Abstract
Background
Human metapneumovirus (hMPV) commonly causes upper and lower respiratory tract infections. Here, we performed long-term retrospective surveillance of hMPV infection among patients hospitalized in South Korea between 2007 and 2016 and investigated seasonal dynamics and clinical characteristics associated with each virus subtype/genotype.
Methods
Patient specimens were tested for hMPV and other respiratory viruses by commercial molecular assays. Medical records of hMPV-positive patients were reviewed, and hMPV subtype/genotype analysis was performed. We also collected meteorological data and analyzed relationships with hMPV activity.
Results
Of 23 694 specimens, 1275 (5.4%) were positive; among them, 94.0% were classified into 5 subtypes (A1, A2a, A2b, B1, and B2). Some clinical manifestations differed according to hMPV genotype; however, there was no correlation between hMPV subtype and clinical outcome. Viral activity peaked at 13–20 weeks (April and May) and was associated with climate-specific factors, including temperature, relative humidity, diurnal temperature variation, wind speed, and sunshine duration.
Conclusions
This large-scale, 10-year study provides valuable information about the clinical characteristics associated with hMPV subtypes and climate factors contributing to virus transmission.
Keywords: human metapneumovirus, genotype, acute respiratory tract infection, epidemiology, meteorological factors
There were differences between characteristics of each human metapneumovirus (hMPV) genotype infection, but disease severity was not altered according to hMPV subtype. Our 10-year surveillance provides valuable information about clinical characteristics of hMPV infections and climate factors contributing to viral transmission.
Human metapneumovirus (hMPV) is a pathogenic virus that causes respiratory tract infections especially among children, older adults, and immunosuppressed persons [1, 2]. hMPV is an antisense single-stranded RNA virus that belongs to the new virus family Pneumoviridae [3]. Phylogenetic analysis of the F and G genes has shown that hMPV can be classified into 2 main antigenic subtypes, A and B, with each subtype further separated into genetic sublineages (A1/A2a/A2b and B1/B2) [4]. It is known that the 2 main subgroups circulate in all parts of the world and concurrent annual circulation is common for all hMPV sublineages, with predominant hMPV subtype switching each year [5, 6].
The clinical manifestations of hMPV infections range from asymptomatic carriage to severe disease including fatal cases [2]. Because of its genetic diversity, previous studies have attempted to associate hMPV subtypes with clinical characteristics. Some studies found that the different clinical characteristics of hMPV infection might be associated with hMPV genotype and that disease severity could be increased with specific hMPV subgroup infections [7, 8]. However, it was also reported that there is no relationship between hMPV sublineages and the severity of illness or clinical manifestations [9, 10]. Therefore, whether there is an association between hMPV genotypes and clinical characteristics is debatable.
The association between climatic factors and hMPV infections is also unclear. Some researchers have assumed that most hMPV infections in humans would occur with strong seasonal predominance patterns [11, 12]. However, the seasonality of hMPV is not fully understood, and there is still very little information available regarding the association between meteorological parameters and hMPV activity.
Here, we conducted a long-term surveillance study of hMPV in South Korea from January 2007 to December 2016. The aim was to compare clinical characteristics including symptoms, diagnoses, laboratory findings, and concurrent pathogen infections and to evaluate the seasonal dynamics of hMPV infections according to hMPV subtypes. Additionally, we investigated the climatic factors that would explain the seasonal prevalence of hMPV infections in South Korea.
MATERIALS AND METHODS
Clinical Samples
This retrospective study was approved by the Institutional Review Board (IRB) of the Chung-Ang University Hospital (IRB number 1807-009-16189). From January 2007 to December 2016, respiratory specimens from 23 694 hospitalized patients with acute respiratory illness were tested by molecular assays for respiratory viral pathogens at the Chung-Ang University Hospital. Among the patients, 21 184 were children (aged <18 years) and 2510 were adults. Respiratory samples were tested for hMPV and other respiratory viral pathogens using Seeplex RV6 or Anyplex II RV16 Detection kits (Seegene, Seoul, South Korea). Specimens were nasal swabs (n = 17 414 [73.5%]), throat swabs (n = 4340 [18.3%]), nasal aspirate (n = 1716 [7.3%]), sputum (n = 145 [0.6%]), and bronchial washing (n = 79 [0.3%]). Seeplex can detect 6 common respiratory viruses including adenovirus, influenza A and B, respiratory syncytial virus (RSV), parainfluenza virus (PIV), and hMPV. With Anyplex, 16 respiratory viruses can be detected and these include adenovirus, influenza A and B, PIV types 1, 2, 3, and 4, rhinovirus A/B/C, RSV A and B, bocavirus, hMPV, enterovirus, and coronavirus 229E, NL63, and OC43. Seeplex was used for routine molecular analysis until December 2012, and Anyplex was employed thereafter.
Clinical and Laboratory Findings
Among the hospitalized patients with acute respiratory illness, 1275 were diagnosed with hMPV infection. We reviewed the medical records for patients with hMPV infection, and the following clinical data were obtained: basic patient characteristics, past medical history, symptoms, and physical examination findings. In addition, we collected the results of routine laboratory testing upon admission. The following analytes were included in admission tests: sodium, potassium, chloride, calcium, phosphorous, glucose, total protein, albumin, total bilirubin, direct bilirubin, blood urea nitrogen, creatinine, uric acid, cholesterol, aspartate aminotransferase, alanine aminotransferase, lactic dehydrogenase, γ-glutamyl transferase, alkaline phosphatase, C-reactive protein, and complete blood cell results. The results of arterial blood gas analysis (n = 458) and procalcitonin (n = 123) were collected when available. To determine the presence of other bacterial and viral infections, we also reviewed various bacterial and tuberculosis culture test results for any specimen, antigen testing (Streptococcus pneumoniae, influenza A and B, RSV, rotavirus, norovirus), antibody testing (Mycoplasma pneumoniae, Legionella pneumophila, cytomegalovirus, Epstein-Barr virus), and molecular tests for respiratory bacterial pathogens (S. pneumoniae, Haemophilus influenzae, Chlamydophila pneumoniae, L. pneumophila, Bordetella pertussis, M. pneumoniae, tuberculosis, and nontuberculous mycobacteria), respiratory viral pathogens, and diarrhea viruses (rotavirus, norovirus, enteric adenovirus, and astrovirus).
Climatic Data
Meteorological data between January 2007 and December 2016 were collected from the database of the Korea Meteorological Administration (KMA; https://data.kma.go.kr/). The surface observation for the KMA was conducted by the Seoul automated synoptic observing system (ASOS station 108), and the 16 meteorological elements, including temperature, relative humidity, wind speed, and atmospheric pressure, among others, were measured every 3 hours [13].
hMPV Genotyping Assays and Sequencing Analysis
A total of 1275 hMPV-positive samples based on respiratory virus polymerase chain reaction (PCR) assays were analyzed by nested PCR–restriction fragment length polymorphism (RFLP) analysis, as described previously [14, 15]. The nested PCR was conducted with 2 primer–probe sets targeting the hMPV fusion (F) gene. Outer primers targeted 450 bp of the F gene, and inner primers were designed to detect 348 bp of the first PCR products. After amplification, PCR-amplified fragments were digested with 10 units of restriction enzyme (Tsp509I, Thermo Fisher Scientific, Pittsburgh, Pennsylvania) for 16 hours at 65°C and electrophoresed.
For the samples that showed negative results or unknown band patterns by RFLP analysis, PCR products were directly sequenced using the inner primer set employed for the nested PCR step. Target nucleotides were analyzed using an ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, California), and the sequences were compared for similarity based on the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/).
Statistical Analysis
Datasets were entered into Microsoft Excel (Microsoft, Washington) and analyzed using R version 3.5.1 (http://www.R-project.org/). For clinical characteristics and laboratory findings, the values were presented as median with interquartile range (continuous variables) or count with percentage (binomial or categorical variables). The Fisher exact test was used to compare differences between binomial variables. Continuous variables were compared by the Kruskal-Wallis test, and post hoc tests were performed using the Dunn multiple comparisons test. Associations between hMPV-positive rates and climate factors were explored by univariate regression analysis based on the Pearson correlation coefficient. Independent associations were analyzed by the multiple linear regression model using the stepwise selection method. A P value <.05 was considered a significant difference. We also utilized principal components analysis (PCA) to determine the contribution of climate variables to hMPV infections. Using PCA, we could reduce high-dimensional data spaces (1 axis per meteorological variable) to 2-dimensional planes for further analysis [16, 17]. Based on the results of PCA, we could select dominant climate factors that affect hMPV infections, and 2-dimensional density plots were generated to evaluate the impact of climate factors in hMPV infections.
RESULTS
Epidemiological Distribution of hMPV Infections
During the study period, 23 694 patients were subjected to molecular testing for respiratory viruses, and 13 871 patients (58.5%) were positive for at least 1 respiratory virus including the following: 3631 cases of rhinovirus (15.3%), 3482 RSV (14.7%), 2652 adenovirus (11.2%), 1842 PIV (7.8%), 1275 hMPV (5.4%), 1223 influenza A (5.2%), 1017 enterovirus (4.3%), 719 influenza B (3.0%), 703 bocavirus (3.0%), and 539 coronavirus (2.3%).
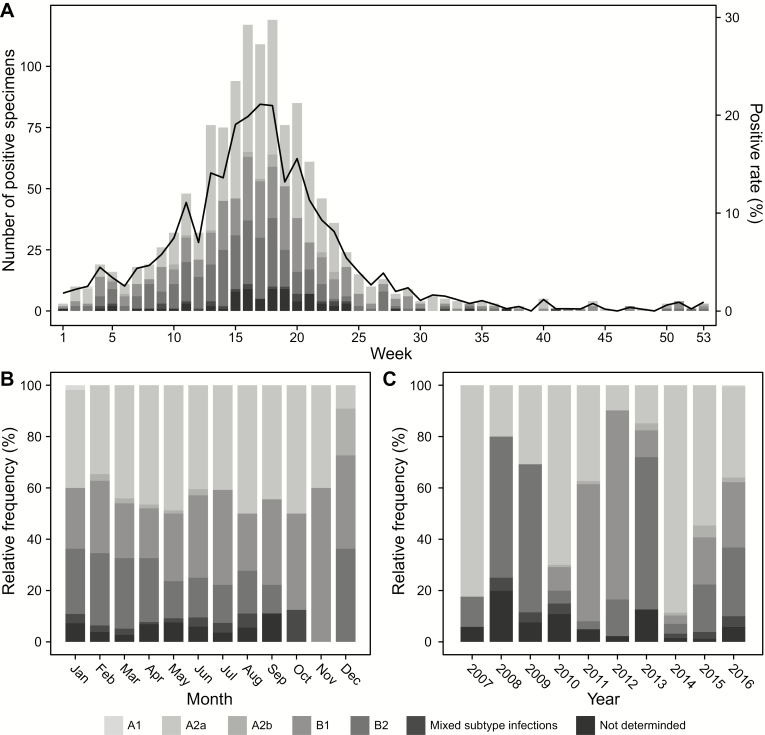
The rate of positivity and weekly distribution of hMPV infections are shown in Figure 1A. These rates for hMPV infections varied from 0% to 21.1% across the weeks. Significant increases in isolation rates occurred between 10 and 25 weeks, and half of the hMPV infections occurred between 13 and 20 weeks (April and May). Of the hMPV-positive specimens, 1199 (94.0%) were subtyped as follows: 570 (44.7%) hMPV A2a, 304 (23.8%) hMPV B1, 279 (21.9%) hMPV B2, 21 (1.6%) hMPV A2b, and 24 (1.9%) mixed-genotype infections. Only 1 patient was infected with hMPV A1 (0.1%). Among mixed-genotype infections, 12 were hMPV A2a/B1 infections, 9 were hMPV A2a/B2, and 3 were hMPV A2a/B2. Specimens from 76 patients could not be genotyped (not determined).
Figure 1.
Number of human metapneumovirus (hMPV)–positive specimens (bars) and hMPV-positive rate (black lines) from 2007 to 2016 (A). Monthly (B) and annual (C) relative frequencies in the numbers of hMPV infections.
The dominant hMPV genotype in peak season was hMPV A2a, and the relative frequencies of hMPV B1 and B2 increased in the winter season (Figure 1B). However, the annual predominance patterns across the study period were complex and unpredictable (Figure 1C).
Clinical Characteristics of hMPV Infections
The clinical characteristics of hMPV infections in pediatric and adult patients are summarized in Tables 1 and 2. The majority of patients with hMPV were children (1200/1275 [94.1%]). Further, clinical manifestations were similar between the pediatric and adult patient groups. Most individuals infected with this virus exhibited upper respiratory symptoms such as cough, fever, sputum, and rhinorrhea, and the median onset of symptoms was 3 days. In both groups, pneumonia was the most common clinical diagnosis associated with hMPV infections. For the pediatric group, all hospitalized patients showed good prognosis and the median length of hospitalization was 5 days. However, a mortality rate of 9.3% was observed in the adult patient group. Most clinical characteristics were not significantly different between hMPV subtypes. In the pediatric group, however, there were significant differences among hMPV subtypes and clinical manifestations (cough, seizure, abdominal pain, and rhonchi) and concurrent infections (M. pneumoniae, adenovirus, and enterovirus). In terms of hMPV subtype infections, there was no significant difference in laboratory findings between children and adults (Supplementary Tables 1 and 2).
Table 1.
Clinical Characteristics of Human Metapneumovirus Infections in Children
| All hMPV Infections | hMPV A2a | hMPV A2b | hMPV B1 | hMPV B2 | Mixed-subtype Infections | P Valueb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | (n = 1200)a | (n = 547) | (n = 16) | (n = 290) | (n = 259) | (n = 23) | |||||||
| Basic patient characteristics | |||||||||||||
| Age, mo, median (IQR) | 25 | (13–39) | 25 | (14–38) | 32 | (20–42) | 24 | (13–39) | 25 | (12–40) | 30 | (12–42) | .9128 |
| Age, mo | |||||||||||||
| <6 | 116 | (9.7) | 49 | (9.0) | 2 | (12.5) | 30 | (10.3) | 25 | (9.7) | 3 | (13.0) | .8456 |
| 6–12 | 163 | (13.6) | 68 | (12.4) | 2 | (12.5) | 36 | (12.4) | 42 | (16.2) | 4 | (17.4) | |
| 12–36 | 572 | (47.7) | 274 | (50.1) | 6 | (37.5) | 145 | (50.0) | 114 | (44.0) | 9 | (39.1) | |
| >36 | 349 | (29.1) | 156 | (28.5) | 6 | (37.5) | 79 | (27.2) | 78 | (30.1) | 7 | (30.4) | |
| Sex, male | 644 | (53.7) | 296 | (54.1) | 8 | (50.0) | 156 | (53.8) | 141 | (54.4) | 12 | (52.2) | .9962 |
| Past medical history | |||||||||||||
| Asthma | 26 | (2.2) | 12 | (2.2) | 1 | (6.3) | 6 | (2.1) | 6 | (2.3) | 0 | (0) | .6601 |
| Previous TB infection | 4 | (0.3) | 2 | (0.4) | 0 | (0) | 0 | (0) | 1 | (0.4) | 1 | (4.3) | .1214 |
| Congestive heart failure | 1 | (0.1) | 0 | (0) | 0 | (0) | 1 | (0.3) | 0 | (0) | 0 | (0) | .5181 |
| Symptoms | |||||||||||||
| Onset (day), median (IQR) | 3 | (2–5) | 4 | (2–5) | 3 | (2–4) | 3 | (2–5) | 3 | (2–5) | 4 | (2–5) | .1000 |
| Cough | 1164 | (97.0) | 536 | (98.0) | 14 | (87.5) | 278 | (95.9) | 256 | (98.8) | 22 | (95.7) | .0193 |
| Fever | 1086 | (90.5) | 494 | (90.3) | 15 | (93.8) | 261 | (90.0) | 239 | (92.3) | 21 | (91.3) | .9011 |
| Sputum | 966 | (80.5) | 449 | (82.1) | 11 | (68.8) | 223 | (76.9) | 215 | (83.0) | 19 | (82.6) | .2039 |
| Rhinorrhea | 870 | (72.5) | 410 | (75.0) | 10 | (62.5) | 218 | (75.2) | 179 | (69.1) | 16 | (69.6) | .2904 |
| Dyspnea | 52 | (4.3) | 24 | (4.4) | 0 | (0) | 10 | (3.4) | 14 | (5.4) | 1 | (4.3) | .7717 |
| Sore throat | 40 | (3.3) | 23 | (4.2) | 0 | (0) | 7 | (2.4) | 4 | (1.5) | 1 | (4.3) | .2341 |
| Headache | 8 | (0.7) | 3 | (0.5) | 0 | (0) | 3 | (1.0) | 1 | (0.4) | 0 | (0) | .6956 |
| Muscle pain | 2 | (0.2) | 2 | (0.4) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | .7535 |
| Seizure | 40 | (3.3) | 26 | (4.8) | 0 | (0) | 4 | (1.4) | 4 | (1.5) | 1 | (4.3) | .0310 |
| Vomiting | 102 | (8.5) | 44 | (8.0) | 2 | (12.5) | 25 | (8.6) | 21 | (8.1) | 3 | (13.0) | .7500 |
| Abdominal pain | 24 | (2.0) | 10 | (1.8) | 1 | (6.3) | 2 | (0.7) | 9 | (3.5) | 1 | (4.3) | .0474 |
| Diarrhea | 72 | (6.0) | 28 | (5.1) | 2 | (12.5) | 16 | (5.5) | 19 | (7.3) | 3 | (13.0) | .1840 |
| Physical examination findings | |||||||||||||
| Body temperature, °C, median (IQR) | 38.3 | (37.6–38.9) | 38.2 | (37.6–38.8) | 38.7 | (38.2–39.0) | 38.4 | (37.8–39.0) | 38.3 | (37.7–39.0) | 38.0 | (37.4–38.8) | .0511 |
| Pulse rate, beats/min, median (IQR) | 126 | (120–132) | 126 | (120–132) | 128 | (112–131) | 128 | (119–132) | 128 | (120–132) | 120 | (120–133) | .8605 |
| Systolic BP, mm Hg, median (IQR) | 90 | (90–100) | 90 | (90–93) | 90 | (80–93) | 90 | (90–100) | 90 | (80–90) | 90 | (90–90) | .9620 |
| Diastolic BP, mm Hg, median (IQR) | 60 | (50–60) | 60 | (50–60) | 60 | (50–60) | 60 | (50–60) | 60 | (50–60) | 60 | (50–60) | .3808 |
| Respiratory rate, breaths/min, median (IQR) | 28 | (26–32) | 28 | (26–32) | 29 | (26–32) | 28 | (26–32) | 30 | (26–32) | 28 | (26–32) | .6153 |
| Rale | 645 | (53.8) | 314 | (57.4) | 9 | (56.3) | 149 | (51.4) | 134 | (51.7) | 13 | (56.5) | .4216 |
| Wheezing | 172 | (14.3) | 82 | (15.0) | 3 | (18.8) | 40 | (13.8) | 39 | (15.1) | 2 | (8.7) | .8932 |
| Rhonchi | 88 | (7.3) | 49 | (9.0) | 0 | (0) | 20 | (6.9) | 10 | (3.9) | 4 | (17.4) | .0199 |
| Chest wall retraction | 23 | (1.9) | 12 | (2.2) | 1 | (6.3) | 2 | (0.7) | 6 | (2.3) | 1 | (4.3) | .1251 |
| Cyanosis | 4 | (0.3) | 2 | (0.4) | 0 | (0) | 1 | (0.3) | 0 | (0) | 0 | (0) | 1 |
| Pharyngeal injection | 496 | (41.3) | 223 | (40.8) | 5 | (31.3) | 127 | (43.8) | 96 | (37.1) | 13 | (56.5) | .2436 |
| TM injection | 54 | (4.5) | 27 | (4.9) | 0 | (0) | 12 | (4.1) | 10 | (3.9) | 1 | (4.3) | .9475 |
| Clinical diagnosis | |||||||||||||
| Pneumonia | 768 | (64.0) | 353 | (64.5) | 12 | (75.0) | 192 | (66.2) | 159 | (61.4) | 16 | (69.6) | .8191 |
| Acute bronchiolitis | 203 | (16.9) | 96 | (17.6) | 1 | (6.3) | 46 | (15.9) | 49 | (18.9) | 4 | (17.4) | |
| Acute bronchitis | 80 | (6.7) | 37 | (6.8) | 1 | (6.3) | 20 | (6.9) | 19 | (7.3) | 1 | (4.3) | |
| Acute pharyngitis | 76 | (6.3) | 30 | (5.5) | 0 | (0) | 16 | (5.5) | 18 | (6.9) | 0 | (0) | |
| Croup | 28 | (2.3) | 16 | (2.9) | 0 | (0) | 5 | (1.7) | 5 | (1.9) | 0 | (0) | |
| Acute gastroenteritis | 17 | (1.4) | 6 | (1.1) | 1 | (6.3) | 3 | (1) | 4 | (1.5) | 1 | (4.3) | |
| Others | 28 | (2.3) | 9 | (1.6) | 1 | (6.3) | 8 | (2.8) | 5 | (1.9) | 1 | (4.3) | |
| Clinical outcome | |||||||||||||
| Length of hospital stay, d, median (IQR) | 5 | (4–6) | 5 | (4–6) | 6 | (4–7) | 5 | (4–7) | 5 | (4–6) | 5 | (4–6) | .8189 |
| ICU admission | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 1.0000 |
| Intubation | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 1.0000 |
| Mortality rate | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 1.0000 |
| Concurrent bacterial infection | |||||||||||||
| Mycoplasma pneumoniae | 167 | (13.9) | 58 | (10.6) | 6 | (37.5) | 46 | (15.9) | 42 | (16.2) | 5 | (21.7) | .0035 |
| Streptococcus pneumoniae | 132 | (11.0) | 64 | (11.7) | 1 | (6.3) | 30 | (10.3) | 25 | (9.7) | 3 | (13.0) | .8749 |
| Haemophilus influenzae | 59 | (4.9) | 24 | (4.4) | 2 | (12.5) | 13 | (4.5) | 15 | (5.8) | 0 | (0) | .3848 |
| Chlamydophila pneumoniae | 4 | (0.3) | 1 | (0.2) | 0 | (0) | 2 | (0.7) | 1 | (0.4) | 0 | (0) | .5294 |
| Other bacteria | 5 | (0.4) | 2 | (0.4) | 0 | (0) | 2 | (0.7) | 1 | (0.4) | 0 | (0) | .8690 |
| Concurrent viral infection | |||||||||||||
| Rhinovirus | 236 | (19.7) | 112 | (20.5) | 3 | (18.8) | 49 | (16.9) | 51 | (19.7) | 4 | (17.4) | .8088 |
| Adenovirus | 107 | (8.9) | 54 | (9.9) | 3 | (18.8) | 11 | (3.8) | 30 | (11.6) | 2 | (8.7) | .0019 |
| Bocavirus | 60 | (5.0) | 28 | (5.1) | 1 | (6.3) | 10 | (3.4) | 18 | (6.9) | 2 | (8.7) | .2547 |
| Parainfluenza viruses | 35 | (2.9) | 12 | (2.2) | 0 | (0) | 10 | (3.4) | 9 | (3.5) | 0 | (0) | .6795 |
| Influenza A virus | 21 | (1.8) | 5 | (0.9) | 1 | (6.3) | 3 | (1.0) | 8 | (3.1) | 0 | (0) | .0697 |
| Enterovirus | 19 | (1.6) | 12 | (2.2) | 1 | (6.3) | 0 | (0) | 4 | (1.5) | 1 | (4.3) | .0125 |
| Influenza B virus | 18 | (1.5) | 7 | (1.3) | 1 | (6.3) | 3 | (1.0) | 4 | (1.5) | 1 | (4.3) | .0697 |
| Respiratory syncytial virus | 13 | (1.1) | 6 | (1.1) | 0 | (0) | 4 | (1.4) | 2 | (0.8) | 0 | (0) | .9119 |
| Coronaviruses | 12 | (1.0) | 4 | (0.7) | 0 | (0) | 3 | (1.0) | 3 | (1.2) | 0 | (0) | .8376 |
| Rotavirus | 8 | (0.7) | 3 | (0.5) | 0 | (0) | 2 | (0.7) | 3 | (1.2) | 0 | (0) | .6866 |
| Noroviruses | 2 | (0.2) | 1 | (0.2) | 0 | (0) | 0 | (0) | 0 | (0) | 1 | (4.3) | .0676 |
| Enteric adenovirus | 2 | (0.2) | 0 | (0) | 0 | (0) | 1 | (0.3) | 0 | (0) | 0 | (0) | .5181 |
| Other viruses | 12 | (1.0) | 4 | (0.7) | 0 | (0) | 3 | (1.0) | 3 | (1.2) | 1 | (4.3) | .3994 |
Data are presented as no. (%) unless otherwise indicated.
Abbreviations: BP, blood pressure; hMPV, human metapneumovirus; ICU, intensive care unit; IQR, interquartile range; TB, tuberculosis; TM, tympanic membrane.
aOne hMPV A1 infection and 64 hMPV infections of undetermined genotype were included.
b P values were calculated by Fisher exact test or Kruskal-Wallis test.
Table 2.
Clinical Characteristics of Human Metapneumovirus Infections in Adults
| All hMPV Infections | hMPV A2a | hMPV A2b | hMPV B1 | hMPV B2 | P Valueb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | (n = 75)a | (n = 23) | (n = 5) | (n = 14) | (n = 20) | ||||||
| Basic patient characteristics | |||||||||||
| Age, y, median (IQR) | 73 | (57–80) | 71 | (65–79) | 56 | (42–79) | 70.5 | (55–80) | 74 | (61–82) | .8109 |
| Sex, male | 31 | (41.3) | 9 | (39.1) | 2 | (40.0) | 5 | (35.7) | 8 | (40.0) | 1.0000 |
| Past medical history | |||||||||||
| Asthma | 18 | (24.0) | 3 | (13.0) | 2 | (40.0) | 3 | (21.4) | 4 | (20.0) | .5124 |
| Hypertension | 28 | (37.3) | 9 | (39.1) | 3 | (60.0) | 8 | (57.1) | 6 | (30.0) | .3866 |
| Renal disease | 12 | (16.0) | 4 | (17.4) | 1 | (20.0) | 2 | (14.3) | 4 | (20.0) | 1.0000 |
| Diabetes mellitus | 14 | (18.7) | 7 | (30.4) | 1 | (20.0) | 2 | (14.3) | 2 | (10.0) | .3788 |
| Neoplastic disease | 8 | (10.7) | 2 | (8.7) | 0 | (0) | 0 | (0) | 3 | (15.0) | .4760 |
| Previous TB infection | 6 | (8.0) | 2 | (8.7) | 0 | (0) | 2 | (14.3) | 2 | (10.0) | .9234 |
| Liver disease | 6 | (8.0) | 3 | (13.0) | 0 | (0) | 0 | (0) | 2 | (10.0) | .5753 |
| COPD | 4 | (5.3) | 4 | (17.4) | 0 | (0) | 0 | (0) | 0 | (0) | .1134 |
| Bronchiectasis | 4 | (5.3) | 1 | (4.3) | 0 | (0) | 1 | (7.1) | 1 | (5.0) | 1.0000 |
| Congestive heart failure | 4 | (5.3) | 2 | (8.7) | 0 | (0) | 0 | (0) | 2 | (10.0) | .7633 |
| Cerebrovascular disease | 1 | (1.3) | 0 | (0) | 0 | (0) | 0 | (0) | 1 | (5.0) | .6290 |
| Symptoms | |||||||||||
| Onset (day), median (IQR) | 3 | (1–6) | 2 | (1–7) | 3 | (1–7) | 3 | (1–4) | 2 | (1–7) | .9304 |
| Cough | 62 | (82.7) | 20 | (87.0) | 4 | (80.0) | 10 | (71.4) | 17 | (85.0) | .6579 |
| Sputum | 61 | (81.3) | 20 | (87.0) | 4 | (80.0) | 10 | (71.4) | 16 | (80.0) | .7043 |
| Fever | 36 | (48.0) | 9 | (39.1) | 3 | (60.0) | 9 | (64.3) | 7 | (35.0) | .3032 |
| Dyspnea | 34 | (45.3) | 9 | (39.1) | 4 | (80.0) | 5 | (35.7) | 9 | (45.0) | .3995 |
| Rhinorrhea | 5 | (6.7) | 0 | (0) | 1 | (20.0) | 1 | (7.1) | 1 | (5.0) | .1627 |
| Muscle pain | 2 | (2.7) | 0 | (0) | 0 | (0) | 1 | (7.1) | 0 | (0) | .3065 |
| Abdominal pain | 2 | (2.7) | 0 | (0) | 0 | (0) | 1 | (7.1) | 1 | (5.0) | .5865 |
| Sore throat | 1 | (1.3) | 0 | (0) | 0 | (0) | 0 | (0) | 1 | (5.0) | .6290 |
| Vomiting | 1 | (1.3) | 0 | (0) | 0 | (0) | 1 | (7.1) | 0 | (0) | .3065 |
| Physical examination findings | |||||||||||
| Body temperature, °C, median (IQR) | 37.4 | (37.0–38.3) | 37.1 | (36.8–37.8) | 39.1 | (37.6–39.2) | 37.3 | (37.0–38.0) | 37.4 | (37.2–38.1) | .2394 |
| Pulse rate, beats/min, median (IQR) | 86 | (78–96) | 86 | (75–92) | 84 | (83–92) | 95.5 | (84–102) | 82 | (74–93) | .0969 |
| Systolic BP, mm Hg, median (IQR) | 120 | (110–130) | 120 | (105–134) | 120 | (110–130) | 115 | (110–120) | 115 | (108–132) | .7400 |
| Diastolic BP, mm Hg, median (IQR) | 70 | (70–80) | 70 | (70–90) | 70 | (70–80) | 70 | (70–75) | 70 | (60–80) | .3205 |
| Respiratory rate, breaths/min, median (IQR) | 20 | (20–22) | 20 | (20–23) | 20 | (20–20) | 20 | (20–20) | 20 | (20–20) | .8387 |
| Rale | 4 | (5.3) | 0 | (0) | 0 | (0) | 0 | (0) | 2 | (10.0) | .3046 |
| Wheezing | 3 | (4.0) | 1 | (4.3) | 1 | (20.0) | 0 | (0) | 0 | (0) | .2041 |
| Pharyngeal injection | 3 | (4.0) | 0 | (0) | 0 | (0) | 0 | (0) | 3 | (15.0) | .0923 |
| Clinical diagnosis | |||||||||||
| Pneumonia | 63 | (84.0) | 19 | (82.6) | 4 | (80.0) | 12 | (85.7) | 18 | (90.0) | .7547 |
| Acute bronchitis | 6 | (8.0) | 2 | (8.7) | 0 | (0) | 1 | (7.1) | 2 | (10.0) | |
| Acute pharyngitis | 1 | (1.3) | 1 | (4.3) | 0 | (0) | 0 | (0) | 0 | (0) | |
| Others | 5 | (6.7) | 1 | (4.3) | 1 | (20) | 1 | (7.1) | 0 | (0) | |
| Clinical outcome | |||||||||||
| Length of hospital stay, d, median (IQR) | 8 | (6–16) | 10 | (7–17) | 6 | (6–11) | 10 | (7–14) | 8 | (6–20) | .6337 |
| ICU admission | 13 | (17.3) | 4 | (17.4) | 1 | (20.0) | 2 | (14.3) | 4 | (20.0) | .6148 |
| Intubation | 11 | (14.7) | 3 | (13.0) | 0 | (0) | 1 | (7.1) | 4 | (20.0) | .6965 |
| Mortality rate | 7 | (9.3) | 1 | (4.3) | 1 | (20.0) | 0 | (0) | 2 | (10.0) | .3028 |
| Concurrent bacterial infection | |||||||||||
| Streptococcus pneumoniae | 36 | (48.0) | 12 | (52.2) | 4 | (80.0) | 5 | (35.7) | 10 | (50.0) | .4192 |
| Mycoplasma pneumoniae | 24 | (32.0) | 7 | (30.4) | 1 | (20.0) | 7 | (50.0) | 7 | (35.0) | .6146 |
| Haemophilus influenzae | 8 | (10.7) | 4 | (17.4) | 0 | (0) | 1 | (7.1) | 3 | (15.0) | .8551 |
| Other bacteria | 2 | (2.7) | 1 | (4.3) | 0 | (0) | 1 | (7.1) | 0 | (0) | .7567 |
| Concurrent viral infection | |||||||||||
| Rhinovirus | 5 | (6.7) | 1 | (4.3) | 1 | (20.0) | 1 | (7.1) | 1 | (5.0) | .5386 |
| Parainfluenza viruses | 3 | (4.0) | 0 | (0) | 0 | (0) | 0 | (0) | 1 | (5.0) | .6290 |
| Enterovirus | 2 | (2.7) | 0 | (0) | 0 | (0) | 0 | (0) | 1 | (5.0) | .6290 |
| Influenza A virus | 1 | (1.3) | 0 | (0) | 0 | (0) | 1 | (7.1) | 0 | (0) | .3065 |
| Respiratory syncytial virus | 1 | (1.3) | 0 | (0) | 1 | (20.0) | 0 | (0) | 0 | (0) | .0806 |
| Coronaviruses | 1 | (1.3) | 0 | (0) | 0 | (0) | 1 | (7.1) | 0 | (0) | .3065 |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: BP, blood pressure; COPD, chronic obstructive pulmonary disease; hMPV, human metapneumovirus; ICU, intensive care unit; IQR, interquartile range; TB, tuberculosis.
aOne hMPV mixed-genotype infection and 12 hMPV infections of undetermined genotype were included.
b P values were calculated by Fisher exact test or Kruskal-Wallis test.
Meteorological Characteristics Associated With hMPV Infections
The meteorological characteristics associated with hMPV infections are shown in Table 3. Comparing 4 hMPV genotypes, the P values associated with the mean temperature, minimum temperature, maximum temperature, ground surface temperature, and relative humidity were all <.05. Specifically, the isolation of hMPV genotype A2a occurred more frequently with higher temperatures and wet weather, whereas hMPV B2 was frequently isolated with relatively low temperatures and dry environments.
Table 3.
Meteorological Variables Related to the Isolation of Human Metapneumovirus
| All hMPV Isolations | hMPV A2a | hMPV A2b | hMPV B1 | hMPV B2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meteorological Variables | (n = 1275)a | (n = 591)b | (n = 21)b | (n = 319)b | (n = 291)b | P Valuec | Difference Between Groups | |||||
| Mean temperature (°C) | 13.5 | (8.0–17.9) | 14.6 | (9.8–18.1) | 13.2 | (7.3–17.3) | 13.5 | (7.2–18.7) | 12.2 | (5.3–16.0) | .0003 | A2a > B2; B1 > B2 |
| Minimum temperature (°C) | 9.0 | (3.3–13.3) | 9.8 | (5.1–13.8) | 7.3 | (3.7–14.1) | 8.8 | (2.7–14.5) | 7.1 | (1.8–12.0) | .0001 | A2a > B2; B1 > B2 |
| Maximum temperature (°C) | 18.8 | (12.7–23.7) | 20.0 | (14.5–24.0) | 18.4 | (11.0–23.8) | 19.1 | (12.4–24.6) | 17.3 | (10.4–21.8) | .0006 | A2a > B2; B1 > B2 |
| Diurnal temperature variation (°C) | 9.9 | (7.4–11.6) | 10.1 | (7.4–11.8) | 10.4 | (8.1–12.3) | 9.7 | (7.4–11.5) | 9.7 | (7.5–11.8) | .5742 | |
| Ground surface temperature (°C) | 15.7 | (9.6–21.3) | 16.8 | (11.5–21.8) | 13.6 | (7.3–19) | 15.9 | (8.0–23.1) | 13.9 | (6.9–18.5) | <.0001 | A2a > B2; B1 > B2 |
| Rainfall (mm) | 0 | (0–0.4) | 0 | (0–0.2) | 0 | (0–1.0) | 0 | (0–0.5) | 0 | (0–0.5) | .7532 | |
| Wind speed (m/s) | 2.7 | (2.2–3.3) | 2.7 | (2.3–3.2) | 2.6 | (2.4–3.4) | 2.7 | (2.2–3.3) | 2.6 | (2.2–3.3) | .9524 | |
| Relative humidity (%) | 55.0 | (43.8–66.5) | 56.9 | (45.9–68.8) | 49.5 | (45.4–66.0) | 53.8 | (42.8–65.9) | 51.3 | (42.6–64.0) | .0015 | A2a > B1; A2a > B2 |
| Atmosphere pressure (mm Hg) | 1005.0 | (999.6–1009.2) | 1005.0 | (999.6–1008.7) | 1004.0 | (1000.3–1008.4) | 1004.0 | (998.1–1009.4) | 1005.0 | (1001.0–1009.8) | .1558 | |
| Sunshine duration (hour) | 13.3 | (12.4–14) | 13.4 | (12.5–14.1) | 13.4 | (11.7–13.8) | 13.3 | (12.3–14.1) | 13.2 | (12.1–13.7) | .0790 | |
Data are shown as median with interquartile range.
Abbreviation: hMPV, human metapneumovirus.
aSeventy-six hMPV infections of undetermined genotype were included.
bMixed-genotype hMPV infections were counted for each of the corresponding hMPV genotype isolations.
c P values were calculated by Kruskal-Wallis test, and post hoc analyses were performed by Dunn test to determine differences between groups.
Correlation Between Climatic Factors and hMPV Infections
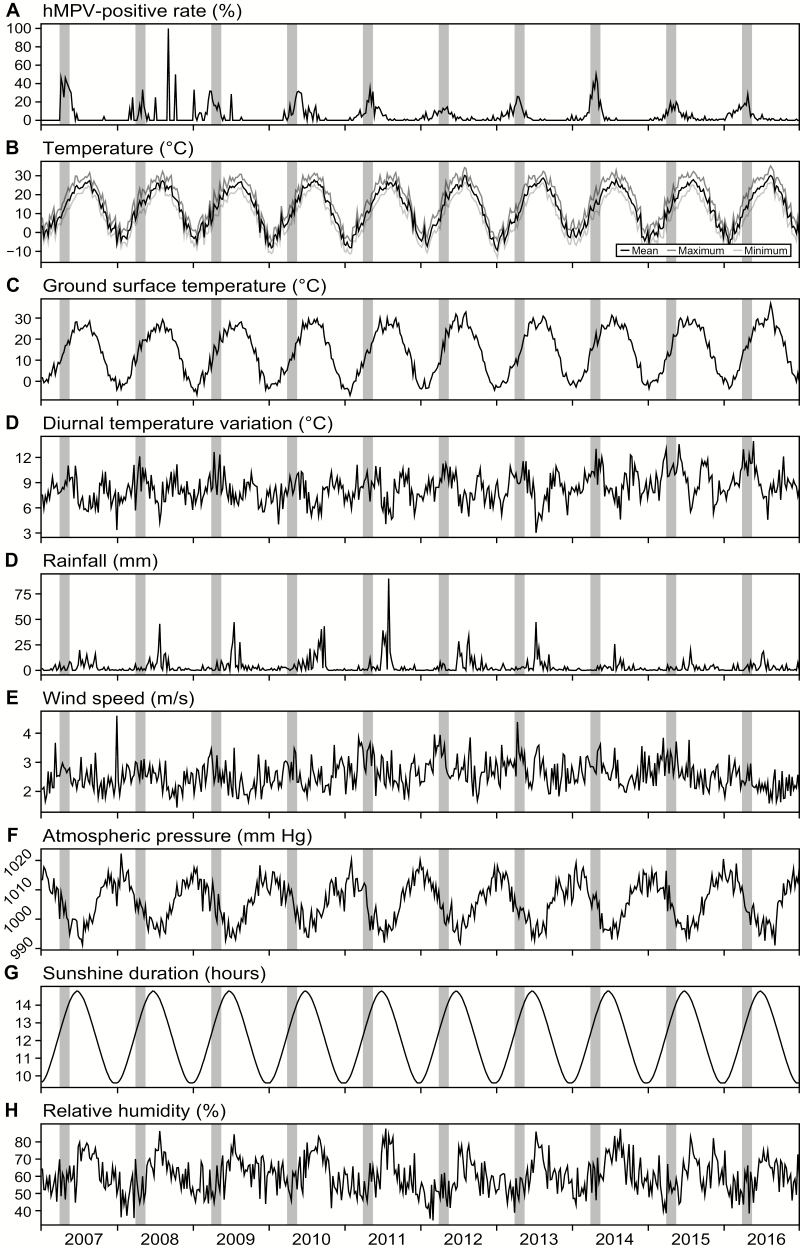
Figure 2 shows annual variations in the rates of hMPV positivity and climatic variables. The intervals between 13 and 20 weeks are marked with orange boxes, and more than half of the infections occurred during these intervals. As shown in Table 4, diurnal temperature variation, wind speed, relative humidity, atmospheric pressure, and sunshine duration were directly correlated with rates of positivity based on univariate regression analyses. When the correlations were further analyzed by multivariate regression analysis, diurnal temperature variation, wind speed, and sunshine duration were found to be independent associative factors.
Figure 2.
A–H, Human metapneumovirus (hMPV)–positive rate and climate variables. Gray bars represent the time interval between 13 and 20 weeks; more than half of all hMPV infections occurred within this interval.
Table 4.
Correlation Between Weekly Rates of Human Metapneumovirus Positivity and Meteorological Variables
| Univariate Regression Analysis | Multivariate Regression Analysis | |||
|---|---|---|---|---|
| Meteorological Variables | Coefficient | P Value | Coefficient | P Value |
| Mean temperature (°C) | −0.0644 | .1436 | −0.0015 | .1790 |
| Minimum temperature (°C) | 0.0253 | .5649 | … | |
| Maximum temperature (°C) | 0.0694 | .1148 | … | |
| Diurnal temperature variation (°C) | 0.2675 | <.0001 | 0.0130 | <.0001 |
| Ground surface temperature (°C) | 0.0736 | .0944 | 0.0009 | .4390 |
| Rainfall (mm) | 0.0366 | .4057 | … | |
| Wind speed (m/s) | 0.2024 | <.0001 | 0.0195 | .0308 |
| Relative humidity (%) | −0.1440 | .0010 | … | |
| Atmosphere pressure (mm Hg) | −0.1262 | .0040 | 0.0024 | .0521 |
| Sunshine duration (hour) | 0.2656 | <.0001 | 0.0457 | <.0001 |
Results of PCA of hMPV infection cases indicated that the first principal component could explain 52.0%–55.8% of the total variances (Supplementary Table 3 and Supplementary Figure 1). The most important contributors to the first principal component were temperature variables including mean, minimum, maximum, and ground surface temperatures. The second principal component, which was dominated by diurnal temperature variation and relative humidity, could explain about 20% of the total variances. The third principal component was dominated by wind speed, which explained approximately 10% of the total variances. From the PCA results, we could assume that temperature, relative humidity, and diurnal temperature variation were the significant climatic factors associated with hMPV infections.
Density plots were created to visualize the relationships between climate factors and hMPV infections (Supplementary Figure 2). Mean temperature with diurnal temperature variation and relative humidity were selected to generate the density plots. Most hMPV infections occurred at a temperature interval of 8°C–22°C, a diurnal temperature difference between 6°C and 14 °C, and a relative humidity interval of 40%–60%.
DISCUSSION
In our study, hMPV was detected in 5.4% of respiratory specimens and was the fifth most commonly isolated respiratory virus. hMPV infections occurred more frequently in children, and a seasonal peak was found to be prominent in the spring season. These results were consistent with previous reports conducted in South Korea [6, 18, 19].
The hMPV subtypes A2a, A2b, B1, and B2 co-circulated throughout the study period, and the annual predominant genotypes were hMPV A2a in 2007, 2010, 2014, 2015, and 2016; B1 in 2011 and 2012; and B2 in 2008 and 2009. hMPV genotype predominance was thus observed both for 1 year and a maximum of 3 consecutive years, whereas the predominant genotype changed every 3–4 years. However, there were no regular cyclic patterns of predominant hMPV genotypes throughout our study period. This circulation complexity was also found in long-term studies from the United States and Germany [11, 20]. In addition, we could not find any similarities in annual predominance patterns between our study and other studies conducted in neighboring counties including China and Japan [21–23]. These results suggest that the varying annual predominance of hMPV subtypes/genotypes is a local phenomenon, and not synchronized across a wide geographic region [11].
The clinical features of hMPV infections in our study were consistent with those described in previous reports [9, 21]. Pediatric patients with hMPV infection showed mild respiratory symptoms before 2–5 days of hospitalization. Most inpatient children were diagnosed with pneumonia and stayed in the hospital for 4–6 days. hMPV infections seemed to cause more severe illness in adult patients; however, it was difficult to draw a conclusion with these results because the proportion of adult patients was too small and more severe cases were more likely to be tested in the adult patient group.
Various studies have focused on the correlation between hMPV subtypes and clinical characteristics in infected patients. Vicente et al found that the clinical severity of hMPV A infections was higher than that of hMPV B infections [8]. Matsuzaki et al indicated that laryngitis was more associated with hMPV B1 infections, whereas wheezing was more prevalent with hMPV B1 and B2 infections [7]. However, there were also many reports suggesting that there are no differences in clinical characteristics caused by various hMPV subtypes [9, 10, 21]. In our study, according to hMPV subtypes, statistical significance was found in terms of symptoms, physical examination findings, and concurrent infections in pediatric patients. However, we found that there were no distinct differences in clinical diagnoses, clinical outcomes, and laboratory findings according to hMPV genotypes. These results support the contentions that certain clinical manifestations are different among hMPV genotypes but that there is no significant relationship between the hMPV subtype and disease severity.
We also attempted to assess the effects of meteorological factors on hMPV infections. hMPV activity was affected by temperature variables, relative humidity, diurnal temperature variation, wind speed, and sunshine duration. Among these factors, the rate of hMPV positivity was positively correlated with 3 variables including diurnal temperature variation, wind speed, and sunshine duration. Our results also showed peaks of hMPV activity at temperatures of approximately 13.5°C and relative humidity of 55.0%; furthermore, this activity tended to be associated with increasing diurnal temperature variation, wind speed, and sunshine duration. Previous studies conducted in other regions suggested that low temperature is the main driver of hMPV seasonality [11, 24–26]. However, based on the results of our study, we could not conclude that temperature variables are not linearly correlated with hMPV epidemics. Because the climate of Seoul is characterized by large seasonal temperature differences, which are not observed in previously studied countries, we hypothesized that hMPV would not be prevalent at low temperatures but would be prevalent at a certain temperature interval. In addition, the amount of rainfall was found to be correlated with the incidence of hMPV infections in subtropical and tropical regions [27–29], whereas hMPV activity was not affected by rainfall in our study. These findings suggest that hMPV infections were affected by climate factors, but that the meteorological drivers of virus activity vary by region and climate group [11, 24].
We also investigated differences in environmental factors that affect the transmission of each hMPV genotype. Our study showed that adaptation to climatic conditions, with respect to the occurrence of human cases of hMPV subgroups, was slightly different. For example, hMPV B2 infections appeared when the temperature was colder, whereas hMPV A2a infections were more prevalent in milder weather with a more humid environment. However, the origins of these differences in adaptation to meteorological conditions among hMPV subtypes were not clear, and further research should be performed to elucidate the link between the transmission abilities of hMPV subgroups and climatic factors.
Although we attempted to minimize errors, there were several limitations to this study. First, 6% of hMPV-positive specimens could not be genotyped due to sample problems such as deficiencies and RNA degradation after long-term storage. Although the proportion of “not determined” specimens was not relatively large, it could have affected the results of our study. Second, the clinical information of hMPV-positive patients was only tracked by medical records due to the nature of our retrospective study. We admit that some information could be misrepresented or omitted, and this possibility could also affect the results of this study. Third, because respiratory molecular testing was routinely used only for pediatric patients, and not for adult patients, the study population was biased for pediatric patients and the number of adult patients was relatively insufficient. Therefore, further long-term prospective surveillance should be performed to solve the present limitations and provide more informative results regarding hMPV infections.
In conclusion, our study provides long-term data on hMPV infections from a tertiary hospital serving a South Korean population. hMPV was the fifth most isolated respiratory virus and patients of various ages were infected, mainly between weeks 13 and 20 (April and May). Among the 5 subtypes of hMPV, hMPV A2a was most frequently isolated and each hMPV subtype showed annual predominance every 3–4 years. There were also significant differences between hMPV genotypes and clinical characteristics; however, disease severity was not altered according to hMPV subtype. Our study also showed that hMPV infections occurred with a seasonal rhythm and were associated with several climate factors including temperature, relative humidity, diurnal temperature variation, wind speed, and sunshine duration. In addition, each hMPV genotype had a different affinity for certain meteorological conditions. The results of our study contribute to the understanding of the clinical characteristics of infections caused by each hMPV subtype, as well as climate factors that contribute to the transmission of hMPV.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the molecular genetics department of Chung-Ang University hospital for their long-term contribution to this study, as well as Ah Ra Cho (Seoul Medical Science Institute) and Jun Hyung Lee (Chonnam National University Hwasun Hospital) for their contributions.
Financial support. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (grant number NRF-2015R1D1A1A01058906).
Potential conflicts of interest. All authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Shahda S, Carlos WG, Kiel PJ, Khan BA, Hage CA. The human metapneumovirus: a case series and review of the literature. Transpl Infect Dis 2011; 13:324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schildgen V, van den Hoogen B, Fouchier R, et al. Human metapneumovirus: lessons learned over the first decade. Clin Microbiol Rev 2011; 24:734–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rima B, Collins P, Easton A, et al. ICTV virus taxonomy profile: Pneumoviridae. J Gen Virol 2017; 98:2912–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim JI, Park S, Lee I, et al. Genome-wide analysis of human metapneumovirus evolution. PLoS One 2016; 11:e0152962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mackay IM, Bialasiewicz S, Jacob KC, et al. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. J Infect Dis 2006; 193:1630–3. [DOI] [PubMed] [Google Scholar]
- 6. Chung JY, Han TH, Kim SW, Hwang ES. Genotype variability of human metapneumovirus, South Korea. J Med Virol 2008; 80:902–5. [DOI] [PubMed] [Google Scholar]
- 7. Matsuzaki Y, Itagaki T, Abiko C, Aoki Y, Suto A, Mizuta K. Clinical impact of human metapneumovirus genotypes and genotype-specific seroprevalence in Yamagata, Japan. J Med Virol 2008; 80:1084–9. [DOI] [PubMed] [Google Scholar]
- 8. Vicente D, Montes M, Cilla G, Perez-Yarza EG, Perez-Trallero E. Differences in clinical severity between genotype A and genotype B human metapneumovirus infection in children. Clin Infect Dis 2006; 42:e111–3. [DOI] [PubMed] [Google Scholar]
- 9. Wei HY, Tsao KC, Huang CG, Huang YC, Lin TY. Clinical features of different genotypes/genogroups of human metapneumovirus in hospitalized children. J Microbiol Immunol Infect 2013; 46:352–7. [DOI] [PubMed] [Google Scholar]
- 10. Agapov E, Sumino KC, Gaudreault-Keener M, Storch GA, Holtzman MJ. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J Infect Dis 2006; 193:396–403. [DOI] [PubMed] [Google Scholar]
- 11. Reiche J, Jacobsen S, Neubauer K, et al. Human metapneumovirus: insights from a ten-year molecular and epidemiological analysis in Germany. PLoS One 2014; 9:e88342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis 2003; 9:628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park M-S, Park S-H, Chae J-H, et al. High-resolution urban observation network for user-specific meteorological information service in the Seoul Metropolitan Area, South Korea. Atmos Meas Tech 2017; 10:1575–94. [Google Scholar]
- 14. Kaida A, Iritani N, Kubo H, Shiomi M, Kohdera U, Murakami T. Seasonal distribution and phylogenetic analysis of human metapneumovirus among children in Osaka City, Japan. J Clin Virol 2006; 35:394–9. [DOI] [PubMed] [Google Scholar]
- 15. Kim HR, Cho AR, Lee MK, Yun SW, Kim TH. Genotype variability and clinical features of human metapneumovirus isolated from Korean children, 2007 to 2010. J Mol Diagn 2012; 14:61–4. [DOI] [PubMed] [Google Scholar]
- 16. Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci 2016; 374:20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Rao Y, Sun Q, et al. Identification of climate factors related to human infection with avian influenza A H7N9 and H5N1 viruses in China. Sci Rep 2015; 5:18094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park JY, Yun KW, Lim JW, Lee MK, Lim IS, Choi ES. Clinical and genetic features of human metapneumovirus infection in children. Pediatr Int 2016; 58:22–6. [DOI] [PubMed] [Google Scholar]
- 19. Jang MS, Shin M. The epidemiology and clinical manifestation of human metapneumovirus infection in children during 2011–2014. Allergy Asthma Respir Dis 2017; 5:269–73. [Google Scholar]
- 20. Yang CF, Wang CK, Tollefson SJ, et al. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virol J 2009; 6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeng SZ, Xiao NG, Zhong LL, Yu T, Zhang B, Duan ZJ. Clinical features of human metapneumovirus genotypes in children with acute lower respiratory tract infection in Changsha, China. J Med Virol 2015; 87:1839–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang C, Du LN, Zhang ZY, et al. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in southwest China. J Clin Microbiol 2012; 50:2714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura M, Hirano E, Ishiguro F, et al. Molecular epidemiology of human metapneumovirus from 2005 to 2011 in Fukui, Japan. Jpn J Infect Dis 2013; 66:56–9. [DOI] [PubMed] [Google Scholar]
- 24. Darniot M, Pitoiset C, Millière L, et al. Different meteorological parameters influence metapneumovirus and respiratory syncytial virus activity. J Clin Virol 2018; 104:77–82. [DOI] [PubMed] [Google Scholar]
- 25. Sundell N, Andersson LM, Brittain-Long R, Lindh M, Westin J. A four year seasonal survey of the relationship between outdoor climate and epidemiology of viral respiratory tract infections in a temperate climate. J Clin Virol 2016; 84:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. du Prel JB, Puppe W, Gröndahl B, et al. Are meteorological parameters associated with acute respiratory tract infections? Clin Infect Dis 2009; 49:861–8. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y, Chen Z, Yan YD, et al. Seasonal distribution and epidemiological characteristics of human metapneumovirus infections in pediatric inpatients in southeast China. Arch Virol 2013; 158:417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nandhini G, Sujatha S, Jain N, et al. Prevalence of human metapneumovirus infection among patients with influenza-like illness: report from a tertiary care centre, southern India. Indian J Med Microbiol 2016; 34:27–32. [DOI] [PubMed] [Google Scholar]
- 29. Chow WZ, Chan YF, Oong XY, et al. Genetic diversity, seasonality and transmission network of human metapneumovirus: identification of a unique sub-lineage of the fusion and attachment genes. Sci Rep 2016; 6:27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.