Abstract
Background: The availability of an early diagnostic tool for severe acute respiratory syndrome (SARS) would have major public health implications. We investigated whether the SARS coronavirus (SARS-CoV) can be detected in serum and plasma samples during the early stages of SARS and studied the potential prognostic implications of such an approach.
Methods: We developed two real-time quantitative reverse transcription-PCR (RT-PCR) assays, one for the polymerase and the other for the nucleocapsid region of the virus genome, for measuring the concentration of SARS-CoV RNA in serum/plasma samples from SARS patients. Plasma samples were obtained from 12 confirmed SARS patients on the day of hospital admission, as well as on days 7 and 14 after fever onset. Serum samples were also obtained from 23 confirmed SARS patients on the day of hospital admission, 11 of whom subsequently required intensive care. Viral RNA was extracted from the plasma/serum samples. The extracted RNA was subjected to analysis by the RT-PCR assays.
Results: The RT-PCR system for the polymerase region detected SARS-CoV RNA in 50% of plasma and 78% of serum samples from SARS patients during the first week of illness. The detection rates for plasma dropped to 25% at day 14 after fever onset. The median serum SARS-CoV concentrations in patients who required and did not require intensive care unit admission during the course of hospitalization were 5800 and 140 copies/mL, respectively (Mann–Whitney test, P <0.005). These data were confirmed by the RT-PCR system for the nucleocapsid region, which showed an even higher detection rate of 87%. The correlation between the results obtained by the two RT-PCR systems was high (Pearson correlation analysis, r = 0.998; P <0.001).
Conclusion: Plasma/serum SARS-CoV quantification represents a potentially useful early diagnostic and prognostic tool for SARS.
Severe acute respiratory syndrome (SARS)1 is a recently emerged infectious disease caused by a novel coronavirus, the SARS coronavirus (SARS-CoV) (1)(2)(3)(4). To date, molecular testing for SARS has focused mainly on reverse transcription-PCR (RT-PCR) analysis of nasopharyngeal aspirates, urine, and stools (5)(6). However, quantitative interpretation of these data is difficult because numerous factors, such as sampling techniques for nasopharyngeal aspirates, urine volume, and variations in bowel transit time (e.g., during diarrhea) and stool consistency, have hindered the standardization of data expression. On the other hand, plasma/serum-based assays may allow the precise and standardized quantitative expression of viral loads, thus enabling the assessment of disease severity and prognosis. Detection of viral nucleic acids in plasma/serum has been well established for viral load studies for numerous other viruses (7)(8). However, there is a paucity of data concerning the detection of SARS-CoV in the plasma/serum of SARS patients. There has been a single report showing the relatively low sensitivity of detecting SARS-CoV RNA in plasma by an ultracentrifugation-based approach, with low concentrations of SARS-CoV detected in the plasma of a patient 9 days after disease onset (1).
Using the publicly released full genomic sequences of SARS-CoV (9)(10)(11), we have developed real-time RT-PCR assays specifically targeting two different regions of the SARS-CoV genome. In this study, we investigated whether SARS-CoV RNA can be detected in serum or plasma samples during the early stages of SARS without ultracentrifugation and studied the potential prognostic implications of such an approach.
Patients and Methods
patients
Peripheral blood samples were obtained from SARS patients admitted to the New Territories East Cluster of Hospital Authority Hospitals in Hong Kong. Samples were recruited between March and May 2003.
In the first part of this study, blood samples were collected from 12 SARS patients on the day of hospital admission, as well as on days 7 and 14 after fever onset. Informed consent was obtained from the patients, and ethics approval was obtained from the Institutional Review Board. In the second part of this study, blood samples were obtained from 23 SARS patients on the day of hospital admission. All studied patients had subsequent serologic evidence of antibodies to SARS-CoV. For the prognostic part of the study, the previously mentioned 23 SARS patients were subdivided into two patient groups: (a) 11 patients who required admission to the intensive care unit (ICU) and (b) 12 patients who did not require ICU admission (non-ICU) during the duration of their hospitalization.
processing of blood samples
Blood samples were collected in EDTA-containing or plain tubes. The plain bottles were allowed to stand at room temperature for 2 h for complete clotting of blood. The blood samples were then centrifuged at 1600g for 10 min at 4 °C. Plasma or serum was then carefully transferred to plain polypropylene tubes. The plasma samples were recentrifuged at 16 000g for 10 min at 4 °C, and the supernatants were collected in fresh polypropylene tubes.
rna extraction
Viral RNA was extracted from 0.28 mL of plasma/serum with use of a QIAamp viral RNA mini reagent set (Qiagen) according to the manufacturer’s recommendations. RNA was eluted with 50 μL of AVE buffer (included in the reagent set) and stored at −80 °C.
real-time quantitative rt-pcr
One-step, real-time quantitative RT-PCR was used for SARS-CoV RNA quantification. We used the publicly released full genomic sequences of SARS-CoV (http://www.ncbi.nlm.nih.gov) to design two RT-PCR systems specifically targeting the SARS-CoV genome. The sequences of the primers and probes showed perfect alignment with all publicly available complete genome sequences of SARS-CoV on GenBank as of July 23, 2003 (accession nos. AY338175, AY338174, NC_004718, AY321118, AY323977, AY283798, AY283797, AY283796, AY283795, AY283794, AY291315, AY279354, AY278490, AY278487, AY278489, AY278554, AY297028, AY274119, AY291451, AY282752, AY278488, AY278741, and AY278491). The SARSPol1 system targeted the polymerase gene (orf1ab polyprotein; nucleotides 15327–15398; accession no. AY278554), and the SARSN system targeted the nucleocapsid gene (N; nucleotides 28758–28823; accession no. AY278554) of the SARS-CoV genome. The SARSPol1 primer sequences were 5′-GAGTGTGCGCAAGTATTAAGTGA-3′ (forward) and 5′-TGATGTTCCACCTGGTTTAACA-3′ (reverse), and the dual-labeled fluorescent probe was 5′-(FAM)ATGGTCATGTGTGGCGGCTCACTA(TAMRA)-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine. The SARSN primer sequences were 5′-TGCCCTCGCGCTATTG-3′ (forward) and 5′-GGCCTTTACCAGAAACTTTGC-3′ (reverse), and the dual-labeled fluorescent probe was 5′-(FAM)TGCTAGACAGATTGAACCAGCTTG(TAMRA)-3′. Calibration curves for SARS-CoV RNA quantification were prepared with serial dilutions of a HPLC-purified single-stranded synthetic DNA oligonucleotide (PROLIGO), with concentrations ranging from 1 × 107 to 1 × 100 copies. The initial concentration of the synthetic oligonucleotide was determined by measuring the absorbance at 260 and 280 nm. The assay was able to detect 5 copies of the calibrator target in the reaction mixture, as indicated in Figs. 1 and 2 in the Data Supplement that accompanies the online version of this article athttp://www.clinchem.org/content/vol49/issue12/. Concentrations of SARS-CoV were expressed as copies/mL of plasma/serum.
Because no recovery experiments had been done, the reported concentrations (copies/mL) were minimum estimates. The sequences of the synthetic DNA oligonucleotides for SARSPol1 and SARSN calibrations were 5′-AACGAGTGTGCGCAAGTATTAAGTGAGATGGTCATGTGTGGCGGCTCACTATATGTTAAACCAGGTGGAACATCATCCGG-3′ and 5′-GAAACTGCCCTCGCGCTATTGCTGCTAGACAGATTGAACCAGCTTGAGAGCAAAGTTTCTGGTAAAGGCCAACAA-3′, respectively.
The RT-PCR reactions were set up according to the manufacturer’s instructions (EZ rTth RNA PCR reagent set; Applied Biosystems) in a reaction volume of 25 μL. The primers and fluorescent probes were used at concentrations of 300 and 100 nM, respectively, and 12 μL of extracted plasma/serum RNA was used for amplification. The thermal profile used for the analysis was as follows: the reaction was initiated at 50 °C for 2 min for the included uracil N-glycosylase to act, followed by reverse transcription at 60 °C for 30 min. After a 5-min denaturation at 95 °C, 40 cycles of PCR were carried out with denaturation at 94 °C for 20 s and annealing/extension for 1 min at 56 °C. Each sample was analyzed in duplicate, and the calibration curve was run in parallel for each analysis. Multiple negative water blanks were also included in every analysis.
statistical analysis
Statistical analysis was performed with SigmaStat 2.03 software (SPSS). The Student t-test was used for the comparison of the ages of the ICU and non-ICU groups, and the Mann–Whitney test was used for the comparison of serum SARS-CoV RNA concentrations between the ICU and non-ICU groups. Pearson correlation analysis was used to assess the correlation of SARS-CoV RNA concentrations between the SARSPol1 and SARSN RT-PCR systems.
Results
development of real-time quantitative rt-pcr
To determine the quantitative performance of the SARS-CoV RT-PCR assays, we used these assays to amplify serially diluted calibrators, which were synthetic DNA oligonucleotides based on the SARS-CoV genomic sequence. The calibration curves for the SARSPol1 (polymerase) and SARSN (nucleocapsid) amplification systems demonstrated a dynamic range from 5 copies to 1 × 107 copies. A semilogarithmic plot of different calibrator concentrations against the threshold cycles yielded correlation coefficients of 0.987 for the SARSPol1 system and 0.993 for the SARSN system. The amplification steps of these assays were sufficient to allow detection of 5 copies of the targets in the reaction mixtures, which corresponded to a detection limit of 74 copies/mL of plasma/serum. The assay consistently demonstrated this low detection limit, as indicated by the amplification of calibrators. In 11 of 15 separate analyses, we were able to detect down to 5 copies of the synthetic oligonucleotide in the reaction mixture, as indicated in Fig. 3 of the online Data Supplement.
To determine the precision of the whole analytical procedure, including RNA extraction, reverse transcription, and amplification, we performed 10 replicate RNA extractions from a sample pooled from plasma obtained from 5 SARS patients and subjected these extracted RNA samples to RT-PCR assays. The CV of the copy number of these replicate analyses for the SARSPol1 and SARSN amplification systems were 16% at 280 copies/mL and 15% at 320 copies/mL, respectively. Because we had not carried out recovery experiments, the actual virus concentrations in the tested samples would be expected to be higher than the actual concentrations measured by real-time RT-PCR. However, this would not affect the interpretation of our data because the reproducibility of the RNA extraction and quantitative RT-PCR steps had been tested as detailed above and our system would serve as a reproducible and valid comparison of the virus concentrations within a patient at different times and between different patients.
detectability of plasma sars-CoVrna at different stages of the disease
To investigate whether SARS-CoV RNA could be detected in plasma and to evaluate the relative usefulness of plasma SARS-CoV measurements at different stages of the disease, we studied 12 serologically confirmed SARS patients. Plasma samples were taken on admission, representing a mean of 3.6 days after fever onset (range, 1–6 days). Additional samples were taken from each of these patients at days 7 and 14 after fever onset. The SARSPol1 real-time RT-PCR system detected plasma SARS-CoV RNA above the detection limit, as determined by the serially diluted calibrator, in six patients on admission (50%). The detection rate remained at 50% (6 of 12) at day 7 and fell to 25% (3 of 12) at day 14 after fever onset (Table 1 ). As negative controls, SARS-CoV RNA was not detected in the plasma samples obtained from 40 healthy individuals.
Table 1.
Serial analysis of plasma SARS-CoV RNA concentrations in SARS patients by the RT-PCR system for the polymerase region.1
| Patients | Plasma SARS-CoV concentration, copies/mL | ||||
|---|---|---|---|---|---|
| Day 1 | Day 7 | Day 14 | |||
| 1 | 298 | Undetectable | 173 | ||
| 2 | 231 | 363 | Undetectable | ||
| 3 | Undetectable | Undetectable | Undetectable | ||
| 4 | Undetectable | Undetectable | 100 | ||
| 5 | 239 | 170 | Undetectable | ||
| 6 | Undetectable | Undetectable | Undetectable | ||
| 7 | Undetectable | 8565 | Undetectable | ||
| 8 | 230 | 819 | Undetectable | ||
| 9 | 125 | 413 | Undetectable | ||
| 10 | Undetectable | Undetectable | Undetectable | ||
| 11 | 404 | 84 | Undetectable | ||
| 12 | Undetectable | Undetectable | 153 | ||
| Detection rate | 50% | 50% | 25% | ||
The detection limit of the assay is 74 copies/mL. Day 1, day 7, and day 14 denote the days after fever onset. The nucleocapsid RT-PCR system has a higher detection sensitivity (see text).
quantitative analysis of sars-CoVrna in sera of sars patients
To test the detectability of SARS-CoV RNA in serum, instead of plasma, during the early stage of SARS, we analyzed 23 serum samples obtained from SARS patients on the day of hospital admission, using the SARSPol1 real-time RT-PCR system. For 22 patients, the serum samples were taken at a mean of 2.6 days (range, 1–6 days) after the onset of fever. One patient did not have fever during his illness. The SARSPol1 system was able to detect SARS-CoV RNA in 18 of the 23 samples (78%), including the patient who did not have fever. The detection rate essentially confirmed the plasma-based results from our first cohort as described above. The median serum SARS-CoV RNA concentration was 752 copies/mL. As negative controls, SARS-CoV RNA was not detected in serum samples obtained from 30 healthy individuals.
corroborative data from a second rt-pcr system
To confirm the data generated by the SARSPol1 RT-PCR system, we used another real-time RT-PCR system (SARSN) targeting the nucleocapsid gene of the SARS-CoV genome to repeat the serum analysis. The SARSN system was able to detect SARS-CoV RNA in 20 of the 23 samples (87%). The median serum SARS-CoV RNA concentration was 3900 copies/mL. The correlation coefficient for SARS-CoV RNA concentrations between the SARSPol1 and SARSN RT-PCR systems was 0.998 (Pearson correlation analysis; P <0.001; Fig. 1 ). As negative controls, SARS-CoV RNA was not detected by the SARSN RT-PCR system in serum samples obtained from 30 healthy individuals.
Figure 1.
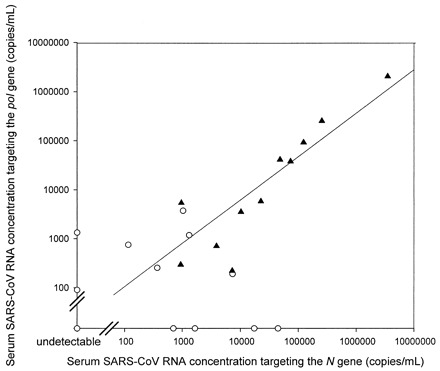
Correlation of serum SARS-CoV RNA concentrations between the SARSPol1 and SARSN RT-PCR systems in SARS patients requiring and not requiring ICU admission.
▴, results for SARS patients requiring ICU admission. ○, results for SARS patients not requiring ICU admission. The double slashes indicate the detection limit (74 copies/mL).
prognostic implications
To determine the potential prognostic implications of the admission serum SARS-CoV concentration, the previously mentioned group of 23 patients was stratified into those who required admission to the ICU (n = 11) and those who did not (n = 12) during the duration of their hospitalization. The mean ages of the two groups of patients were 56 and 47 years, respectively (Student t-test, P = 0.188). The male-to-female ratios were 5:6 for the ICU group and 6:6 for the non-ICU group. For the SARSPol1 RT-PCR assay, the median serum SARS-CoV concentrations in the ICU and non-ICU groups were 5800 and 140 copies/mL, respectively (see Fig. 4 in the online Data Supplement). The difference in the serum SARS-CoV concentrations between these groups was statistically significant (Mann–Whitney test, P <0.005). For the SARSN RT-PCR assay, the median serum SARS-CoV concentrations in the ICU and non-ICU groups were 23 000 and 870 copies/mL, respectively (see Fig. 4 in the online Data Supplement). The difference in serum SARS-CoV concentrations between these groups was also statistically significant (Mann–Whitney test, P <0.007).
Discussion
In this report we have demonstrated that SARS-CoV RNA is detectable in the plasma and serum of patients during the early stages of SARS, with detection rates up to 78% for the SARSPol1 system and 87% for the SARSN system. The findings demonstrate that plasma/serum SARS-CoV measurement is a sensitive method for detecting SARS-CoV infection during the first week after fever onset. This sensitivity, especially for serum, is much higher than the sensitivities reported for other clinical specimen types at a similar stage of infection. For example, RT-PCR analysis of nasopharyngeal aspirates had been reported to have a sensitivity of 32% in the first week after symptom onset (6).
The data presented here also demonstrated that the median concentrations of serum SARS-CoV RNA in patients who required ICU admission during the course of hospitalization were 30- and 26-fold higher than the median concentrations in those who did not require intensive care, as measured by the SARSPol1 and SARSN RT-PCR systems, respectively. Our results showed that there is a strong correlation between the serum SARS-CoV RNA concentrations obtained by the SARSPol1 and those obtained by the SARSN RT-PCR system, although the median serum SARS-CoV concentrations differed. The differences observed in the quantitative values may be a result of the coexistence of subgenomic fragments of the N gene with the full virus genome in serum. This could be a subject of further investigation.
We envision that plasma/serum SARS-CoV measurements can function in a synergistic manner with existing diagnostic strategies for SARS. Thus, plasma/serum RT-PCR can be performed with high sensitivity during the first week of the disease, RT-PCR analysis of stool and respiratory samples can be performed during the second week, and serologic testing for antibodies against SARS-CoV can be used from day 21 onward (6). The availability of a diagnostic test for the early identification of SARS patients could potentially be useful in the public health control of SARS. Furthermore, our data show that serum SARS-CoV measurement is a prognostic marker that can be used even on the first day of hospital admission. Apart from its obvious clinical significance, this observation also suggests that a high systemic viral load may lead to more severe tissue damage either directly or indirectly through the activation of a potentially damaging immune reaction. Elucidation of the latter possibility would be fertile ground for future research.
Supplementary Material
Acknowledgments
This work is supported by the Hong Kong Research Grants Council Special Grants for SARS Research (CUHK 4508/03M). We thank C.H. Tung, Chief Executive of the Hong Kong Special Administrative Region, for encouraging us to undertake this study. We thank Profs. Ambrose King and Sydney Chung for support during the course of this work.
Footnotes
Nonstandard abbreviations: SARS, severe acute respiratory syndrome; CoV, coronavirus; RT-PCR, reverse transcription-PCR; and ICU, intensive care unit.
References
- 1.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, et al. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 2003;423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon LL, Wong OK, Luk W, Yuen KY, Peiris JS, Guan Y. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin Chem 2003;49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003;361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damond F, Descamps D, Farfara I, Telles JN, Puyeo S, Campa P, et al. Quantification of proviral load of human immunodeficiency virus type 2 subtypes A and B using real-time PCR. J Clin Microbiol 2001;39:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White PA, Pan Y, Freeman AJ, Marinos G, Ffrench RA, Lloyd AR, et al. Quantification of hepatitis C virus in human liver and serum samples by using LightCycler reverse transcriptase PCR. J Clin Microbiol 2002;40:4346-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. The genome sequence of the SARS-associated coronavirus. Science 2003;300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 10.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003;300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 11.Tsui SK, Chim SS, Lo YMD, . and The Chinese University of Hong Kong Molecular SARS Research Group. Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N Engl J Med 2003;349:187-188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


