Abstract
Background
Respiratory virus–laden particles are commonly detected in the exhaled breath of symptomatic patients or in air sampled from healthcare settings. However, the temporal relationship of detecting virus-laden particles at nonhealthcare locations vs surveillance data obtained by conventional means has not been fully assessed.
Methods
From October 2016 to June 2018, air was sampled weekly from a university campus in Hong Kong. Viral genomes were detected and quantified by real-time reverse-transcription polymerase chain reaction. Logistic regression models were fitted to examine the adjusted odds ratios (aORs) of ecological and environmental factors associated with the detection of virus-laden airborne particles.
Results
Influenza A (16.9% [117/694]) and influenza B (4.5% [31/694]) viruses were detected at higher frequencies in air than rhinovirus (2.2% [6/270]), respiratory syncytial virus (0.4% [1/270]), or human coronaviruses (0% [0/270]). Multivariate analyses showed that increased crowdedness (aOR, 2.3 [95% confidence interval {CI}, 1.5–3.8]; P < .001) and higher indoor temperature (aOR, 1.2 [95% CI, 1.1–1.3]; P < .001) were associated with detection of influenza airborne particles, but absolute humidity was not (aOR, 0.9 [95% CI, .7–1.1]; P = .213). Higher copies of influenza viral genome were detected from airborne particles >4 μm in spring and <1 μm in autumn. Influenza A(H3N2) and influenza B viruses that caused epidemics during the study period were detected in air prior to observing increased influenza activities in the community.
Conclusions
Air sampling as a surveillance tool for monitoring influenza activity at public locations may provide early detection signals on influenza viruses that circulate in the community.
Keywords: influenza and respiratory viruses, airborne particles, human density, temporal pattern, surveillance
Influenza viruses were detected at higher frequencies than other respiratory viruses in air sampled from a university campus. Increased crowdedness and higher indoor temperature were associated with detection of influenza airborne particles by multivariate analyses.
Respiratory viruses are among the leading causes of morbidity and mortality. A substantial global disease burden is attributed to influenza virus and respiratory syncytial virus (RSV), which cause lower respiratory infections and cardiopulmonary complications [1–4]. While most infections are associated with acute respiratory illnesses, respiratory viruses can also infect without causing symptoms [5–7], although the infectivity of asymptomatic cases is uncertain. A common feature of respiratory viruses is their ability to spread via multiple non–mutually exclusive modes, by direct contact or indirectly via large droplets, fine droplet nuclei, or fomites [8–11]. The predominant transmission mode may vary between viruses and be influenced by environmental conditions; however, such information remains a major knowledge gap [8–11]. Airborne transmission via fine droplet nuclei is less confined by the spatiotemporal restriction posed by gravity and has been the most challenging transmission mode for infection control considerations [8].
Previous studies have reported detection of influenza and other respiratory virus–laden particles in the exhaled breath of symptomatic patients [12–15] or in air sampled at healthcare facilities [16–19]. It is currently unknown if people with mild or subclinical respiratory infections, who are capable to continue their daily activities in the community without substantial impediment, may also release virus-laden particles with airborne transmission potential. A recent study reported molecular detection of respiratory viruses in nasal swabs among 6.2% of adult tourists in New York City, of which >65% of positive cases were asymptomatic [6, 7]. Public locations where infected and susceptible hosts converge may serve as hotspots within a transmission network during epidemics. However, the temporal detection frequency of common respiratory viruses at public locations has not been systematically assessed and correlated with surveillance data obtained by conventional methods.
To evaluate the airborne transmission potential of respiratory viruses at public locations, we performed weekly surveillance from October 2016 to June 2018 to determine the quantity and airborne particle size distribution of influenza A viruses (IAVs), influenza B viruses (IBVs), human rhinoviruses (HRVs), RSV, and human coronaviruses (HCoVs) 229E and OC43 in ambient air sampled from a university campus in Hong Kong. Parameters including relative humidity (RH), temperature, crowdedness, and ventilation that may be associated with detection of airborne virus particles were recorded and analyzed by logistic regression. We compared the temporal relationship between the detection of influenza airborne particles at the university campus and the detection of symptomatic influenza infections in the community under conventional surveillance.
METHODS
Air Sample Collection
Air was sampled weekly from a university campus in Hong Kong from October 2016 to June 2018 except during the summer holidays in 2017 (weeks 19–35) and on public holidays. Samples were collected from canteens, lecture halls, shuttle buses, and the University Health Service (UHS) during peak hours of human flow, using 6–13 air samplers per week. Air was sampled for 30 minutes using the National Institute for Occupational Safety and Health (NIOSH) bioaerosol sampler (BC251) that separates particles into >4 μm, 1–4 μm, and <1 μm size fractions [20] using air pumps (XR5000, SKC) calibrated to 3.5 L/minute. Samplers were set up at 1.2-m height, and samplers not connected to pumps served as negative controls. After collection, the samplers were transported on ice to the laboratory for processing.
Recording of Ecological and Environmental Factors
Indoor temperature (°C), indoor RH (%), and indoor/outdoor carbon dioxide (CO2, ppm) were measured by an indoor air quality meter (IAQ-CALC 7525, TSI). Outdoor temperature and RH were obtained from the Hong Kong Observatory [21]. Numbers of occupants on site were visually counted during each sampling event except within canteens, where the mobile population was approximated by the number of transactions recorded during the sampling period. The ventilation rate (q, L/second) was calculated using indoor (Cindoor) and outdoor (Coutdoor) CO2 concentrations (ppm), CO2 production rate per person (a; 0.005 L/second for a person with a light intensity activity of 1.2 metabolic equivalents), and numbers of occupants (Np) on site [22]:
Detection and Quantification of Viral RNA Genomes From Airborne Particles
Sampled airborne particles were resuspended with 1 mL of minimal essential medium containing 0.3% bovine serum albumin (Sigma) [23]. Viral RNA (vRNA) was extracted from 140 μL of resuspended media using the QIAamp Viral RNA Mini Kit (Qiagen) and eluted into 60 μL water. Respiratory viruses were detected using quantitative real-time reverse-transcription polymerase chain reaction (rRT-qPCR) by the ViiA 7 Real-time PCR System (ThermoFisher) using 5 μL eluent, AgPath-IDTM One-step RT-PCR Reagents (Life Technology), and specific primers and probes (Supplementary Table 1) [24–28]. IAV M gene–positive samples were further subtyped (H1 and H3) by rRT-qPCR [25]. A sample was considered positive when an exponential amplification curve crossed the threshold line within 40 cycles. Quantification of influenza viruses was based on standard curves constructed using 10-fold serial-diluted plasmids; the minimum linear range of quantification (LoQ) was 2 genome copies per reaction (8163 copies/m3 air).
Infectivity of Airborne Virus–laden Particles In Vitro
To test if rRT-qPCR–positive samples contain infectious viruses, 250 μL of resuspended airborne particles was inoculated on Madin-Darby canine kidney cells (MDCK; influenza-positive samples), human epithelial (Hep)-2 cells (RSV-positive samples), or human lung fibroblasts Medical Research Council cell strain 5 cells (MRC-5; HRV/HCoV-positive samples) in 24-well plates. Air samples were incubated with (i) MDCK/Hep-2/MRC-5 cells with media, and (ii) with media only. After 48 hours, 250 μL of the supernatant from (i) and (ii) was blindly transferred and incubated with and without fresh cells, respectively, for another 48 hours. Subsequently, viral copy numbers were determined using rRT-qPCR. A sample was considered infectious if a higher viral genome copy was detected after 2 blind passages in cells when compared to the control wells without cells.
Statistical Analysis
Statistical analyses and graphic production were performed using Microsoft Excel and R version 3.3.1 software programs. Absolute humidity (AH; kg/m3), as a better predictor than RH for influenza infectivity [29], was calculated by Clausius-Clapeyron equation using the R package [30, 31]. Fisher exact and χ2 tests were used to determine the difference of categorical data, and Kruskal-Wallis test was used for numeric data. Factors associated with detection of influenza airborne particles were analyzed by logistic regression. Numbers of occupants on site, ventilation rate, and indoor CO2 concentration were categorized as high (upper 33%), intermediate (middle 33%), and low (lower 33%) based on the numeric data distribution (Supplementary Figure 1). Factors that were significantly associated (P < .05) with detection of influenza airborne particles were further investigated in multivariable logistic regression models. The final models were determined using backward model selection based on Akaike information criterion (AIC) (StepAIC function in R-package MASS) [32]. Factors known to be associated with airborne particle formation though failing to meet P < .05 were further investigated in competing models, and adjusted odds ratios (aORs) were examined.
Ethics Statement
The Institutional Review Board of the University of Hong Kong (HKU/HA HKW IRB) reviewed the study protocol and concluded that human ethics approval was not needed, as human subjects were not involved in this study.
RESULTS
Air Sampling and Detection of Respiratory Virus–laden Particles in Air by rRT-qPCR
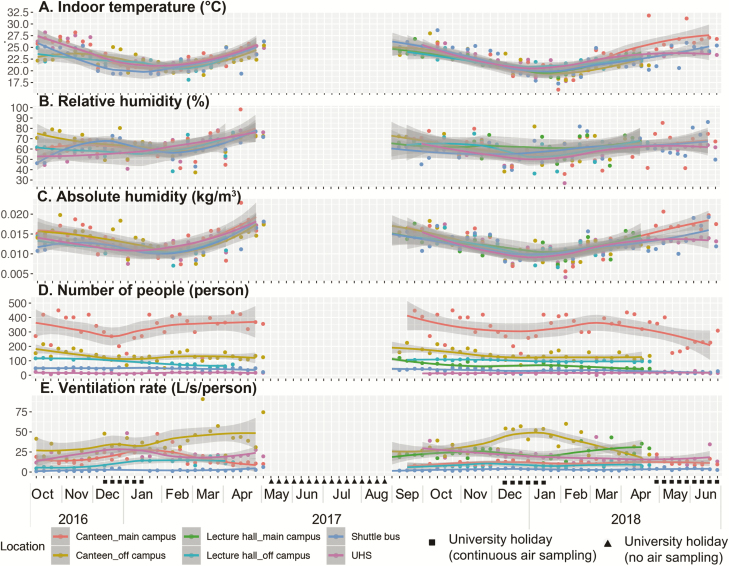
A total of 334 sampling events were conducted at canteens (n = 135), lecture halls (n = 61), shuttle buses (n = 68), and UHS (n = 70). A total of 1028 air samples were collected by NIOSH samplers; 694 were with functional samplers and 334 with negative control samplers. Environmental factors including indoor temperatures (range, 16.0°C–31.8°C) and AH (4.2–22.9 g/m3) were influenced by outdoor temperatures and followed seasonal changes (Figure 1A and 1B). Relative humidity varied across sampling months and locations (27.1%–98.3%; Figure 1C). The numbers of occupants on site, indoor CO2 concentration, and the derived ventilation rate varied significantly across different sampling locations (Kruskal-Wallis test, P < .001). More occupants were recorded at canteens (50–450) than at UHS (2–30) (Figure 1D). Higher ventilation was recorded at canteens (4.4–90.9 L/second/person) than at shuttle buses (1.1–6.4 L/second/person) (Figure 1E), with the highest CO2 concentrations recorded in shuttle buses (1234–4837 ppm) and the lowest at UHS (559–948 ppm) (Figure 1F). The numbers of occupants and ventilation rate on site were highly dependent on the teaching activity and the university academic calendar (χ2 test, P < .001).
Figure 1.
Temporal changes in environmental and ecological parameters at sampling sites from October 2016 to June 2018. Air sampling was conducted weekly except during the summer holiday in 2017 (weeks 19–35). The recorded mean (temperature, humidity, carbon dioxide [CO2] concentration) or absolute (numbers of occupants on site) values at different sampling sites are shown in dots of different colors. Locally estimated scatterplot smoothing regression fitted 95% confidence interval ranges are shown in gray. Temporal changes in indoor temperature (A), indoor relative humidity (B), indoor absolute humidity (C), numbers of occupants on site (D), and ventilation rate (E), are shown. Abbreviation: UHS, University Health Service.
Influenza vRNA was detected from 20.6% (143/694) functional NIOSH samplers. Specifically, IAV, IBV, and both IAV and IBV RNA were detected from 16.9% (117/694), 4.5% (31/694), and 0.7% (5/694) NIOSH samplers, respectively (Table 1). The false-positive rate for IAV detection from negative control NIOSH samplers was 3.3% (11/334) (χ2 test, P < .001). The majority of the IAV-positive samples (75.2% [88/117]) were not subtypable because of low viral load, whereas H1, H3, or both H1 and H3 vRNA was detected from 6.8% (8/117), 16.2% (19/117), and 1.7% (2/117) IAV-positive samples, respectively. Low copies of vRNA (below LoQ) were detected from 80.7% (71/88) of the nonsubtypable samples and from 65.5% (19/29) of the H1 or H3 subtypable samples by rRT-qPCR (χ2 test, P = .154). Similarly, most IBV-positive samples (80.6% [25/31]) contained low copies of viral genome that were below the LoQ.
Table 1.
Quantity and Size Distribution of Respiratory Virus–laden Particles in Air Sampled From a University Campus
| Detection of Respiratory Virus RNA in the 3 Size Fractions of NIOSH Samplers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Measurements | Total | >4 μm | 1–4 μm | <1 μm | >4 μm and 1–4 μm | >4 μm and <1 μm | 1–4 μm and <1 μm | All Size Fractions | Undetermined |
| Influenza A virus | |||||||||
| No. of rRT-qPCR–positive samples | 117/694 | 31 | 31 | 34 | 3 | 12 | 5 | 1 | 0 |
| No. of quantifiable samples | 27/117 | 8 | 3 | 11 | 0 | 4 | 1 | 0 | … |
| Median gene copies/m3 | 20 400 | 20 878 | 12 910 | 23 318 | … | 20 043 | 8378 | … | … |
| Influenza B virus | |||||||||
| No. of rRT-qPCR–positive samples | 31/694 | 11 | 11 | 7 | 1 | 1 | 0 | 0 | 0 |
| No. of quantifiable samples | 6/31 | 4 | 1 | 1 | 0 | 0 | … | … | … |
| Median gene copies/m3 | 14 696 | 17 057 | 9877 | 10 071 | … | … | … | … | … |
| Human rhinovirus | |||||||||
| No. of rRT-qPCR–positive samples | 6/270 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| No. of quantifiable samples | 0/3 | 0 | … | … | … | … | … | … | 0 |
| Median gene copies/m3 | … | … | … | … | … | … | … | … | … |
| Respiratory syncytial virus | |||||||||
| No. of rRT-qPCR–positive samples | 1/270 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| No. of quantifiable samples | 0/1 | … | … | … | … | … | … | … | 0 |
| Median gene copies/m3 | … | … | … | … | … | … | … | … | … |
| Human coronavirus 229E/OC43 | |||||||||
| No. of rRT-qPCR–positive samples | 0/270 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. of quantifiable samples | … | … | … | … | … | … | … | … | … |
| Median gene copies/m3 | … | … | … | … | … | … | … | … | … |
Abbreviations: NIOSH, National Institute for Occupational Safety and Health; rRT-qPCR, quantitative real-time reverse-transcription polymerase chain reaction.
Detection of HRV, RSV, and HCoV in air was limited. Among 270 NIOSH samplers applied from week 41 of 2016 to week 18 of 2017, vRNA of HRV, RSV, and HCoV was detected in 2.2% (6/270; cycle threshold [Ct] >37.4), 0.4% (1/270; Ct = 39.3), and 0% (0/270). During the same period, IAV, IBV, or both were detected at 20.7% (56/270), 3.0% (8/270), and 1.1% (3/270), respectively. Although the detection frequency for HRV, RSV, and HCoV were significantly lower than that of IAV in air (χ2 test, P < .001), HRV was detected at higher frequency than other respiratory viruses among clinical samples collected from symptomatic patients in Hong Kong [33] (Supplementary Figure 2). Detection of multiple respiratory viruses per air sampler was rare, with 0.7% (2/270) samplers testing positive for both IAV and HRV. Taken together, the results suggest that low concentrations of influenza, HRV, and RSV vRNA can be detected from air sampled at the university campus. Influenza vRNA was detected in air at a higher frequency than other respiratory viruses.
Size Distribution of Influenza Virus–laden Particles
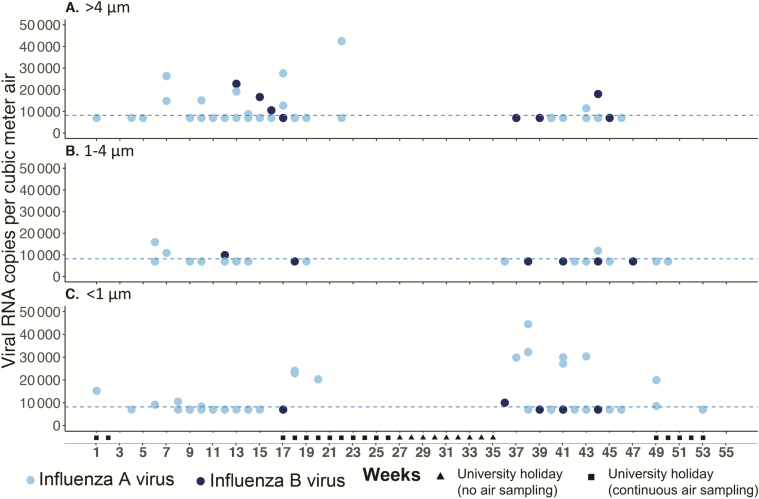
The detection frequencies of IAV vRNA from >4 μm, 1–4 μm, and <1 μm size fractions were 6.8% (47/694), 5.8% (40/694), and 7.5% (52/694), respectively (χ2 test, P = .432). The detection frequencies of IBV were also comparable among >4 μm, 1–4 μm, and <1 μm size fractions at 1.9% (13/694), 1.7% (12/694), and 1.2% (8/694), respectively (χ2 test, P = .524). Higher copies of influenza viral genome were detected from airborne particles >4 μm in the spring weeks (weeks 7–18) and <1 μm in the autumn weeks (weeks 36–45) (Figure 2). Specifically, 62.5% (10/16) of quantifiable samples detected in spring were from particles >4 μm, whereas 70.0% (7/10) of quantifiable samples detected in autumn were from particles <1 μm (Fisher exact test, P = .039). The RH levels were comparable in spring and autumn (Kruskal-Wallis test, P = .645), whereas indoor temperature and AH were significantly higher in autumn than in spring (P < .001). There was no difference in crowdedness (P = .091) or CO2 concentration (P = .070) between spring and autumn.
Figure 2.
Temporal changes in the detection of influenza airborne particles by size. Air sampling was conducted weekly except during the summer holiday in 2017 (weeks 19–35) using National Institute for Occupational Safety and Health bioaerosol samplers that collect airborne particles into 3 size fractions. Influenza A virus (IAV; light blue dots) and influenza B virus (IBV; dark blue dots) viral RNA (vRNA) was detected and quantified by quantitative real-time reverse-transcription polymerase chain reaction (rRT-qPCR). The dotted line indicated the linear range of quantification threshold at 8163 M gene copies/m3 air. Dots between zero and the quantification threshold indicate rRT-qPCR–positive samples that cannot be quantified. Copies of IAV and IBV vRNA detected from >4 μm (A), 1–4 μm (B), and <1 μm (C) airborne particles are shown.
Infectivity of Influenza Virus–laden Particles In Vitro
Among samples positive for IAV by rRT-qPCR, increased IAV RNA copies were detected in 5.1% (6/117) samples after 2 serial passages in MDCK cells (mean ∆Ct = 1.8 [standard deviation, 1.2]). Among samples positive for IBV by rRT-qPCR, increased IBV RNA copies were detected in 3.2% (1/31) samples after 2 passages (∆Ct = 1.5; Supplementary Table 2). Infectious influenza viruses were mainly detected from particles <1 μm (5/6 for IAV, 1/1 for IBV).
Factors Associated With Detection of Influenza Airborne Particles
Univariate logistic regression was used to examine if environmental parameters (temperature, RH, AH, CO2 concentration), ecological factors (holiday vs teaching days, sampling locations, numbers of occupants, ventilation), or influenza activities in the community (weekly laboratory-based influenza surveillance data in Hong Kong from the Centre for Health Protection [CHP]) [33] were associated with the detection of influenza airborne particles at the university campus. Increased risk of detecting influenza airborne particles was associated with sampling on teaching days (vs holiday), greater number of occupants on site (crowdedness), lower ventilation rate, higher indoor CO2 concentration, higher indoor temperature, negative difference in indoor-outdoor temperature, and negative difference in indoor-outdoor AH. Influenza activity by conventional surveillance was negatively associated with detection of influenza airborne particles in the university campus (Table 2 and Supplementary Table 3).
Table 2.
Univariate Analyses on Factors Associated With the Detection of Influenza Viral RNA in Air
| Variable | Analyzed Events, No. | Positive Events, No. (%) | Unadjusted OR (95% CI) | P Value |
|---|---|---|---|---|
| Sampling date | ||||
| Holiday | 184 | 17 (9.2) | Reference | Reference |
| Teaching date | 510 | 126 (24.7) | 3.22 (1.93–5.70) | < .001 |
| Sampling location | ||||
| Canteen | 296 | 66 (22.3) | Reference | Reference |
| Lecture hall | 122 | 27 (22.1) | 0.99 (.59–1.63) | .970 |
| Shuttle bus | 136 | 21 (15.4) | 0.64 (.36–1.08) | .101 |
| UHS | 140 | 29 (20.7) | 0.91 (.55–1.48) | .709 |
| Crowdedness (No. of occupants on site)a | ||||
| Low | 215 | 31 (14.4) | Reference | Reference |
| Intermediate | 229 | 40 (17.5) | 1.26 (.76–2.11) | .382 |
| High | 250 | 72 (28.8) | 2.40 (1.51–3.88) | < .001 |
| Ventilation rate (L/sec/person)b | ||||
| Low | 233 | 55 (23.6) | Reference | Reference |
| Intermediate | 222 | 50 (22.5) | 0.94 (.61–1.46) | .784 |
| High | 239 | 38 (15.9) | 0.61 (.38–.97) | .036 |
| Indoor CO2 level (ppm)c | ||||
| Low | 228 | 40 (17.5) | Reference | Reference |
| Intermediate | 220 | 39 (17.7) | 1.01 (.62–1.65) | .959 |
| High | 246 | 64 (26.0) | 1.65 (1.06–2.59) | .027 |
| Indoor temperature (°C) | ||||
| Numeric | 694 | 143 (20.6) | 1.15 (1.06–1.24) | < .001 |
| Indoor RH (%) | ||||
| Numeric | 694 | 143 (20.6) | 1.00 (.98–1.02) | .752 |
| Indoor AH (kg/m3) | ||||
| Numeric (per 0.002 increase) | 694 | 143 (20.6) | 1.1 (.99–1.27) | .083 |
| Temperature difference (indoor-outdoor) (°C) | ||||
| Numeric | 694 | 143 (20.6) | 0.92 (.87–.98) | .006 |
| RH difference (indoor-outdoor) (%) | ||||
| Numeric | 694 | 143 (20.6) | 1.00 (.99–1.01) | .895 |
| AH difference (indoor-outdoor) (kg/m3) | ||||
| Numeric (per 0.002 increase) | 694 | 143 (20.6) | 0.89 (.81–.97) | .012 |
| Flu activity: influenza detection rate (%) | ||||
| Numeric | 694 | 143 (20.6) | 0.97 (.94–1.00) | .033 |
Abbreviations: AH, absolute humidity; CI, confidence interval; CO2, carbon dioxide; OR, odds ratio; RH, relative humidity; UHS, University Health Service.
aNumber of occupants was categorized into high, intermediate, and low based on the numeric data distribution: high (upper 33% percentile), intermediate (middle 33% percentile), and low (lower 33% percentile).
bVentilation was categorized into high, intermediate, and low based on the numeric data distribution: high (upper 33%), intermediate (middle 33%), and low (lower 33%).
cIndoor CO2 concentration was categorized into high, intermediate, and low based on the numeric data distribution: high (upper 33% percentile), intermediate (middle 33% percentile), and low (lower 33% percentile).
The best-fit multivariable logistic regression model (AIC, 687.1) suggested that increased crowdedness and higher indoor temperatures were independent risk factors for the detection of influenza airborne particles on campus (Table 3 and Supplementary Figure 3). AH as a predictor for influenza infectivity [29] and seasonal influenza epidemics [34] was not included in the best-fit multivariable model; therefore, competing models were built to separately examine the effect of indoor AH (AIC, 687.6), outdoor AH (AIC, 688.6), or indoor-outdoor AH difference (AIC, 687.3). However, none of the AH parameters was significantly associated with detection of influenza airborne particles (Table 3).
Table 3.
Multivariable Analyses on Factors Associated With the Detection of Influenza Viral RNA in Air
| Risk Factor | Best Fit Model | Competing Model 1 | Competing Model 2 | Competing Model 3 |
|---|---|---|---|---|
| aOR (95% CI); P Value | aOR (95% CI); P Value | aOR (95% CI); P Value | aOR (95% CI); P Value | |
| Crowdedness (No. of occupants on site)a | ||||
| Low | Reference | Reference | Reference | Reference |
| Intermediate | 1.21 (.73–2.04); P = .461 | 1.22 (.73–2.05); P = .452 | 1.18 (.70–2.00); P = .530 | 1.16 (.69–1.95); P = .585 |
| High | 2.29 (1.44–3.71); P < .001 | 2.33 (1.46–3.79); P < .001 | 2.19 (1.36–3.60); P = .002 | 2.13 (1.33–3.49); P = .002 |
| Indoor temperature (°C) | ||||
| Numeric | 1.14 (1.05–1.23); P = .001 | 1.19 (1.07–1.33); P = .001 | 1.12 (1.02–1.23); P = .014 | 1.13 (1.04–1.23); P = .003 |
| Indoor AH (kg/m3) | ||||
| Numeric (per 0.002 increase) | … | 0.89 (.74–1.07); P = .213 | … | … |
| Outdoor AH (kg/m3) | ||||
| Numeric (per 0.002 increase) | … | … | 1.03 (.95–1.12); P = .475 | … |
| AH difference (indoor–outdoor) (kg/m3) | ||||
| Numeric (per 0.002 increase) | … | … | … | 0.94 (.86–1.03); P = .181 |
Sampling performed on teaching days vs holidays was highly associated with numbers of occupants on site (χ2 test, P < .001); as such, crowdedness (number of occupants on site) was included in the multivariable analysis.
Abbreviations: AH, absolute humidity; aOR, adjusted odds ratio; CI, confidence interval.
aNumber of occupants was categorized into high, intermediate, and low based on the numeric data distribution: high (upper 33%), intermediate (middle 33%), and low (lower 33%)
Detection of Influenza Virus–laden Particles in Air Over Influenza Epidemics in the Community
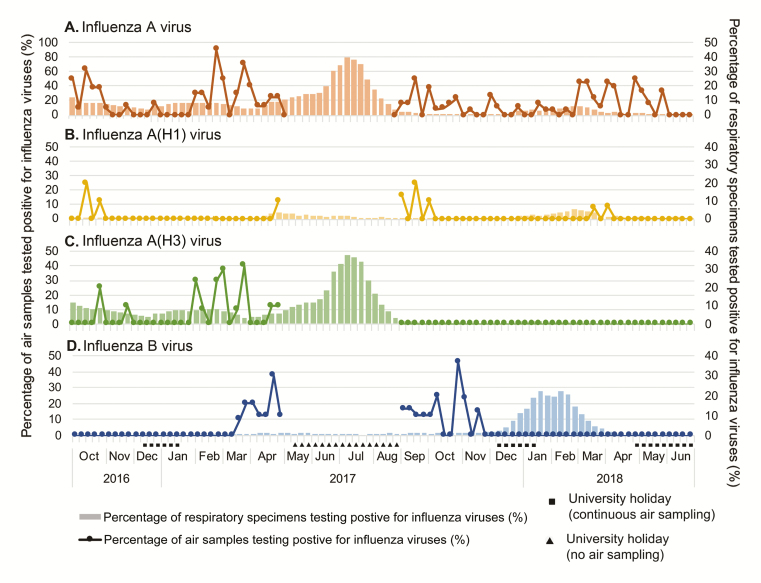
Hong Kong is located within the subtropical region where influenza epidemics may occur more than once per year [35]. We analyzed the temporal relationship between detection of influenza airborne particles at university campus vs detection of symptomatic influenza virus infections in the community, approximated by the weekly influenza positive rate among patients with respiratory infections (CHP). A weekly positive rate above a 10.7% threshold would suggest increased influenza activity in the community [33]. During the study period, an A(H3N2) epidemic and an influenza B epidemic were recorded in 2017 summer and 2017–2018 winter months, respectively.
IAV-laden particles were mainly detected in air on teaching days than on holidays (Figure 3A). A(H1N1) RNA was detected sporadically in air during the study period while there was also limited A(H1N1) activity in the community (Figure 3B). A(H3N2) RNA was detected in air on campus prior to the 2017 summer epidemic of A(H3N2), but we did not sample during the peak of the community epidemic, and no A(H3N2) RNA was detected after the epidemic (Figure 3C). Similarly, IBV RNA was detected in airborne particles on campus prior to the 2017–2018 winter epidemic of IBV but not afterwards (Figure 3D). We continued to sample air on campus during the IBV epidemic in winter 2017–2018, but IBV RNA was only detected at low frequency on campus, probably due to reduced student activities during the winter holidays.
Figure 3.
Temporal relationship between detection of influenza airborne particles at a university campus vs detection of symptomatic influenza infections in the community. Air was sampled weekly at the university campus except during the summer holiday in 2017 (weeks 19–35), and the weekly positive rates for influenza A virus (IAV) and influenza B virus (IBV) in airborne particles are shown in dotted lines. The percentages of respiratory specimens testing positive for IAV and IBV from symptomatic inpatients and outpatients in Hong Kong are shown in bars (data source: Centre for Health Protection). The weekly detection frequency of IAV (A), A(H1) (B), A(H3) (C), and IBV (D) in airborne particles sampled on campus and the weekly percentage of respiratory specimens testing positive for IAV or IBV are shown.
Discussion
Transmission of respiratory viruses may be mediated by symptomatic as well as presymptomatic, mildly infected, or subclinically infected people who are capable of continuing their daily activities. We report temporal detection frequency for common respiratory viruses in air sampled from a university campus in Hong Kong, where 28 000 undergraduate and postgraduate students as well as 7000 staff interact daily. IAVs and IBVs were detected at higher frequencies than other respiratory viruses. A recent study that collected nasopharyngeal swabs of 2685 tourists in New York City reported a higher detection frequency for HRV and HCoV than IAV at 3.2%, 2.4%, and 0.2%, respectively [6, 7]. To attain a comprehensive understanding on the potential transmission modes for respiratory viruses, future studies should sample concurrently from humans and their surrounding environment.
Influenza A(H3N2) and B viruses that caused epidemics in Hong Kong during the study period were detected in air sampled on campus prior to the peak influenza activity in the community but not afterwards, which may reflect changes in susceptibility of the population to the epidemic strain. However, the detection frequency during the epidemics may be underestimated as no or limited samples were collected during the period, which coincided with the university holidays. A previous study reporting data from bioaerosol sampling at a Barcelona subway also reported a higher detection frequency for influenza-laden aerosols prior to the peak of symptomatic cases in 2013–2014 winter [36]. These results highlight the potential value of applying air sampling as a practical surveillance tool that can be easily implemented at public locations for early detection of impending influenza epidemics.
Increased crowdedness was a significant risk factor for detection of influenza airborne particles on campus. Overcrowding plays a critical role in shaping the indoor air microbiome [37] and has been speculated to contribute to influenza outbreaks [38]. Increased indoor temperature and indoor CO2 concentration were significant risk factors that may also be affected by crowdedness. Although humidity may modulate influenza virus survival and affect transmission [29], the competing models showed a null effect for indoor or outdoor AH in the detection of influenza airborne particles. Collectively, the identified risk factors may provide insights for improving environmental interventions to reduce the airborne transmission risk of influenza.
The majority of IAV (73.5% [86/117]) or IBV (64.5% [20/31]) virus–laden particles were ≤4 µm, which is comparable to those detected in air sampled at healthcare facilities [18, 19, 39]. Notably, limited infectious influenza-laden particles were <1 µm. The results support the airborne transmission potential of influenza viruses via particles within respirable fraction. Higher copies of influenza viral genome were detected from airborne particles >4 μm in spring and <1 μm in autumn, with higher indoor temperatures recorded in autumn than spring. Further studies are needed to understand the effect of environmental parameters on airborne particles in indoor environments.
Our study is limited by the specific demographic features of the study population and their behavior, which may limit the extrapolation of the results to other public locations. A small volume of 105 L of air sampled in 30 minutes may have limited our ability to detect respiratory viruses more frequently. Our data argue for the need to extend such investigations to other venues where humans congregate, such as transport systems. Studies would also be strengthened if respiratory samples of individuals at the sampling locations can be collected to link with detections of viral RNA in air. Characterizing the host immune response and the viruses circulating among mildly or subclinically infected individuals prior to epidemics may provide insights on the emergence of influenza epidemics.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank William G. Lindsley and the National Institute for Occupational Safety and Health (NIOSH) for providing the NIOSH bioaerosol samplers; Daniel K. W. Chu for helpful discussions; and Man Kuen Cheung, Elizabeth Wong, Sam Poon, Karen Yip, Tracy Lam, and Daniel Wong for facilitating air sample collection on campus.
Financial support. This study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (contract number HHSN272201400006C) and the Theme-Based Research Scheme (T11-705/14N) and Collaborative Research Fund (C7025-16G) from the Research Grants Council, Hong Kong Special Administrative Region, China.
Potential conflicts of interest. B. C. has received honoraria from Sanofi and Roche for advisory boards. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Shi T, McAllister DA, O’Brien KL, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017; 390:946–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ackerson B, Tseng HF, Sy LS, et al. Severe morbidity and mortality associated with respiratory syncytial virus versus influenza infection in hospitalized older adults [manuscript published online ahead of print 19 November 2018]. Clin Infect Dis 2018. doi: 10.1093/cid/ciy991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Troeger C, Forouzanfar M, Rao PC, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 2017; 17:1133–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iuliano AD, Roguski KM, Chang HH, et al. Global Seasonal Influenza-associated Mortality Collaborator Network Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 2018; 391:1285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ip DKM, Lau LLH, Chan KH, et al. The dynamic relationship between clinical symptomatology and viral shedding in naturally acquired seasonal and pandemic influenza virus infections. Clin Infect Dis 2016; 62:431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birger R, Morita H, Comito D, et al. Asymptomatic shedding of respiratory virus among an ambulatory population across seasons. mSphere 2018; 3. doi: 10.1128/mSphere.00249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaman J, Morita H, Birger R, et al. Asymptomatic summertime shedding of respiratory viruses. J Infect Dis 2018; 217:1074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Transmission of influenza A in human beings. Lancet Infect Dis 2007; 7:257–65. [DOI] [PubMed] [Google Scholar]
- 9. Tellier R. Review of aerosol transmission of influenza A virus. Emerg Infect Dis 2006; 12:1657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tellier R. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface 2009; 6(Suppl 6):S783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kutter JS, Spronken MI, Fraaij PL, Fouchier RA, Herfst S. Transmission routes of respiratory viruses among humans. Curr Opin Virol 2018; 28:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindsley WG, Blachere FM, Beezhold DH, et al. Viable influenza A virus in airborne particles expelled during coughs versus exhalations. Influenza Other Respir Viruses 2016; 10:404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Milton DK, Fabian MP, Cowling BJ, Grantham ML, McDevitt JJ. Influenza virus aerosols in human exhaled breath: particle size, culturability, and effect of surgical masks. PLoS Pathog 2013; 9:e1003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bischoff WE, Swett K, Leng I, Peters TR. Exposure to influenza virus aerosols during routine patient care. J Infect Dis 2013; 207:1037–46. [DOI] [PubMed] [Google Scholar]
- 15. Yan J, Grantham M, Pantelic J, et al. EMIT Consortium Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc Natl Acad Sci U S A 2018; 115:1081–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leung NH, Zhou J, Chu DK, et al. Quantification of influenza virus RNA in aerosols in patient rooms. PLoS One 2016; 11:e0148669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan M, Bonny TS, Loeb J, et al. Collection of viable aerosolized influenza virus and other respiratory viruses in a student health care center through water-based condensation growth. mSphere 2017; 2. doi: 10.1128/mSphere.00251-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blachere FM, Lindsley WG, Pearce TA, et al. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis 2009; 48:438–40. [DOI] [PubMed] [Google Scholar]
- 19. Lindsley WG, Blachere FM, Davis KA, et al. Distribution of airborne influenza virus and respiratory syncytial virus in an urgent care medical clinic. Clin Infect Dis 2010; 50:693–8. [DOI] [PubMed] [Google Scholar]
- 20. Lindsley WG, Green BJ, Blachere FM, et al. Sampling and characterization of bioaerosols. NIOSH manual of analytical methods. 5th ed. Atlanta, GA: National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention,2017. [Google Scholar]
- 21. Hong Kong Observatory. Climatological information services Available at: http://www.hko.gov.hk/cis/climat_e.htm. Accessed 21 December 2018.
- 22. The American Society of Heating, Refrigerating and Air-Conditioning Engineers (ASHRAE). Standard 62.2–2016—ventilation and acceptable indoor air quality in low-rise residential buildings (ANSI approved). 2016. Available at: https://basc.pnnl.gov/resources/ashrae-standard-622-2016-ventilation-acceptable-indoor-air-quality-low-rise-residential. Accessed 8 May 2019. [Google Scholar]
- 23. Zhou J, Wu J, Zeng X, et al. Isolation of H5N6, H7N9 and H9N2 avian influenza A viruses from air sampled at live poultry markets in China, 2014 and 2015. Euro Surveill 2016; 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization. CDC protocol of realtime RT-PCR for influenza A(H1N1) Available at: https://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf?ua=1. Accessed 8 May 2019.
- 25. Munro SB, Kuypers J, Jerome KR. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J Clin Microbiol 2013; 51:1124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Osterback R, Tevaluoto T, Ylinen T, et al. Simultaneous detection and differentiation of human rhino- and enteroviruses in clinical specimens by real-time PCR with locked nucleic acid probes. J Clin Microbiol 2013; 51:3960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y, Cui D, Zheng S, et al. Simultaneous detection of influenza A, influenza B, and respiratory syncytial viruses and subtyping of influenza A H3N2 virus and H1N1 (2009) virus by multiplex real-time PCR. J Clin Microbiol 2011; 49:1653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Elden LJ, van Loon AM, van Alphen F, et al. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J Infect Dis 2004; 189:652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci U S A 2009; 106:3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wallace JM, Hobbs PV.. Atmospheric science: an introductory survey.2nd ed. Amsterdam/Boston: Academic Press/Elsevier, 2006. [Google Scholar]
- 31. Cai J. Humidity: calculate water vapor measures from temperature and dew point. R package version 0.1.4. Available at: https://www.rdocumentation.org/packages/humidity/versions/0.1.4. Accessed 8 May 2019. [Google Scholar]
- 32. Venables WN, Ripley BD.. Modern applied statistics with S. 4th ed. New York: Springer, 2002. [Google Scholar]
- 33. Center for Health Protection. Flu Express Available at: https://www.chp.gov.hk/en/resources/29/304.html. Accessed 21 December 2018.
- 34. Shaman J, Pitzer VE, Viboud C, Grenfell BT, Lipsitch M. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol 2010; 8:e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu P, Goldstein E, Ho LM, et al. Excess mortality associated with influenza A and B virus in Hong Kong, 1998–2009. J Infect Dis 2012; 206:1862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Triadó-Margarit X, Veillette M, Duchaine C, et al. Bioaerosols in the Barcelona subway system. Indoor Air 2017; 27:564–75. [DOI] [PubMed] [Google Scholar]
- 37. Prussin AJ 2nd, Vikram A, Bibby KJ, Marr LC. Seasonal dynamics of the airborne bacterial community and selected viruses in a children’s daycare center. PLoS One 2016; 11:e0151004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oxford JS, Sefton A, Jackson R, Innes W, Daniels RS, Johnson NP. World War I may have allowed the emergence of “Spanish” influenza. Lancet Infect Dis 2002; 2:111–4. [DOI] [PubMed] [Google Scholar]
- 39. Yang W, Elankumaran S, Marr LC. Concentrations and size distributions of airborne influenza A viruses measured indoors at a health centre, a day-care centre and on aeroplanes. J R Soc Interface 2011; 8:1176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.