Abstract
Background
Human bocavirus (HBoV) is a parvovirus that has been recently detected in patients with respiratory illness.
Objectives
We developed a sensitive, specific, and quantitative real-time PCR assay based on the TaqMan method for HBoV detection and quantification in respiratory specimens.
Study design
Three individual real-time PCR assays were designed to amplify HBoV NS1, NP-1, and VP1 genes. For clinical evaluation, 506 nasal aspirates obtained from patients with acute respiratory tract infections during December 2006 to May 2007 were tested.
Results
Each assay had a broad dynamic range (50 × 107 to 5 × 107 copies of plasmid DNA) and high inter- and intra-assay reproducibility. The detection limit of each assay was 10 genome copies per reaction, and no crossreactivity with other major respiratory viruses or bacteria was detected. Clinical evaluation revealed that 11 (2.1%) of 506 patients diagnosed with upper respiratory tract infections, pneumonia, bronchitis, pharyngitis, or sinusitis had HBoV detected by all three assays, with viral loads ranging from 8.2 × 104 to 8.1 × 109 copies/ml of specimen.
Conclusions
The three assays for HBoV diagnosis and quantification are highly sensitive, specific real-time tools for the reliable epidemiological and pathogenetic study of HBoV infection.
Abbreviations: HBoV, human bocavirus; LRTIs, lower respiratory track infections; PCR, polymerase chain reaction; MGB, minor groove binder; FAM, 6-carboxyfluorescein; UNG, uracil-N-glycosylase; ORFs, open reading frames; PIV, parainfluenza virus; RSV, respiratory syncytial virus
Keywords: Human bocavirus, Respiratory diseases, Real-time PCR, Quantification
1. Introduction
In 2005, using random amplification methods, human bocavirus (HBoV) was identified from pooled respiratory specimens of children who had lower respiratory tract infections (Allander et al., 2005).
HBoV is classified into the family Parvoviridae, subfamily Parvovirinae, and genus Bocavirus. Recently, after the initial detection of HBoV in respiratory secretions, several prevalence studies in children with respiratory tract infection reported a worldwide prevalence ranging from 1.5% to 10.3% (Arnold et al., 2006, Bastien et al., 2006, Choi et al., 2006, Kesebir et al., 2006, Ma et al., 2006, Sloots et al., 2006, Weissbrich et al., 2006).
At present, the applicable identification methods for HBoV are conventional and real-time PCR, because HBoV culture and serological methods have not yet been established. In general, real-time PCR has the advantage that amplification and analysis are completed in a closed system, so it is less prone to contamination, and it is more sensitive and specific than conventional PCR, can quantify gene copy numbers, and a large number of samples can be processed in a single step without laborious postanalysis (Espy et al., 2006). A previous study introduced a real-time PCR method that targeted NS1 and NP-1 genes with high sensitivity and specificity (Lu et al., 2006).
In this article, we present a highly sensitive and specific TaqMan-based real-time PCR assay for the rapid detection and quantitation of HBoV in clinical specimens.
2. Methods
2.1. Design of primers and probes
The full sequence of HBoV was retrieved from the ST1 strain (GenBank accession no. DQ000495) to design primers and probes. HBoV NS1, NP-1, and VP1 genes were analyzed for target sites that were applicable to TaqMan real-time PCR requirements, using Primer Express software, version 2.0 (Applied Biosystems, Foster City, CA). The TaqMan MGB probes were labeled with FAM at the 5′ end and with Black Hole Quencher at the 3′ end (Table 1 ).
Table 1.
Primers and probes used in real-time PCR assay for HBoV
| Gene | Primer–probe | Sequence (5′–3′) | Positiona |
|---|---|---|---|
| NS1 | Forward | TGA ACC TGA AGA GGC TGA CAT ATT TC | 656–681 |
| Reverse | CAT GTT GCC GCC AGT AAC TC | 718–737 | |
| Probe | CAC CCC ATG CCT CTC G | 702–717 | |
| NP-1 | Forward | CAG CCA CCT ATC GTC TTG CA | 2513–2532 |
| Reverse | CCC TCG TCT TCA TCA CTT GGT | 2551–2571 | |
| Probe | CTG CTT CGA AGA CCT C | 2533–2548 | |
| VP1 | Forward | AGC TGT CAC TTC TCA CCA CAA G | 3679–3700 |
| Reverse | TTG CTT TAG GTC TGA AGC GCT TAT | 3725–3748 | |
| Probe | ATT GGC AGC GCC TTA C | 3701–3716 | |
Nucleotide position was designated according to HBoV strain ST1 (GenBank accession no. DQ000495)
2.2. Construction of the plasmid DNA standard
About 5.2 kbp of the HBoV full genome was amplified with forward primer (5′-GGATTCCAAGATGGCGTCTGT-3′) and reverse primer (5′-ACACATTAAAAGATATAGAGTTTC-3′) from a PCR-positive specimen. The product was cloned into plasmid vector pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and verified by sequencing. The pCR2.1-HBoV plasmid was purified with a QIamp mini prep kit (Qiagen, Hilden, Germany) and quantified using UV spectroscopy (2.5 × 107 DNA copies/μl). For standard curve generation, serial 10-fold dilutions of the pCR2.1-HBoV plasmid were prepared in 10 mM Tris–EDTA buffer (pH 8.0) and stored at −20 °C.
2.3. Real-time assay for human bocavirus
The real-time PCR assay was performed in a 20-μl reaction mixture containing 2 μl extracted DNA or standard plasmid, 10 μl TaqMan Universal PCR Master Mix containing ROX as a passive reference dye (Applied Biosystems), 900 nM each of forward and reverse primer, and 250 nM probe. Amplification and detection were performed in an ABI Prism 7900HT sequence detection system (Applied Biosystems) under the following conditions: uracil-N-glycosylase (UNG) was activated at 50 °C for 2 min, followed by PCR activation at 95 °C for 10 min and 40 cycles of amplification (15 s at 95 °C and 1 min at 60 °C). Analysis of each assay was performed using Sequence Detector software version 2.1 (Applied Biosystems). The sensitivity of the assays was determined by using 10, 5 and 1 copies of pCR2.1-HBoV plasmid per reaction. For the specificity test, genomes from major causative agents of respiratory illness including DNA/RNA viruses and bacteria were used (Table 2 ). Each intra- and inter-assay was performed in triplicate on three different days to evaluate reproducibility.
Table 2.
List of human pathogens used for real-time PCR specificity test
| Viruses and bacteriaa | Sourceb | Titer |
|---|---|---|
| RNA viruses | ||
| Influenza virus A | 2 × 107 TCID50/ml | |
| Influenza virus B | 4.2 × 105 TCID50 | |
| PIV1/2/3 | >5 × 105 TCID50/ml | |
| RSV | ATCC VR-26 | 4 × 107 TCID50/ml |
| HCoV 229E | ATCC VR-740 | 3 × 104 TCID50/ml |
| HCoV OC43 | ATCC VR-1558 | 4 × 105 TCID50/ml |
| Rhinovirus 13 | ATCC VR-1123 | 106 TCID50/ml |
| Enterovirus | 5.2 × 105 TCID50/ml | |
| DNA viruses | ||
| Parvovirus B19 | 104 copies/ml | |
| Adenovirus 11 | 5 × 105 pfu/ml | |
| HSV-2 | ATCC VR-540 | 2.5 × 104 TCID50/ml |
| HHV-6 | ATCC VR-1480 | 3 × 104 TCID50/ml |
| VZV | ATCC VR-1503 | 4.5 × 104 TCID50/ml |
| Bacteria | ||
| Haemophilus influenzae | 2 × 105 CFU | |
| Legionella pneumophila | 3 × 105 CFU | |
| Streptococcus pneumoniae | 1.5 × 105 CFU | |
| Mycoplasma pneumoniae | 3 × 106 CFU | |
| Chlamydia pneumoniae | 103 IFU/ml | |
HSV-2, herpes simplex virus 2; HHV-6, human herpesvirus 6; VZV, varicella-zoster virus; PIV1/2/3, parainfluenza virus 1/2/3; RSV, respiratory syncytial virus; HCoV, human coronavirus.
These bacteria were provided by Division of Bacterial Respiratory Infections, Center for Infectious Diseases, Korea National Institute of Health.
If not specified, strains are clinical isolates.
2.4. Conventional PCR for human bocavirus
We compared the sensitivity of real-time PCR with conventional PCR performed by amplifying a 346 bp fragment of the NS1 gene (640–986 bp) using the following primer set: F3 (5′-TGGCTACACGTCCTTTTGAACC-3′) and R2 (5′-GACTTCGTTATCTAGGGTTGCG-3′). The PCR assays were performed in a 50-μl reaction volume containing 4 μl DNA extract, 5 μl 10× PCR buffer, 2 μl dNTP mix (final concentration of 400 μM of each dNTP), 1 μl SP Taq (Cosmo Gentec, Seoul, Korea), 1 μl of each primer (10 pM), and nuclease-free water to 50 μl. The PCR reaction was carried out at 95 °C for 15 min, 35 cycles of amplification (30 s at 94 °C; 30 s at 55 °C; 30 s at 72 °C), and a final extension step at 72 °C for 10 min. The PCR products were run on a 2% agarose gel, stained with SYBR safe DNA gel stain dye (Invitrogen) and visualized under UV light.
2.5. Clinical specimens
From December 2006 to May 2007, 506 nasal aspirates were collected from patients with acute respiratory infections who visited local clinics; their median age was 36 months. Specimen collection and study were carried out as part of acute respiratory tract infection surveillance by the Korea Center for Disease Control (KCDC). Informed consent was obtained from the children's parents and patients. The age distribution of the 506 patients was 83 (16.4%) <12 months; 252 (48.8%) 1–5 years; 48 (9.5%) 6–9 years; 37 (7.3%) 10–19 years; 86 (17%) > 20 years. The male to female ratio was 1.0. The diagnoses of the patients were upper respiratory infection in 154; pharyngitis in 112; bronchitis in 49; croup in 3; pneumonia in 36; otitis media in 43; sinusitis in 109. After specimen collection, total nucleic acid was extracted from 200 μl aliquots of each specimen within 24 h using an automated MagNa pure LC total nucleic acid extraction kit (Roche Diagnostics, Mannheim, Germany). All specimens were tested for common respiratory viruses (influenza viruses A/B, RSV, PIV 1/2/3, coronaviruses, rhinovirus, and adenoviruses) using a modified version of a previously described multiplex PCR and reverse transcription PCR (Na et al., 2002, Paul et al., 2004). The rest of the specimens were kept at −70 °C.
3. Results
3.1. Real-time PCR standard curve and dynamic range
For quantification of the HBoV, full genome size of HBoV plasmid DNA was used to generate a standard curve. Serial 10-fold dilutions spanning from 50 × 107 to 5 × 107 copies of plasmids were used per reaction mixture and each reaction was performed in triplicate. The standard curves generated in each assay showed amplification efficiencies greater than 89% (NS1, 96.8%; NP-1, 90.5%; VP1, 89.5%) and linear regressions better than 0.99 in all three assays. The dynamic range of the assays was analyzed by means of C T values in 7 log units with corresponding C T values.
3.2. Real-time PCR detection limit, specificity, and reproducibility
The detection limit of the assays was determined using 12 replicates of each primer–probe set with 10, 5 and 1 copies of plasmid DNA. Ten copies resulted in a detection rate of 100% (NP-1), 100% (NS1), and 100% (VP1). At five copies the analytical detection limit was decreased to 75% (NP-1), 75% (NS1), and 25% (VP1). At the lowest level of one copy template, NP-1 gene detection (53.3%) was higher than the other target genes, NS1 (33.3%) and VP1 (6.7%).
The crossreactivity with other DNA viruses such as human parvovirus B19, adenovirus 11, herpes simplex virus 2 and human herpes virus 6, varicella-zoster virus, and respiratory pathogenic bacteria, including Haemophilus influenzae, Legionella pneumophila, Streptococcus pneumoniae, Mycoplasma pneumoniae, and Chlamydia pneumoniae, was assessed by applying each of the three assays to 200 ng of DNA from each species. In addition, the cDNA of respiratory RNA viruses (influenza virus A/B, parainfluenza viruses 1/2/3, RSV, coronavirus 229E/OC43, rhinovirus 13, and enterovirus) was evaluated to exclude possible amplification by reverse transcription. No nonspecific amplification was detected in these samples by any of the three assays.
The intra- and inter-assay reproducibility was assessed using 10-fold serial dilutions of standard HBoV plasmid DNA ranging from 50 × 107 to 5 × 107 copies in triplicate on three different days. Intra-assay variation ranged from 0.03% to 1.12% and inter-assay variation from 0.03% to 3.43% and showed high assay reproducibility and precision (Table 3 ).
Table 3.
Intra- and inter-assay reproducibility of the HBoV real-time PCRa
| Gene | DNA copies | Intra-assayb |
Inter-assayc |
||||
|---|---|---|---|---|---|---|---|
| Mean CT | S.D. | CV (%) | Mean CT | S.D. | CV (%) | ||
| NS1 | 5 × 107 | 13.4 | 0.02 | 0.13 | 13.42 | 0.15 | 1.0 |
| 5 × 106 | 17.31 | 0.02 | 0.10 | 17.41 | 0.36 | 2.05 | |
| 5 × 105 | 21.08 | 0.02 | 0.11 | 21.14 | 0.20 | 0.94 | |
| 5 × 104 | 24.5 | 0.04 | 0.15 | 24.55 | 0.14 | 0.56 | |
| 5 × 103 | 28.11 | 0.04 | 0.14 | 38.24 | 0.26 | 0.93 | |
| 5 × 102 | 31.77 | 0.26 | 0.81 | 31.52 | 0.06 | 0.20 | |
| 5 × 10 | 35.48 | 0.40 | 1.12 | 35.47 | 0.34 | 0.95 | |
| NP-1 | 5 × 107 | 13.7 | 0.06 | 0.46 | 14.00 | 0.48 | 3.42 |
| 5 × 106 | 17.65 | 0.04 | 0.25 | 18.16 | 0.62 | 3.43 | |
| 5 × 105 | 21.52 | 0.02 | 0.10 | 21.94 | 0.49 | 2.23 | |
| 5 × 104 | 25.07 | 0.01 | 0.03 | 25.44 | 0.44 | 1.72 | |
| 5 × 103 | 28.78 | 0.06 | 0.19 | 29.16 | 0.53 | 1.81 | |
| 5 × 102 | 32.48 | 0.24 | 0.73 | 32.46 | 0.40 | 1.23 | |
| 5 × 10 | 36.51 | 0.36 | 0.98 | 36.90 | 0.01 | 0.02 | |
| VP1 | 5 × 107 | 13.83 | 0.03 | 0.23 | 13.82 | 0.19 | 1.38 |
| 5 × 106 | 17.58 | 0.03 | 0.16 | 17.67 | 0.26 | 1.47 | |
| 5 × 105 | 21.43 | 0.21 | 0.09 | 21.36 | 0.19 | 0.89 | |
| 5 × 104 | 24.82 | 0.03 | 0.12 | 24.73 | 0.19 | 0.78 | |
| 5 × 103 | 28.39 | 0.11 | 0.39 | 28.38 | 0.24 | 0.85 | |
| 5 × 102 | 31.97 | 0.14 | 0.44 | 31.69 | 0.20 | 0.64 | |
| 5 × 10 | 35.82 | 0.20 | 0.55 | 35.94 | 0.01 | 0.03 | |
Tenfold serial dilutions of HBoV plasmid DNA were assayed in triplicate and on three different days. The mean values of CT, standard deviations (S.D.s) and coefficient of variations (CVs) were calculated.
Each value was determined from three replicates within a assay.
Assays were performed three times on different days in an independent manner.
3.3. Comparison of real-time PCR and conventional PCR assay
To assess the sensitivity of real-time PCR compared with conventional PCR, 10-fold serial dilutions of standard HBoV plasmid DNA ranging from 1 × 102 to 1 × 108 copies as a template were used. The detection limit of the gel-based conventional PCR assay that amplifies 346-bp of the NS1 gene was 1000 copies of HBoV plasmid DNA (Fig. 1 ). On the other hand, as shown above, the detection limit of three real-time PCR assays obtained from plasmid DNA samples was as low as 10 copies. The sensitivity of the real-time PCR was equivalent to an increase of two log units over the conventional PCR assay.
Fig. 1.
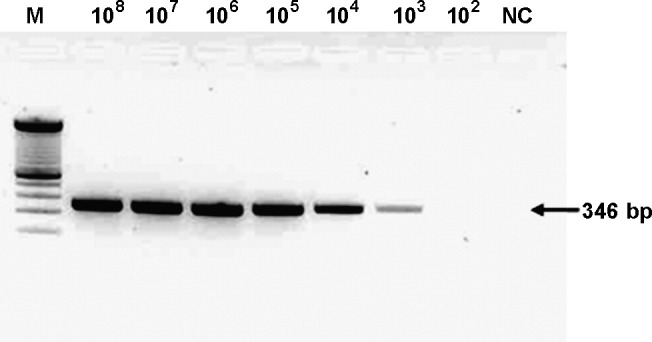
Sensitivity of the conventional PCR assay for HBoV NS1 gene. Lane M, 100 bp size marker (Invitrogen); lanes 1–7, 10-fold serial dilutions of HBoV plasmid standard; lane 8, negative control. HBoV plasmid standard DNA (2.5 × 107 DNA copies/μl) 4 μl was subjected on each assay.
3.4. Detection of HBoV in clinical specimens
For clinical evaluation, the three newly developed real-time PCR assays were used to detect HBoV in nasal aspirates collected from 506 patients with acute respiratory tract infections. Among them, 11 (2.1%) specimens were positive for three targets, three were positive for two targets (NS1 and VP1), and three were positive for only a single target (two for NP-1 and one for VP1). Positive criteria defined that all three primer–probe sets were positives with more than 10 copies of each target. RT-PCR results for other common respiratory viruses indicated that 80 (15.8%) specimens were positive for rhinovirus, 42 (8.3%) for influenza virus A, 19 (3.7%) for RSV, 4 (0.8%) for adenovirus, 2 (0.4%) for influenza virus B, 1 (0.2%) for parainfluenza virus, and 1 (0.2%) for coronavirus OC43. Among 11 patients who gave HBoV-positive specimens, two patients were coinfected with RSV and one patient with rhinovirus. Interestingly, one sample, which was not positive by conventional PCR, contained a mean of 8.2 × 104 copies/ml of HBoV in real-time PCR assays. Considering all positive assays, the clinical specimen detection limit was around 10 genome copies.
The clinical characteristics of positive patients (eight infants and two adults) who were diagnosed with upper respiratory infection, bronchitis, pneumonia, pharyngitis, or sinusitis are described in Table 4 .
Table 4.
Characteristics of 11 HBoV real-time PCR-positive patients
| Patients | Age | Sex | Symptom(s) | Diagnosis | Viral loada |
|---|---|---|---|---|---|
| KNIH0027 | 9 months | Male | Cough, congestion, rhinorrhea | URTIb | 5.8 × 108 |
| KNIH1068 | 25 year | Male | Rhinorrhea, sore throat | URTI | 8.2 × 104 |
| KNIH2673 | 47 years | Female | Rhinorrhea | URTI | 1.5 × 107 |
| KNIH3345 | 1 year | Male | Fever, cough, congestion, rhinorrhea | Pharyngitis | 4.9 × 106 |
| KNIH3380 | 1 year | Male | Cough, rhinorrhea | Bronchitis | 3 × 107 |
| KNIH3550 | 1 year | Female | Fever, cough, rhinorrhea, sputum | Pneumonia | 4.7 × 107 |
| KNIH3631 | 1 year | Female | Cough, congestion, rhinorrhea | Sinusitis | 1.4 × 105 |
| KNIH3755 | 11 months | Male | Cough, rhinorrhea | URTI | 1.2 × 109 |
| KNIH3850 | 1 year | Female | Fever, cough, rhinorrhea | Pharyngitis | 2.3 × 108 |
| KNIH3996 | 8 months | Female | Fever, rhinorrhea | Bronchitis | 8.1 × 109 |
| KNIH4392 | 1 year | Female | Fever, cough | Bronchitis | 1.6 × 106 |
Mean viral load of per milliliter of specimen in three primer–probe sets calculated from three replicated tests.
Upper respiratory track infection.
4. Discussion
Our quantitative real-time PCR assays provide sensitive and specific diagnostic tools targeting three putative ORFs and allow quantification of HBoV in clinical specimens. Each assay was performed over a wide dynamic range with low intra- and inter-assay variation, and showed no crossreactivity with other respiratory viruses or bacteria. The sensitivity of the three assays was significantly higher than conventional PCR assay.
To define positive criteria that minimized false positives, we tested clinical specimens in triplicate, and defined positive results as those where all three independent assays (NS1, NP-1, and VP1) were positive. For practical laboratory application, however, the assay targeting NS1 gene seems to be the most efficient and reliable because the NS1 gene is highly conserved.
As in a previous report, we were careful to prevent possible contamination of the PCR, so preparation of the PCR premix, control plasmid DNA, and clinical sample processing were performed in a separate room (Lu et al., 2006). Moreover, the real-time PCR is performed in a closed system, and UNG was added to the PCR master mix as another level of control for amplicon contamination (Espy et al., 2006). However, we could not exclude other factors that might lead to false positive results, such as cross-contamination during specimen handling and nucleic acid extraction.
As mentioned above, applicability of these assays for clinical diagnosis was estimated using respiratory specimens from patients with acute respiratory tract infections, and a broad range of HBoV genome copy number was detected. Interestingly, one specimen with a lower viral load among these positives was missed by conventional PCR, reflecting the sensitivity of our real-time assays. Moreover, to the best of our knowledge, this is the first report to identify HBoV infection in adults in Korea. Previously, Lu et al. (2006) reported that HBoV PCR-positive patients with pneumonia had a relatively low viral load. Although more clinical data should be required to interpret correlation between pathogenesis and the viral load, the real-time assays described in this study would provide useful tools for analysis of clinical and molecular epidemiological investigation of HBoV infection.
Acknowledgment
This work was supported by an intramural research fund from the National Institute of Health, Korea.
References
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.C., Singh K.K., Spector S.A., Sawyer M.H. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin Infect Dis. 2006;43:283–288. doi: 10.1086/505399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N., Brandt K., Dust K., Ward D., Li Y. Human bocavirus infection. Can Emerg Infect Dis. 2006;12:848–850. doi: 10.3201/eid1205.051424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.H., Lee H.J., Kim S.J., Eun B.W., Kim N.H., Lee J.A. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clin Infect Dis. 2006;43:585–592. doi: 10.1086/506350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesebir D., Vazquez M., Weibel C., Shapiro E.D., Ferguson D., Landry M.L. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006;194:1276–1282. doi: 10.1086/508213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Chittaganpitch M., Olsen S.J., Mackay I.M., Sloots T.P., Fry A.M. Real-time PCR assays for detection of bocavirus in human specimens. J Clin Microbiol. 2006;44:3231–3235. doi: 10.1128/JCM.00889-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Endo R., Ishiguro N., Ebihara T., Ishiko H., Ariga T. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44:1132–1134. doi: 10.1128/JCM.44.3.1132-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na B.K., Kim J.H., Shin G.C., Lee J.Y., Lee J.S., Kang C. Detection and typing of respiratory adenoviruses in a single-tube multiplex polymerase chain reaction. J Med Virol. 2002;66:512–517. doi: 10.1002/jmv.2174. [DOI] [PubMed] [Google Scholar]
- Paul G., Afina S.G., Mirjam D., Willem B.V., Jan-Willem P., Sylvia M.B. Practical implementation of a multiplex PCR for acute respiratory tract infections in children. J Clin Microbiol. 2004;42:5596–5603. doi: 10.1128/JCM.42.12.5596-5603.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots T.P., McErlean P., Speicher D.J., Arden K.E., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrich B., Neske F., Schubert J., Tollmann F., Blath K., Blessing K. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect Dis. 2006;6:109. doi: 10.1186/1471-2334-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]


