ, Thomas G, Simmen T. The subcellular distribution of calnexin is mediated by PACS-2. Mol Biol Cell. 2008;19:2777–2788. doi: 10.1091/mbc.e07-10-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werneburg NW, Bronk SF, Guicciardi ME, Thomas L, Dikeakos JD, Thomas G, Gores GJ. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein-induced lysosomal translocation of proapoptotic effectors is mediated by phosphofurin acidic cluster sorting protein-2 (PACS-2) J Biol Chem. 2012;287:24427–24437. doi: 10.1074/jbc.M112.342238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data in the present study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO (https://www.ncbi.nlm.nih.gov/geo) series (accession no. GSE134769).