Abstract
Background
A novel coronavirus of zoonotic origin (2019-nCoV) has recently been identified in patients with acute respiratory disease. This virus is genetically similar to SARS coronavirus and bat SARS-like coronaviruses. The outbreak was initially detected in Wuhan, a major city of China, but has subsequently been detected in other provinces of China. Travel-associated cases have also been reported in a few other countries. Outbreaks in health care workers indicate human-to-human transmission. Molecular tests for rapid detection of this virus are urgently needed for early identification of infected patients.
Methods
We developed two 1-step quantitative real-time reverse-transcription PCR assays to detect two different regions (ORF1b and N) of the viral genome. The primer and probe sets were designed to react with this novel coronavirus and its closely related viruses, such as SARS coronavirus. These assays were evaluated using a panel of positive and negative controls. In addition, respiratory specimens from two 2019-nCoV-infected patients were tested.
Results
Using RNA extracted from cells infected by SARS coronavirus as a positive control, these assays were shown to have a dynamic range of at least seven orders of magnitude (2x10−4-2000 TCID50/reaction). Using DNA plasmids as positive standards, the detection limits of these assays were found to be below 10 copies per reaction. All negative control samples were negative in the assays. Samples from two 2019-nCoV-infected patients were positive in the tests.
Conclusions
The established assays can achieve a rapid detection of 2019n-CoV in human samples, thereby allowing early identification of patients.
Introduction
Coronaviruses can be classified into 4 genera (α, β, γ, and δ) and these viruses are detected in a wide range of animal species, including humans (1). There are 6 previously known human coronaviruses that can be transmitted between humans. Human alphacoronaviruses, 229E and NL63, and betacoronaviruses, OC43 and HKU1, are common respiratory viruses usually causing mild upper respiratory diseases. In contrast to these, the other 2 human beta-coronaviruses, SARS and MERS coronaviruses, are highly pathogenic in humans. The case fatality rates of SARS and MERS are about 10% and 35%, respectively (2, 3). SARS and MERS can be transmitted between humans and may cause major outbreaks in health care settings or in community settings. Both SARS and MERS coronaviruses are of zoonotic origins. MERS coronavirus is widely circulated in dromedary camels. Over 90% of dromedary camels in Middle East countries are seropositive for MERS coronavirus (4, 5). Because of such a high prevalence in this animal species, sporadic zoonotic transmissions of MERS coronavirus from camels to humans have been repeatedly detected in the affected region since its discovery in 2012. For the SARS outbreak in 2003, exotic mammals in wet markets, such as palm civets and raccoon dogs, are believed to be the animal sources of the virus (6). The detection of SARS coronavirus in these species led to a temporary ban of selling of these animals in 2004, thereby preventing additional spill-over events from animals to humans. But further surveillance studies indicated that these animals are rarely positive for SARS coronavirus in non-market settings, suggesting that they only served as intermediate hosts in the SARS event (1, 7). Subsequent virus surveillances in wild animals identified a vast number of coronaviruses in bats (8). Interestingly, several bat coronaviruses that are genetically similar to human SARS coronavirus were detected in horseshoe bats. This group of SARS-like bat coronaviruses, together with SARS coronavirus, form a unique clade under the subgenus Sarbecovirus. It is now generally believed that SARS coronavirus is a recombinant virus between several bat SARS-like beta-coronaviruses (9). Some of these bat coronaviruses are experimentally capable of infecting human cells in cultures (9), suggesting that these animal viruses and their recombinants can pose threats to human health.
In December 2019, a cluster of atypical pneumonia patients epidemiologically linked to a wholesale market in Wuhan was detected (10). A novel beta-coronavirus named as 2019-novel coronavirus (2019-nCoV) has been identified in some of these patients. This virus is genetically similar to a bat virus in the subgenus Sarbecovirus (11). As of 22 January 2020, over 200 cases, including 4 fatal ones, have been detected in Wuhan and other provinces (12). In addition, confirmed patients with a recent travel history to Wuhan have been detected in multiple countries (Japan: 1, South Korea: 1, USA: 1, and Thailand: 2). Some of the confirmed cases have no epidemiological link to wet markets, suggesting the possibility of human-to-human transmissions and/or multiple spill-over events in different settings. At the time of preparing this article, a hospital outbreak had been reported (13). The spectrum of this disease in humans is yet to be fully determined. Signs of infection are highly nonspecific, and these include respiratory symptoms, fever, cough, dyspnea, and viral pneumonia. Thus, diagnostic tests specific for this infection are urgently needed for confirming suspected cases, screening patients and conducting virus surveillance.
In this study, we report the development of RT-PCR assays to detect this novel virus in human clinical specimens.
Materials and Methods
Primer and Probe Sequences
Two monoplex real-time RT-PCR assays targeting the ORF1b and N gene regions of 2019-nCoV were designed based on the first publicly available sequence in Genbank (Accession number: MN908947). The downloaded sequence and those for SARS coronavirus, bat SARS-like coronaviruses, and other representative coronaviruses were edited and aligned. Phylogenetic analyses of these sequences were performed using the neighbor joining method in MEGA X (14). Two sequence regions (ORF1b and N) that are highly conserved amongst sarbecoviruses were selected for primer and probe designs. The primer and probe sequences for the ORF1b gene assay are: 5’-TGGGGYTTTACRGGTAACCT-3’ (Forward; Y = C/T, R = A/G), 5’-AACRCGCTTAACAAAGCACTC-3’ (Reverse; R = A/G) and 5′-TAGTTGTGATGCWATCATGACTAG-3′ (Probe in 5’-FAM/ZEN/3’-IBFQ format; W = A/T), whereas the primer and probe sequences for the N gene assay are: 5′-TAATCAGACAAGGAACTGATTA-3′ (Forward), 5′-CGAAGGTGTGACTTCCATG-3′ (Reverse) and 5′-GCAAATTGTGCAATTTGCGG-3′ (Probe in 5’-FAM/ZEN/3’-IBFQ format). The expected amplicon sizes of ORF1b and N gene assays are 132 bp and 110 bp, respectively. All primers and probes were purchased from a commercial source (Integrated DNA Technologies). The primer and probe sequences were subsequently confirmed to have perfect matches with other 2019-nCoV genome sequences available from Global Initiative on Sharing All Influenza Data (GISAID; https://www.gisaid.org/; Accession numbers: EPI_ISL_402119, EPI_ISL_402120, EPI_ISL_402121, EPI_ISL_402123 and EPI_ISL_402124; Accessed 12 Jan 2020).
Quantitative RT-PCR Assay Condition
A typical 20 µL monoplex RT-PCR assay contained 5 μL of 4X master reaction mixture (TaqMan Fast Virus 1-Step Master Mix, ThermoFisher), 0.5 µmol/L of forward primer, 0.5 μmol/L of reverse primer, 0.25 μmo/L of probe, and 4 μl of RNA sample. RT-PCR reactions were conducted by a thermal cycler (ViiA7 Real-Time PCR system, ThermoFisher) with the following conditions: reverse transcription at 50°C for 5 min, inactivation of reverse transcriptase at 95°C for 20 s, 40 cycles of PCR amplification (Denaturing at 95°C for 5 s; Annealing/Extending at 60°C for 30 s). The time for each RT-PCR run was about 1 h and 15 min.
Viral RNA purification kit (QIAamp Viral RNA Mini Kit, Qiagen) and DNA plasmid purification kit (QIAprep Spin Miniprep Kit, Qiagen) were used for RNA and DNA extractions, respectively, as instructed by the manufacturer. For all RNA extractions, RNA was extracted from 140 µL of sample and eluted in 60 µL elution buffer containing poly(A) carrier RNA.
Tested Samples
Viral RNA extracted from SARS coronavirus-infected cells was used for positive controls. In addition, the RT-PCR products of SARS coronavirus generated by the ORF1b and N gene assays were cloned into plasmids. Serially diluted RNA or DNA samples were used to evaluate the performance of these assays. To determine the specificity of these assays, negative control samples were tested. These negative control samples were: 1) RNA extracted from cultured viruses: human coronaviruses (229E, OC43, and MERS), camel coronavirus (HKU23), human influenza A viruses (H1N1, H3N2, H5N1, and H7N9 subtypes), avian influenza (H1, H4, H6, and H9 subtypes), influenza B viruses (Yamagata and Victoria lineages), and adenovirus, 2) RNA extracted from retrospective human respiratory specimens previously tested positive for coronavirus (229E, HKU1, NL63, OC43), influenza A viruses (H1N1 and H3N2 subtypes), influenza B viruses (Yamagata and Victoria lineages), adenovirus, enterovirus, human parainfluenza virus (PIV1, 2, 3 and 4), respiratory syncytial virus, human metapneumovirus, rhinovirus and human bocavirus, and 3) RNA extracted from sputum samples from patients without respiratory viral infections (n = 9). Culture supernatants of infected cells or clinical samples stored in standard virus transport media were subjected to a brief centrifugation (15,170 × g, 2 min) and 140 µL of supernatant from each sample was used for RNA extraction.
Two patients (Patients 1 and 2) suspected to be infected by 2019-nCoV in Beijing were included in this study. For Patient 1, a sputum sample was collected at day 5 post-onset of symptoms. The sputum sample stored in standard virus transport media was treated with an equal volume of Sputasol (ThermoFisher) before RNA extraction. For Patient 2, a throat swab sample was collected at day 3 post-onset of symptoms and stored in standard virus transport media. 140 µL of each aqueous sample was used for viral RNA extraction. Relevant clinical and epidemiological data for both patients were not accessible to this study. RNA extracted from these samples were serially diluted and tested by the assays.
Results
Specificity of the Assays
At the time of preparing this assay, there was only one viral sequence available from the public domain. With such a limited sequence information, we decided to develop two RT-PCR assays that can react with multiple coronaviruses that are in the subgenus Sarbecovirus (Fig. 1A; see discussion). Using additional sequence information from this clade of coronaviruses, we selected highly conserved ORF1b and N gene regions as our targets (Fig. 1B). These designs allow the use of nucleic acids derived from SARS coronavirus as positive controls.
Fig. 1.
Sequence analysis of 2019-nCoV. (A) Phylogenetics of 2019-nCoV and related sarbecoviruses. Representative α-, β-, γ-coronaviruses were included in the analysis. Bootstrap values ≥70% are shown. Gene accession numbers in GenBank for corresponding viral sequences are provided. Scale bar indicates estimated genetic distance. Human coronaviruses are in bold and 2019-nCoV is underlined. (B) Sequence alignment of ORF-1b (top) and N (bottom) amplicons generated from 2019-nCoV and SARS-CoV. Arrows indicate the regions targeted by the studied primers. Sequence variations between these viruses are shown. The expected amplicon sizes generated from ORF1b and N gene assays are 132 and 110 bp, respectively.
Evaluation of the Assays Using Viral RNA Extracted from Positive and Negative Control Samples
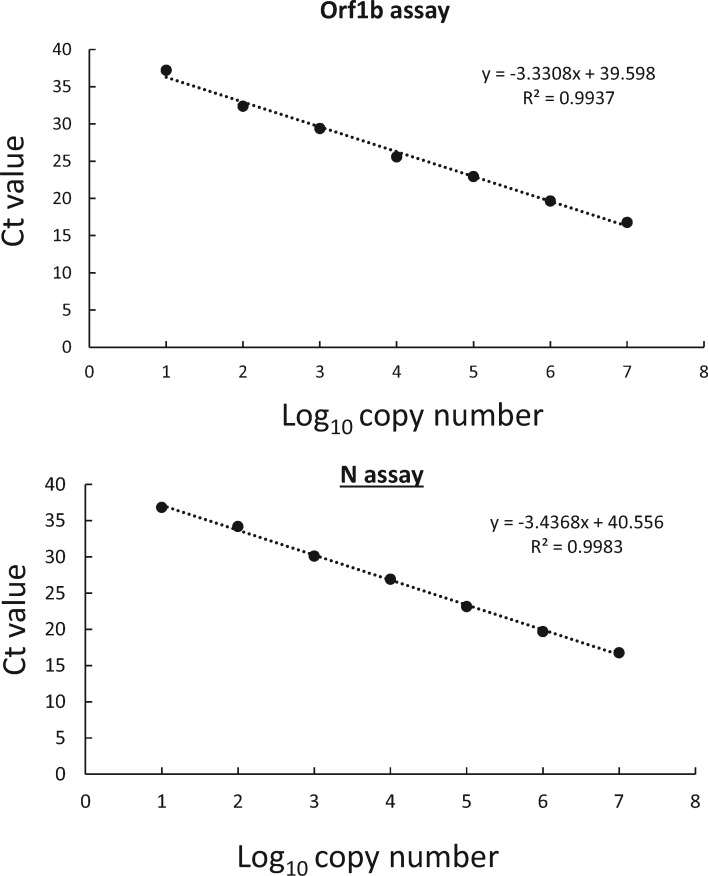
Viral RNA from cells infected by SARS coronavirus or DNA plasmids containing the target sequences were positive in the assays as expected. Using viral RNA extracted from virus cultures with known titers, both assays have a dynamic range of at least seven orders of magnitude (2 x 10−4-2000 TCID50/reaction; data not shown). To accurately determine the detection limits of these assays, we also tested serially diluted positive control plasmids in these assays. In our preliminary trial, reactions with ≥10 copies of this positive control plasmid were consistently positive in both of the assays (n = 5). The DNA amplifications of these newly established assays were found to be efficient. The amplification efficiencies of ORF1b and N gene assays were 99.6% and 95.4%, respectively (Fig. 2, R2 > 0.99 in both cases).
Fig. 2.
Dynamic range of ORF1b and N gene assays.
All negative controls were found to be negative in these assays (data not shown). In addition, we also spiked known amounts of SARS coronavirus into representative negative control samples (10-fold serially diluted virus sample; range: 2 x 10−4-2000 TCID50/reaction; Number of tests per concentration: 1). RNA extracted from these spiked samples were all positive in the tests, with no significant RT-PCR inhibition detected.
Detection of 2019-nCoV in Patient Samples
Respiratory specimens from two suspected patients were tested by both RT-PCR assays. Both patients were positive in the tests (Table 1). Preliminary results from serially diluted RNA samples suggested that the N gene assay is about 10 times more sensitive than the ORF-1b gene assay in detecting positive clinical specimens. As these samples could only be tested qualitatively by these assays at the testing site, the exact viral copy numbers in these specimens cannot be determined.
Table 1.
Detection of 2019-nCoV in patient samples.
| Dilution | Assay (Ct value) |
|
|---|---|---|
| Orf1b a | N a | |
| Patient 1 | ||
| Neat | 28.79 | 30.96 |
| 10x | 31.935 | 33.83 |
| 100x | Neg | 36.9 |
| 1000x | Neg | Neg |
| 10,000x | Neg | Neg |
| Patient 2 | ||
| Neat | 22.10 | 24.14 |
| 10x | 25.66 | 28.02 |
| 100x | 29.33 | 31.33 |
| 1000x | 32.55 | 34.54 |
| 10,000x | Neg | 37.70 |
Single test results at each dilution.
Discussion
In this study, two assays targeting the viruses in the subgenus Sarbecovirus were made and evaluated. We intentionally made these assays to be reactive to multiple viruses in this subgenus. This is because there is insufficient public information about the genetic diversity of 2019-nCoV in humans and animals. In 2003, SARS coronaviruses found in palm civets and humans were genetically highly similar, but distinct. For example, a partial deletion was detected in SARS coronavirus found in palm civets (6). Similar subtle sequence variations were also observed in MERS coronavirus found in camels and humans (15). To avoid the possible scenario in which the genetic diversity of 2019-nCoV is much more diverse than appreciated, our primer and probe sets were designed to react with this clade of coronaviruses.
We evaluated our assays using a panel of positive and negative control samples. The assays were found to be sensitive and specific to only sarbecoviruses. We further used respiratory specimens from patients infected by 2019-nCoV to demonstrate the potential use of these tests. The tested clinical samples were different in nature (sputum vs. throat swab) and they were collected at different onset times (day 5 vs. day 3). Thus, it is premature to use these data to determine the viral replication kinetics in humans. Further systematic investigations on clinical specimens collected from 2019-nCoV-infected patients at different post-onset time points will be needed. Nonetheless, the results demonstrated the clinical value of these respiratory samples for molecular detection of 2019-nCoV. In addition, the N gene RT-PCR assay was found to be more sensitive in detecting 2019-nCoV RNA in the studied clinical samples. It is possible that these clinical samples might contain infected cells expressing subgenomic mRNA (16), resulting in more N gene copies in the samples.
With the exception of 2019-nCoV and SARS coronavirus, none of the sarbecoviruses have been previously detected in humans. SARS was eliminated in humans and the last reported human SARS case was detected in 2004 (17). Individuals with samples that are positive in these RT-PCR assays should be considered to be infected by the 2019-nCoV or its related animal coronaviruses. Based on their detection performances, the N gene RT-PCR is recommended as a screening assay, and the Orf1b assay is recommended as a confirmatory one. Using a diagnostic algorithm similar to MERS, an N gene positive/Orf1b negative result should be regarded as indeterminate and the case is recommended to be referred to a WHO reference lab for further testing (18). In the event of having positive PCR results, sequence analyses of positive amplicons can help to confirm the result and to distinguish between 2019-nCoV and other genetically related coronaviruses (e.g., SARS coronavirus).
The biosafety classification of 2019-nCoV is yet to be fully defined. Clinical samples for molecular tests are recommended to be handled using biosafety level 2 practices (19). As SARS coronavirus is classified as a biosafety level 3 (BSL3) pathogen, we therefore intentionally used DNA plasmids as positive controls in these tests. This can avoid distributing SARS coronavirus genomic RNA which has the potential of generating infectious clones via reverse genetics. In addition, this can also help to distribute positive controls to laboratories at different geographical regions in a more cost-effective and robust manner. These DNA plasmid controls are available upon request.
During the reviewing process of this manuscript, Corman and colleagues reported the use synthetic constructs and SARS virus to develop RT-PCR assays for 2019-nCoV detection (20).
Glossary
Nonstandard Abbreviations
- TCID50
Median tissue culture infectious dose
- SARS
Severe Acute Respiratory Syndrome
- MERS
Middle East Respiratory Syndrome
Disclosures
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
K. Hui, provision of study material or patients; L. Poon, financial support, provision of study material or patients.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest: Employment or Leadership: None declared. Consultant or Advisory Role: None declared. Stock Ownership: None declared. Honoraria: None declared. Research Funding: RGC Theme-based research grant (Grant T11-705/14N) and National Institutes of Allergy and Infectious Diseases, National Institutes of Health (USA) (contract HHSN272201400006C). Expert Testimony: None declared. Patents: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments
We thank staff members of Daxing District Center for Disease Control and Prevention for their help with field investigation and sampling. We also thank colleagues for making 2019-nCoV viral sequences available for public access.
References
- 1. Cui J, Li F, Shi ZL.. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Donnelly CA, Malik MR, Elkholy A, Cauchemez S, Van Kerkhove MD.. Worldwide reduction in MERS cases and deaths since 2016. Emerg Infect Dis 2019;25:1758–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poon LL, Guan Y, Nicholls JM, Yuen KY, Peiris JS.. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis 2004;4:663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hemida MG, Perera RA, Wang P, Alhammadi MA, Siu LY, Li M, et al. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill 2013;18:20659.. [DOI] [PubMed] [Google Scholar]
- 5. Reusken CB, Ababneh M, Raj VS, Meyer B, Eljarah A, Abutarbush S, et al. Middle East Respiratory Syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill 2013;18:20662.. [DOI] [PubMed] [Google Scholar]
- 6. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003;302:276–8. [DOI] [PubMed] [Google Scholar]
- 7. de Wit E, van Doremalen N, Falzarano D, Munster VJ.. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14:523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan RW, Poon LL.. The emergence of human coronavirus emc: How scared should we be? mBio 2013;4:e00191–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 2017;13:e1006698.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Novel Coronavirus (2019-nCoV), Situation report- 1. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf? sfvrsn=20a99c10_4 (Accessed January 2020).
- 11. Zhou P, Yang X, Wang X, Hu B, Zhang L, Zhang W, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Preprint at https://www.biorxiv.org/content/10.1101/2020.01.22.914952v1 (2020).
- 12.World Health Organization. Novel Coronavirus (2019-nCoV), Situation report- 2. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf? sfvrsn=4d5bcbca_2 (Accessed January 2020).
- 13.South China Morning Post. Wuhan virus kills fourth patient, infects hospital staff amid fear of ‘super-spreader’https://www.scmp.com/news/china/society/article/3046908/new-china-virus-likely-human-transmission-stage-infections. (Accessed January 2020).
- 14. Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu DKW, Hui KPY, Perera R, Miguel E, Niemeyer D, Zhao J, et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci USA 2018;115:3144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simons FA, Vennema H, Rofina JE, Pol JM, Horzinek MC, Rottier PJ, Egberink HA.. mRNA PCR for the diagnosis of feline infectious peritonitis. J Virol Methods 2005;124:111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang M, Yan M, Xu H, Liang W, Kan B, Zheng B, et al. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis 2005;11:1860–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. Detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR. https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf? sfvrsn=af1aac73_4/ (Accessed January2020).
- 19.World Health Organization. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-(2019-nCoV)-in-suspected-human-cases (Accessed January 2020).
- 20. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25:pii=2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]