Abstract
Responding to the worldwide outbreak of SARS in 2003, virus removal performance and mechanism of a SMBR were investigated by employing phage T4 as a model virus. Two membrane modules were compared in continuous operation for about 75 days. During stable operation, SMBR achieved almost complete phage removal for both membrane modules. For the 0.22 μm module, the cake layer, the gel layer and the membrane contributed 6.3 log, 3.1 log and 1.7 log, respectively to phage removal, confirming the importance of the cake/gel layer formed on the surface of membrane. The damage of the cake/gel layer resulted in the decrease of phage removal. As for the 0.1 μm one, the membrane alone played a major role in phage removal. Inactivation by activated sludge and adsorption by cake/gel layer contributed about 3.6 log to phage removal everyday so that there was no phage accumulation in bulk solution. The results demonstrated that SMBR was an efficient system and recommended for treatment of virus-bearing wastewater.
Keywords: Submerged membrane bioreactor (SMBR), Phage removal, Phage T4, Removal mechanism, Wastewater treatment
1. Introduction
Hospital wastewater often contains a wide variety of microbial pathogens and viruses. However, this wastewater has long been treated with the conventional wastewater treatment processes. Even in well-functioning biological plants, as many as 103 CFU ml−1 resistant coliform bacteria were found in its effluent [1], [2], [3], to say nothing of much smaller viruses.
As for other disinfection methods, such as chlorination, chlorine dioxide, ozone and UV radiation etc., the mutagenic/carcinogenic and toxic disinfection by-products, which are potentially harmful to humans and aquatic organisms, are often accompanied with the disinfection treatment [4]. Moreover, the presence of suspended solids and organic compounds in wastewater often lower disinfection efficiency drastically [5].
SMBR, which is characterized by its ability of complete suspended solids removal from effluent, low/zero sludge production, compact size and lower energy consumption, has gained more and more attention [6], [7], [8], [9]. Some of above characteristics make SMBR have a potential ability to remove virus more effectively and safely. In last decades, several researches on viral removal by MBR had been carried out and gained some achievements, at the same time, present a few deficiencies. Chiemchaisri et al. [10] put forth that gel layer formed on the membrane surface could reject 4–6 log coliphage Qβ but did not gain the complete phage removal. Then Urase et al. [11] demonstrated that the cake/gel layer of membrane surface made a major contribution to reject virus in activated sludge by batch experiments. However, there was still 3–4 log of phages remained in effluent. Afterwards, Kawamura et al. [12] fulfilled the complete removal of phage Qβ and T1 by using ultramembrane unit but little phage removal mechanism was considered. Otaki et al. [13] also employed microfiltration and ultrafiltration process to the virus removal of the water supply. Recently, Wen et al. [8] investigated the performance of a SMBR for treatment of hospital wastewater but no considerations was given to rival removal. To date, however, more detailed and systematic reports on viral removal efficiency and mechanism of SMBR are still scarce [14].
In this study, a SMBR for treatment of virus-bearing wastewater was investigated using phage T4 as a tracer focusing on: (1) the removal efficiency of well-running SBMR to virus that suddenly surged into wastewater; (2) the effects of pore size of membrane modules on viral removal performance; (3) the effects of cake/gel layer disintegration on rival removal efficiency; and (4) the mechanisms of viral removal by SMBR equipped with different membrane modules. Based on above experiments, the feasibility of SMBR to remove SARS coronavirus was evaluated.
2. Materials and methods
2.1. System description
A bench-scale SMBR with an effective volume of 12 l was applied to treat municipal wastewater (Fig. 1 ). Two different membrane modules (pore size of 0.22 μm and 0.1 μm) were mounted in each membrane compartment. The membrane flux was driven by the difference of water head between the liquid level in the bioreactor and the effluent pipe (8.5 kPa). Prior to the injection of phage T4 into wastewater, the SMBR had been continuously operated for 34 days to make it work well. Then, T4 was fed to wastewater and the removal efficiency of well-running SBMR to virus was estimated. The operation parameters of SMBR were as below: temperature 14.5 °C, pH 6.4, DO 7.4 mg l−1, MLSS 4.5 g l−1, COD load 1.05 kgCOD/m3 d and HRT 10.8 h.
Fig. 1.
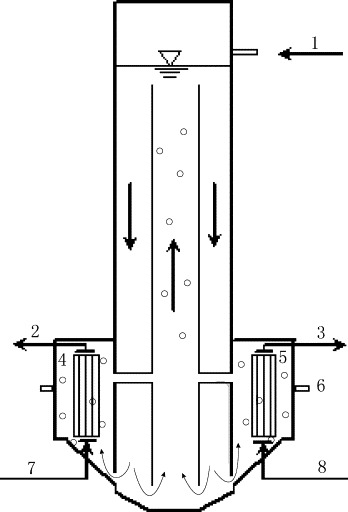
Schematic diagram of the SMBR. (1) Influent; (2 and 3) effluent of 0.22 μm and 0.1 μm; (4) membrane module no. 1 (0.22 μm, hollow fiber membrane, PVDF, membrane area 0.18 m2); (5) membrane module no. 2 (0.1 μm, hollow fiber membrane, PP, membrane area 0.18 m2); (6) sampling outlet of bulk solution; (7 and 8) compressed air inlet.
2.2. Preparation for phage T4
Phage T4 was selected as a model virus in this study because: (1) its size is similar to that of the SARS coronavirus [15]; (2) it is harmless to humans; (3) it can be seeded with a high concentration in tracer experiments; and (4) the assay method is relatively easy and simple [16]. T4 stock solution (1010 PFU ml−1) was prepared in advance and it was added to wastewater to make the phage concentration in a range from 105 to 108 PFU ml−1.
T4 in wastewater was viewed under Atomic Force Microscope (NanoScope IIIa Multimode Scanning Probe Microscopy Instruments, Digital Instruments, Santa Barbara, CA, USA).
The surfaces of a new membrane and a long-time used one were viewed under Scanning Electron microscope (FEI QUANTA 200).
2.3. Sample collection and analysis
The COD, NH4 +–N, and suspended solids (SS) of effluent from 0.22 μm membrane were determined by methods described by the literature [17]. For phage assay, samples were taken from the influent tank and outlet of each module at the same time everyday. Phage concentration was assayed according to the double-layer-agar method described by Adams [18] with E. coli B as host bacteria. In order to estimate the role activated sludge played on viral removal, the phage concentration of bulk solution was sampled and assayed also.
2.4. Data presentation
r overall, r c, r g, r m were employed to represent the virus removal efficiency by overall membrane, cake layer, gel layer and membrane alone, respectively. The equations were as follows:
| (1) |
| (2) |
| (3) |
| (4) |
where C in and C out, and , and were used to represent the phage concentration of the influent and effluent at stage a/b (operation stage with/without sludge discharge), c and d (operation stage with membrane cleansed by tap water and chemical solutions, respectively) during the whole operation (see Fig. 3A). C b was the phage concentration of bulk solution. It should be noted that the treatment procedure for bulk solution samples was different from one described by Ueda et al. [14]. In this study, the sample was directly collected from bulk solution without centrifugation, and vigorously shaken before dilution and assay. The phage removal efficiency of inactivation by activated sludge (r AS) and adsorption by cake/gel layer could be calculated statistically by the differential between phages added to the SMBR and ones retained in bulk solution.
Fig. 3.
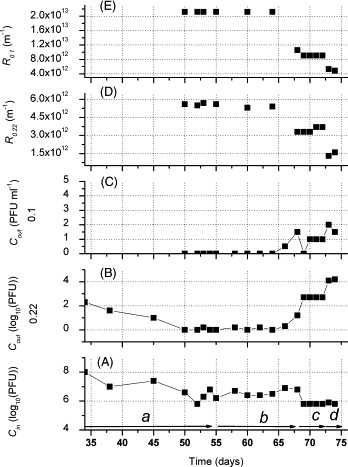
(A) Phage concentration of the influent; (B and C) phage concentration of the effluent of 0.22 μm and 0.1 μm membrane; (D and E) filtration resistance of 0.22 μm and 0.1 μm membrane; (a and b) operation stage with/without sludge discharge; (c and d) operation stage with membrane cleansed by tap water/chemical solutions (0.7% sodium hydroxide; 2% sodium hypochlorite; 12 h), respectively.
Results of the preliminary experiments have demonstrated that the phage concentration in the influent tank was kept constant for at least 24 h after seeding. Moreover, microorganisms in activated sludge did not interfere with the enumeration of the phage plaque because views were finished before they could grow obviously.
In all practical membrane filtration applications, as the resistance increases the flux will decline. The following equation can be used to describe the overall characteristics of membrane fouling:
| (5) |
where R (m−1) is the filtration resistance of the membrane, ΔP (kPa) is the transmembrane pressure across the membrane, μ (mPa s) is the absolute viscosity of water, and J (m d−1) is the permeate flux.
3. Results
3.1. Removal efficiency of SMBR for COD, NH4+–N and SS
The COD, NH4 +–N and SS of treated wastewater by SMBR were 26.2 ± 10.6 mg l−1, 2.5 ± 1.0 mg l−1 and not detectable, respectively. It is clear that the SMBR could ensure a very low and stable effluent COD, NH4 +–N and SS. Replacement of secondary sedimentation tank by membrane unit makes very low or zero SS possible for SMBR. Previous studies showed that pathogenic bacteria and viruses were usually adsorbed onto the surfaces of suspended solids regardless of the surface properties, which makes them more stable [2], [13], [16].Therefore, SMBR has superiority in phage removal over other techniques.
3.2. Overall removal efficiency of the two membrane modules for T4
According to AFM image, the average size of T4 was about 107.9 ± 12.9 nm (see Fig. 2 ). Two membrane modules with different pore sizes (0.22 μm and 0.1 μm), one larger and the other smaller than T4 size, were used in this study to discuss the relationship between the membrane pore size and virus removal performance. For 0.22 μm membrane module, 2 log concentration of T4 was detected in the effluent at the beginning (see Fig. 3B, Stage a). The results indicated that the cake/gel layer formed on the 0.22 μm membrane surface was not adequate to remove all unexpected phages surged into feed wastewater. But, as time past, this value decreased gradually and stabilized at 0.2–0.3 log till day 50. It was inferred that cake layer formed on the membrane surface further reduced the effective pore size. The SEM images of the membrane surface show the significant difference between a new membrane and a long-term used one (Fig. 4A and B). It is clear that the used membrane was covered with bacteria and biopolymers, making it difficult for the phages to pass through.
Fig. 2.
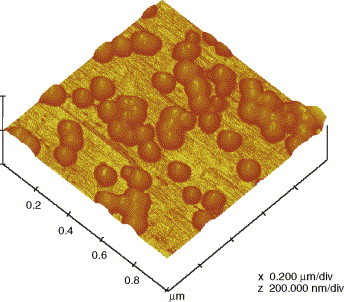
AFM image of the phage T4 added into wastewater.
Fig. 4.
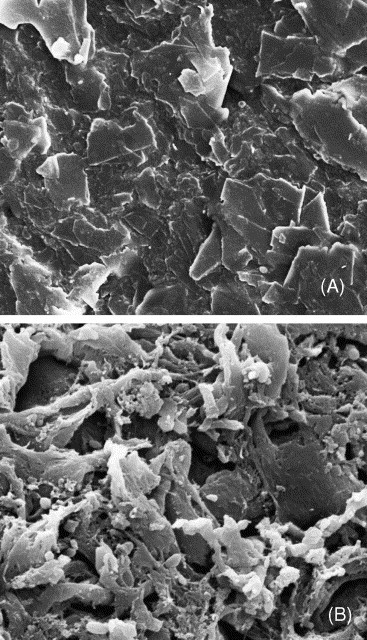
SEM images of membrane surface (PVDF, 0.22 μm). (A) The surface of a new membrane, ×10000; (B) The surface of the membrane after long-term SMBR operation, ×10000).
In comparison, as for 0.1 μm membrane, T4 was not detectable in the effluent from the beginning to day 65 (Fig. 3C, Stages a and b).This phenomenon implied that by selecting a membrane with a mean pore size slightly smaller than that of the target virus, complete viral removal could be expected in the well-running SMBR.
3.3. Variation of T4 concentrations in bulk solution
Variations of phage concentrations of influent and bulk solution from day 54 to 68 (Fig. 3, Stage b) were presented in Fig. 5 . During this period, no sludge was discharged and C b remained at almost a constant value (6.3 log). It was clear that there was no accumulation of phage in bulk solution under the experimental conditions in spite of continuous injection of T4 into the reactor. According to statistic calculation, about 11.0 log phages were fed into the system during this period without excess sludge discharge, which would at least allow C b value to reach 9.9 log. So it can be speculated that about 3.6 log of phages were possibly removed inside SMBR compartment everyday. Two main factors, inactivation by activated sludge (r AS) and adsorption by cake/gel layer might account for it. Inactivation by extra cellular enzyme, phagocytosis by bacteria and protozoans, and lysis in activated sludge might lead to loss of phages in bulk solution. During the aeration, phages collided with the cake/gel layer and some were adsorbed by it. However, the contribution of each mechanism to phage removal is required further study.
Fig. 5.
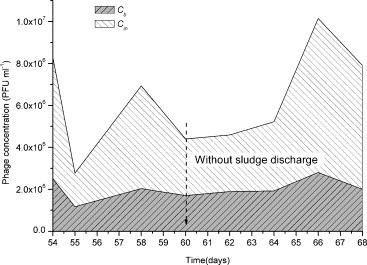
Comparison of phage concentrations of influent and bulk solution.
3.4. Phage removal mechanisms of SMBR
In order to investigate the role of cake/gel layer as well as membrane pore size in phage removal, these two modules were taken out on day 68 and washed with tap water to destruct the cake layer (Fig. 3, Stage c). Five days later, chemical cleansing was carried out for the removal of the gel layer on the membrane surface (Fig. 3, Stage d).As shown in Fig. 3B and C, when the cake layer was present, almost all phages (about 6.3 log) in bulk solution were intercepted by both membrane modules. When the cake layer was removed but the gel one were present, the phage removal efficiency of the 0.22 μm and 0.1 μm membranes were 3.1 log (r c 0.22) and 5.8 log (r c 0.1), respectively under an influent phage concentration of 5.8 log. When the gel layer was removed by chemical cleansing, the phage removal rates for the two modules were 1.7 log (r m 0.22) and 5.8 log (r m 0.1) for corresponding membrane modules. It can be concluded that the cake layer and gel layer play a significant role on the phage removal for the 0.22 μm membrane module. If cake/gel layer was decomposed by any factors, 2.7–4.1 log of phage would permeate to effluent. For the 0.1 μm module, on the other hand, membrane alone could block most phages from leaking by direct membrane interception. 1–2 PFU ml−1 of phage leakage is likely because that the pore size is not absolutely uniform. Consequently, the formation of cake layer and gel layer on 0.1 μm membrane would permit a safer disinfection effect.
Fig. 3D and E shows the variations of the filtration resistance of the two modules. The filtration resistance decreased with the removal of cake and gel layers through hydraulic and chemical cleansing on the membrane. For each membrane module, there was a positive correlation between the filtration resistance and the phage removal, which was more evident for the 0.22 μm membrane.
4. Discussion
In most of the previous studies, model viruses with a diameter smaller than the pore size of membrane have been used for investigating virus removal mechanisms [10], [11], [14]. Urase et al. [11] investigated the membrane interception performance using a new membrane system by batch experiments. Results showed that there was still 3–4 log of phages in the effluent which was likely due to the loose cake/gel layer formed on membrane since the experimental period was quite short. Chiemchaisri et al. [10] referred to phage removal during organic stabilization and nitrogen removal in an MBR system, but little information on mechanism was discussed. In this paper, viral removal in an MBR equipped with two different membranes (pore size: 0.22 μm and 0.1 μm, respectively), one larger and the other smaller than phage T4 size, was evaluated under different operational stages. Although both of the two membranes demonstrated almost complete removal of phage T4 during stable operation, phage removal mechanisms for the two membranes were quite different. For the 0.1 μm membrane, direct interception by membrane played a key role for T4 removal. For the 0.22 μm membrane, on the other hand, the cake/gel layer formed on the membrane played an important role.
So far, limited information was reported on the accumulation of phage in bulk solution [14]. By measuring the phage concentration of mixed liquor rather than supernatant of bulk solution, we demonstrated that there was no phage accumulation occurred in bulk solution. It was thought that inactivation by activated sludge and adsorption by cake/gel layer might contribute to the phage removal in bulk solution.
In conclusion, data in this study showed that the well-running SMBR had a good ability to intercept/inactivate the virus that surged suddenly into municipal wastewater. In order to preclude any virus leakage, the membrane module, with a pore size slightly smaller than diameter of the target virus, would be a safe option. This is easy to achieve. However, how to cope with wastewater when different types of viruses coexist, and how to dispose the virus-bearing excess sludge remain to be studied further.
Acknowledgments
This research was supported by funds (no. 20510076, 20477057) from the National Science Foundation of China.
References
- 1.Koivunena J., Siitonenb A., Heinonen-Tanski H. Elimination of enteric bacteria in biological–chemical wastewater treatment and tertiary filtration units. Water Res. 2003;37:690–698. doi: 10.1016/s0043-1354(02)00305-6. [DOI] [PubMed] [Google Scholar]
- 2.Chitnis V., Chitnis S., Vaidya K., Ravikant S., Patil S., Chitnis D.S. Bacterial population changes in hospital effluent treatment plant in central India. Water Res. 2004;38:441–447. doi: 10.1016/j.watres.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 3.Reinthaler F.F., Posch J., Feierl G., Wust G., Haas D., Ruckenbauer G. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003;37:1685–1690. doi: 10.1016/S0043-1354(02)00569-9. [DOI] [PubMed] [Google Scholar]
- 4.Monarca S., Feretti D., Collivignarelli C., Guzzella L., Zerbini I., Bertanza G. The influence of different disinfections on mutagenicity and toxicity of urban wastewater. Water Res. 2000;34(17):4261–4269. [Google Scholar]
- 5.Sehulster L.M., Hollinger F.B., Dreesman G.R., Melnick J.L. Immunological and biophysical alternation of hepatitis B virus antigen by sodium hypochlorite disinfection. Appl Environ Microbiol. 1981;42:762–767. doi: 10.1128/aem.42.5.762-767.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephenson T., Judd S., Jefferson B., Brindle K. IWA Publishing; Alliance House (London): 2000. Membrane bioreactors for wastewater treatment. [Google Scholar]
- 7.Yeom I.-T., Nah Y.-M., Ahn K.-H. Treatment of household wastewater using an intermittently aerated membrane bioreactor. Desalination. 1999;124:193–204. [Google Scholar]
- 8.Wen X., Ding H., Huang X., Liu R. Treatment of hospital wastewater using a submerged membrane bioreactor. Process Biochem. 2004;39(11):1427–1431. [Google Scholar]
- 9.Till S.W., Judd S.J., Mcloughlin B. Reduction of faecal coliform bacteria in sewage effluent using a microporous polymeric membrane. Water Res. 1998;32(5):1417–1422. [Google Scholar]
- 10.Chiemchaisri C, Wong YK, Urase T, Yamamoto K. Organic stabilization and nitrogen removal in membrane separation bioreactor for domestic wastewater treatment. Water Sci Tech 1992;25:(10)231–40. [PubMed]
- 11.Urase T., Yamamoto K., Ohgaki S. Evaluation of virus removal in membrane separation processes using coliphage Qb. Water Sci Tech. 1993;28(7):9–15. [Google Scholar]
- 12.Kawamura K., Nishimura K., Magara Y. Coliphage rejection under ultramembrane filtration. Desalination. 1996;106:89–97. [Google Scholar]
- 13.Otaki M., Yano K., Ohgaki S. Virus removal in a membrane separation process. Water Sci Tech. 1998;37(10):107–116. [Google Scholar]
- 14.Ueda T., Horan N.J. Fate of indigenous bacteriophage in a membrane bioreactor. Water Res. 2000;34(7):2151–2159. [Google Scholar]
- 15.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 16.Sakoda A., Sakai Y., Hayakawa K., Suzuki M. Adsorption of viruses in water environment onto solid surfaces. Water Sci Technol. 1997;35(7):107–114. [Google Scholar]
- 17.Committee of State of Environmental Protection Agency. Water and wastewater monitoring and analysis, 4th ed. China Construction Industry Press, china, 1998 (in Chinese).
- 18.Adams M.H. Interscience Publishers, Inc.; New York: 1959. Bacteriophages. p. 443–451. [Google Scholar]


