Abstract
The recognition of nucleic acids is a general strategy used by the host to detect invading pathogens. Many studies have established that MITA/STING is a central component in the innate immune response to cytosolic DNA and RNA derived from pathogens. MITA can act both as a direct sensor of cyclic dinucleotides (CDNs) and as an adaptor for the recruitment of downstream signaling components. In both roles, MITA is part of signaling cascades that orchestrate innate immune defenses against various pathogens, including viruses, bacteria and parasites. Here, we highlight recent studies that have uncovered the molecular mechanisms of MITA-mediated signal transduction and regulation, and discuss some notable issues that remain elusive.
Keywords: MITA, cGAMP, Innate immune response, DNA sensor
1. Introduction
Recently, tremendous advances have been made in our understanding of the innate immune response to infectious pathogens. Host germ line-encoded pattern-recognition receptors (PRRs) of the innate immune system recognize pathogen-associated molecular patterns (PAMPs) generated by invading pathogens, such as lipids, lipoproteins, proteins and nucleic acids. Among these PAMPs, recognition of pathogen-derived nucleic acids is a general strategy used by host cells to detect infectious agents, a subject of intense study in past decades.
RNA viruses produce RNA during the viral life cycle that can be recognized by the host as danger signal to trigger innate immune responses. Viral RNAs are typically recognized by two classes of PRRs, membrane-bound Toll-like receptors (TLRs) and cytosolic RIG-I-like receptors (RLRs). While TLRs such as TLR3 recognize viral RNA in the endosome of certain immune cells, RLRs, including RIG-I and MDA5, are essential for the recognition of cytosolic viral RNA in most cell types [1]. Upon recognition of viral RNAs, RIG-I and MDA5 are recruited to the mitochondrial adaptor protein VISA (also known as MAVS, IPS-1, and Cardif) [2], [3], [4], [5], which triggers a series of signaling cascades that lead to the activation of transcription factors IRF3 and NF-κB. Activated IRF3 and NF-κB work synergistically to induce the production of type I interferons (IFNs) and proinflammatory cytokines, leading to innate antiviral responses.
The presence of DNA in endosome or the cytosol is also a danger signal for the innate immune system. These DNA molecules, including exogenous DNA derived from invading pathogens and endogenous inappropriately aggregated self-DNA, can be recognized by DNA sensing systems to initiate innate immune responses [6], [7], [8]. Compared to the well-studied RNA-induced innate immune responses, our understanding of DNA-triggered signaling is relatively limited. Exhilaratingly, the discovery of many DNA sensors and downstream adaptors, especially the discovery of MITA, has shed new light on cytosolic DNA-triggered signaling pathways. Using expression cloning, several groups independently identified MITA (Mediator of IRF3 Activation, also known as STING, MPYS, ERIS and TMEM173) as a critical mediator of the innate immune response to cytosolic nucleic acid ligands [9], [10], [11], [12]. Subsequently, a series of studies have established the essential roles of MITA in innate immune responses to DNA viruses [9], [13], [14], [15], some RNA viruses [9], [10], [11], [16], [17], retroviruses [18], [19], bacteria [9], [13], [20], [21], [22] and protozoan parasites [23]. Additionally, MITA has a central role in the pathogenesis of inflammatory and autoimmune diseases triggered by recognition of self-DNA that inappropriately accumulates in the cytoplasm [6], [24], which has been reviewed elsewhere [25]. Although most studies have focused on nucleic acid-triggered signaling, MITA has also been proposed to sense virus-cell membrane fusion events [26].
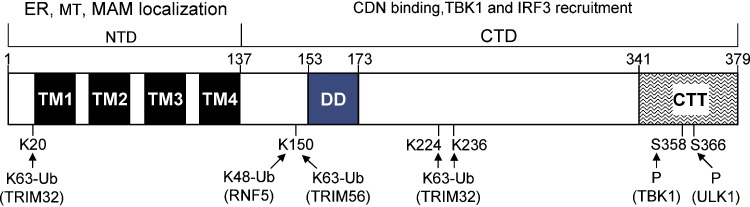
Human and murine MITA contain 379 and 378 amino acids, respectively, and share 81% similarity and 69% identity in sequence. Homologs in other species, including Sus scrofa, Bos Taurus, Rattus norvegicus, Xenopus, Drosophila, and Danio rerio, also exhibit high sequence similarity [11], [27], [28], [29]. MITA contains four transmembrane motifs in the N-terminus (aa1-137), which predominantly anchors itself in the endoplasmic reticulum (ER) and partially in the mitochondria and mitochondria-associated membrane (MAM) [9], [10], [11], [13]. The C-terminal domain (CTD, aa138-379) extends into the cytosol to bind the cytosolic CDNs and recruits downstream factors (Fig. 1 ). Among tissues surveyed, MITA showed high expression in the heart, spleen, peripheral leukocytes, placenta and lung, and moderate expression in the thymus, small intestines, liver and kidney, but almost undetectable expression in the brain, skeletal muscle and colon. Among transformed cell lines, MITA is highly expressed in THP-1, U937, L929 and Raw264.7 cells, making them sensitive to DNA stimulation, but is poorly expressed in HEK293T, HeLa, and Huh-7 cells [11], [30]. This expression pattern suggests that MITA might function in the immune system.
Fig. 1.
Schematic presentation of MITA structure. Human MITA contains four N-terminal transmembrane (TM) domains, which anchor itself in ER, mitochondria (MT) and mitochodria-associated membrane (MAM). Its C-terminal domain (CTD) hangs in the cytosol, which is responsible for CDN binding and recruitment of downstream components including TBK1 and IRF3. The CTD also contains a dimerization domain (DD) and a flexible C-terminal tail (CTT). MITA is post-translationally modified with K48- and K63-linked polyubiquitination by the indicated E3 ubiquitin ligases and with phosphorylation (P) by the indicated kinases.
MITA might act in two different ways: as a downstream adaptor of RNA and DNA sensors or by direct binding of CDNs secreted by bacteria or endogenously generated by the DNA sensor cGAS. Both pathways lead to the production of type I IFNs and proinflammatory cytokines. In this review, we summarize recent advances in the understanding of MITA-mediated signal transduction and regulation in response to cytosolic nucleic acids.
2. The MITA-mediated signaling pathways
2.1. MITA-mediated signaling in response to cytosolic DNA
Genetic evidence has established the requirement of MITA in type I IFN induction in innate immune response to cytosolic DNA and CDNs. The most important remaining questions were how DNA pathogens are recognized and how MITA links DNA sensing to downstream signaling. In the past years, great efforts have been made by many groups to identify DNA sensors, and several candidates are listed in Table 1 .
Table 1.
Mechanisms of cytosolic DNA sensing.
| Sensor | Ligand | Cell type | Signaling | Knockout confirmed | Reference |
|---|---|---|---|---|---|
| TLR9 | CpG DNA | pDCs | MyD88- IRF7 MyD88- NF-κB |
Essential | [36], [37] |
| DAI | dsDNA | Most cell types | TBK1-IRF3 RIP3 |
Not essential | [38], [60] |
| AIM2 | dsDNA | Most cell types | ASC-Caspase-1 Inflammasome |
Essential | [31], [32], [33], [34] |
| NLRP3 | dsDNA | Most cell types | ASC-Caspase-1 Inflammasome |
Essential | [35] |
| RNA Pol III | AT-rich dsDNA | Most cell types | RIG-I-VISA-IRF3 | ND | [39] |
| MITA | c-di-GMP, c-di-AMP cGAMP |
Most cell types | TBK1-IRF3 | Essential | [12], [30], [50], [51] |
| IFI16 | dsDNA, ssDNA | THP1, Raw264.7, MEF, BMDM | MITA-TBK1-IRF3 ASC-caspase-1 |
ND | [40] |
| DDX41 | dsDNA c-di-GMP |
mDCs BMDCs |
MITA-TBK1-IRF3 | ND | [15] |
| LSm14A | dsRNA, dsDNA | Most cell types | RIG-I-VISA MITA |
ND | [41] |
| cGAS | dsDNA RTI |
BMDM, DC Fibroblast |
cGAMP-MITA- TBK1-IRF3 |
Essential | [18], [44], [46], [47], [59] |
ND, not determined.
Among these DNA sensors, AIM2 and NALP3 initiate the ASC-caspase-1 signaling pathway, leading to inflammation but not type I IFN production [31], [32], [33], [34], [35]. TLR9 is known to recognize CpG-DNA in plasmacytoid dendritic cells (pDCs), and transduce signals through the adaptor protein MyD88 to induce type I IFNs and inflammatory cytokines [36], [37]. Genetic evidence suggests that DAI is not required for the sensing of cytosolic DNA in the cell types examined, including mouse embryonic fibroblasts (MEFs), bone marrow derived dendritic cells (BMDCs) and macrophages from DAI-deficient mice [38]. RNA polymerase III only responds to AT-rich dsDNA and signals through the RIG-I-mediated signaling pathway [39]. DDX41 was reported to sense cytosolic DNA in human myeloid dendritic cells (mDCs), BMDCs and monocytes. DDX41 binds to poly(dA:dT) or HSV-1 DNA via its DEADc domain, resulting in its interaction with MITA and activation of TBK1 [15]. Another reported DNA sensor, IFI16, can bind to both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA), leading to IFN production and inflammation through MITA- and ASC-mediated pathways, respectively [40]. LSm14A, a member of the LSm family involved in RNA processing in the processing bodies, has recently been demonstrated as a sensor of viral nucleic acids [41]. LSm14A can bind to poly(I:C), poly(dA:dT) and viral DNA. Depletion of LSm14A markedly reduces both SeV- and HSV-1-induced IFN-β, suggesting that LSm14A mediates innate immune responses to both RNA and DNA viruses [41]. It has also been reported that MITA can associate with both ssDNA and dsDNA, thereby directly acting as a DNA sensor [42]. However, none of these candidates seems to be a universally required DNA sensor for detecting viral DNA in distinct cell types or at the animal level.
Recently, a new DNA sensor, cGAS (cyclic GAMP synthase, also known as MB21D or C6orf150), was identified [43], [44], [45]. Expression analysis showed that the expression of murine cGAS was high in Raw264.7 cells and BMDMs, but low in immortalized MEFs, suggesting a role for cGAS in the immune system. Genetic studies suggested that cGAS is required for the responses to all the DNA or DNA viruses examined, including HA-DNA, E. coli DNA, poly(dA:dT), ISD, HAV-1 and VACA, to induce type I IFNs in primary fibroblasts, macrophages, and DCs [46]. Additionally, cGAS is required for innate immune control of DNA virus in mice, such as HSV-1, MHV68 and vaccinia virus [46], [47]. A recent study established cGAS as the dominant cytosolic DNA sensor responsible for the detection of internalized adenovirus [48]. Furthermore, cGAS is essential for retrovirus-triggered innate immune responses by sensing reverse-transcribed DNA [18]. Based on these advances, cGAS seems to be a general sensor of cytosolic DNA in most immune cells [47].
cGAS can bind to DNA in the cytoplasm and subsequently catalyze the synthesis of cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) from GTP and ATP. The intracellularly generated cGAMP is similar to the bacterial second messengers c-di-AMP and c-di-GMP, which were previously found to be potent inducers of innate immune responses [14], [49], [50]. Thus, cGAMP in metazoans functions as an endogenous second messenger that triggers IFN production in response to cytosolic DNA [30].
It has been demonstrated that MITA can directly bind to CDNs such as c-di-GMP, c-di-AMP and cGAMP [51], [52], [53], [54], [55], [56], [57], [58]. Additionally, c-di-GMP and c-di-AMP induce innate immune responses in a MITA-dependent manner [12], [14]. These findings suggest that, the cGAS-induced, cGAMP-mediated innate immune response might also require MITA. Indeed, overexpression of cGAS in HEK293T cells which naturally lacks MITA expression failed to induce IFN-β. Consistently, delivery of cGAMP failed to induce IFN-β in MITA-deficient cells, indicating an essential role for MITA in cGAS-induced innate immune responses [30], [44], [59].
Collectively, MITA is generally involved in many aspects of cytosolic DNA-triggered innate immune responses. First, MITA functions downstream of some essential ssDNA/dsDNA sensors such as DDX41 and IFI16 to induce type I IFNs. Second, MITA acts as a direct sensor for CDNs. Finally, the established DNA sensor cGAS, which is generally involved in the recognition of DNA from DNA viruses, bacteria, parasites, and retroviruses, initiates downstream signaling in a MITA-dependent manner.
In all these scenarios, activated MITA initiates signaling cascades leading to production of type I IFNs and proinflammatory cytokines. The mechanisms of these processes are illustrated in Fig. 2 and will be further discussed below. Briefly, MITA recruits both TBK1 and IRF3, which facilitates the phosphorylation and activation of IRF3 by TBK1, leading to the induction of type I IFNs. However, how MITA couples signaling to NF-κB activation remains unclear.
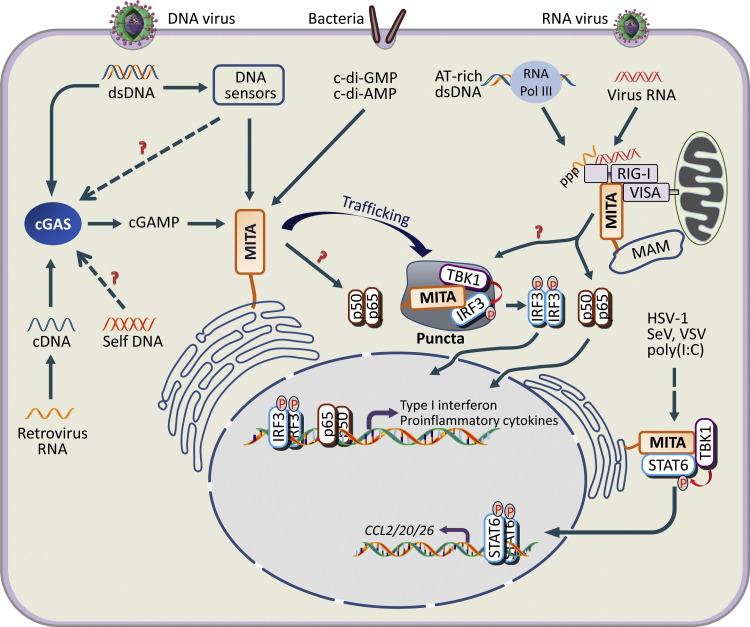
Fig. 2.
MITA-mediated signaling pathways in response to cytosolic nucleic acids. MITA is involved in both RNA and DNA pathogen-triggered signaling. MITA functions downstream of RIG-I and VISA in response to infection by some RNA viruses, leading to activation of the transcription factors IRF3 and NF-κB, which induces type I IFNs and inflammatory cytokines. It is likely that mitochondria- or MAM-associated MITA is involved in this process. DNA from pathogens such as DNA viruses, bacteria and parasites can be detected by cGAS, which synthesizes noncanonical cGAMP that subsequently binds to ER-localized MITA to initiate downstream signaling. Cytosolic DNA can also be detected by other DNA sensors to activate MITA, but whether cGAS is involved in these processes has not been determined. Bacteria can also trigger innate immune defense by secreting c-di-GMP and c-di-AMP that directly bind to MITA. Retroviruses generate cDNA during reverse transcription that can also be sensed by cGAS, which then produces cGAMP to activate MITA. Furthermore, aberrantly aggregated self-DNA can also trigger innate immune responses in a MITA-dependent manner, but whether cGAS or other DNA sensors are involved remains unclear. Upon binding to CDNs or activation by upstream DNA sensors, MITA traffics to the perinuclear region and forms punctate structures that contain IRF3 and TBK1, facilitating TBK1-IRF3 interactions and phosphorylation of IRF3 by TBK1. Activated IRF3 collaborates with NF-κB to induce type I IFNs and inflammatory cytokines. However, the molecular mechanism that links MITA to NF-κB activation remains unclear. Additionally, MITA can recruit STAT6 to the ER upon virus infection, leading to TBK1-dependent phosphorylation, dimerization and subsequent translocation of STAT6 into the nucleus where it induces transcription of CCL2, CCL20 and CCL26. MAM, mitochondria associated membrane; Puncta, perinuclear punctate structure; MITA, also known as STING, MPYS, ERIS and TMEM173; VISA, also known as MAVS, IPS-1 and Cardif.
2.2. MITA-mediated signaling in response to RNA viruses
Several studies have established the critical roles of MITA in the innate immune responses to some RNA viruses. Knockdown of MITA impaired Sendai virus (SeV)- and vesicular stomatitis virus (VSV)-induced type I IFN production in human transformed cell lines, such as HEK293, HeLa and Huh7 cells, as well as in human primary macrophage and DCs [10], [11], [61]. Furthermore, genetic studies suggested that MITA deficiency rendered MEFs highly susceptible to VSV, but less susceptibility was observed in BMDCs or BMDMs, implicating MITA is involved in innate antiviral response in specific cell types to certain RNA viruses [9]. In vivo, MITA-deficient mice were defective in type I IFN production and highly susceptible to lethal infection with VSV but not encephalomyocarditis virus (EMCV) [13]. Compared to the universal requirement for MITA in cytosolic DNA-triggered signaling, MITA seems to be involved in innate immune responses against RNA viruses in a virus- and cell type-specific manner.
Several lines of evidence suggest that MITA is only involved in RIG-I, but not MDA5 signaling. First, MITA interacts with RIG-I but not MDA5; second, MITA does not mediate signaling triggered by high molecular weight poly(I:C), which is known to be sensed by MDA5; third, MITA is involved in SeV-, VSV-, newcastle disease virus (NDV)-, Japanese encephalitis virus (JEV)-, but not EMCV-induced innate immune responses, most likely because all of these viruses, except EMCV, are detected by RIG-I [9], [10], [11], [17].
Mechanistically, MITA interacts with VISA, most likely at mitochondria or MAM [10], [13]. The ability of RIG-I and VISA to induce IFN-β was diminished in MITA-deficient cells, whereas the ability of MITA to induce IFN-β was not affected in VISA-deficient cells, indicating that MITA functions downstream of VISA. MITA seems to act as an accessory adaptor to recruit TBK1 and IRF3 to the VISA-associated complex after viral infection, facilitating activation of IRF3 and NF-κB (Fig. 2) [10]. To date, the roles of MITA in RNA virus-induced innate immune response have mainly been investigated in transformed human cell lines. Additional studies using primary cells or in vivo studies will provide more definitive insights into the roles of MITA in innate immune responses against RNA viruses.
3. Molecular mechanisms of MITA-mediated signal transduction
Although some puzzles still remain to be solved, great progress has been made to advance our understanding of the molecular mechanisms of MITA-mediated responses to cytosolic DNA. MITA either functions downstream of DNA sensors or acts as a direct sensor of CDNs, acting in both roles to initiate signaling cascades that activate the transcription factors IRF3 and NF-κB, leading to type I IFN and proinflammatory cytokine production. Although MITA participates in both cytoplasmic RNA- and DNA-triggered signaling pathways which converge on the TBK1-IRF3 axis, the molecular mechanisms of these two pathways appear to be different. For example, NEMO is thought to be required for RNA-triggered IRF3 activation, but is dispensable for DNA-triggered, MITA-mediated activation of IRF3 [62]. Additionally, different cellular fractions of MITA appear to participate in these two pathways. The MAM- or mitochondria-localized MITA is important for RNA-triggered signaling, whereas the ER-localized MITA is responsible for DNA-triggered signaling [10], [13].
As a pivotal factor in DNA-triggered signaling, how MITA is activated to initiate downstream signaling is a central question that has been extensively studied in the past years. Certain critical events have been demonstrated to contribute to MITA activation and subsequent downstream signaling, such as stimuli-induced MITA dimerization and oligomerization, MITA-mediated signaling complex assembly, and membrane system-associated translocation of MITA to the perinuclear regions.
3.1. MITA is a scaffold protein
It has been reported that MITA can associate with both TBK1 and IRF3, and thereby serves as a scaffold protein that facilitates interactions between TBK1 and IRF3 [10], [62]. Specifically, the C-terminus of MITA (aa341-379) is required and sufficient for interactions with TBK1 and IRF3 that facilitate IRF3 phosphorylation by TBK1 [62]. Crystal structures of MITA also suggest the role of the MITA C-terminus in mediating protein interactions [53], [54]. Approximately 40 residues at the C-terminus of MITA could not be modeled in crystal structures, suggesting that the C-terminus of MITA is highly flexible and most likely protrudes from the concave cavity to interact with downstream proteins, such as TBK1 and IRF3 [53], [54]. Surprisingly, a truncated MITA protein that only contained aa139-344 could strongly interact with TBK1 in the presence of c-di-GMP, suggesting the presence of an additional TBK1 binding region of MITA [54]. However, another group found that MITA did not directly facilitate the activation of IRF3 by TBK1 in an in vitro kinase assay because TBK1 alone was sufficient to phosphorylate IRF3. This discrepancy most likely resulted from the use of excess protein in vitro [47].
3.2. MITA undergoes dimerization upon ligand binding
Several studies indicated that most of MITA is monomeric in cells under physiological conditions, and stimulation with cytosolic dsDNA and dsRNA induced MITA dimerization, which is thought to be important for MITA activation and subsequent downstream signaling [11], [54]. However, purified MITA in solution exists as a dimer, and crystallography studies showed that the CTD of MITA exists as a symmetrical V-shaped dimer both in the presence and absence of a ligand [52], [53], [54], [55], [56], [57], [58]. There are at least two possible reasons for this discrepancy. Most likely, MITA exists as a weak dimer under physiological conditions, which is easily disturbed by the conditions used for normal detection methods. Upstream activators or ligand binding can strengthen MITA dimerization, thereby making it easier to detect the dimeric form of MITA. This hypothesis is supported by a report that c-di-GMP binds to the MITA dimer interface in a perfectly symmetrical manner, thereby acting as a ‘glue’ to reinforce MITA homodimer by increasing the dimer interface area [53]. Alternatively, it remains possible that dimerization of MITA is signal-induced.
It has also been reported that MITA can form high molecular weight aggregates after IFN stimulatory DNA (ISD) stimulation, indicating that MITA may form oligomers or polymers for its signaling complex assembly [62]. Such a mechanism would be similar to VISA and ASC, which are activated by polymerization-mediated signalosome assembly [63], [64].
3.3. MITA translocates to the perinuclear region to activate IRF3
Viral nucleic acids trigger the translocation of MITA from the ER to the perinuclear regions, which is essential for signal transduction. Artificial addition of an ER retention signal to MITA hampers its ability to induce antiviral responses [65]. Many membrane-containing organelles and structures have been implicated in this process, such as Golgi apparatuses, endosomes, exocysts, microsomes, and autophagy-like puncta [9], [13], [62], [65].
Several lines of evidence suggest that dynamic membrane trafficking mediates the sequential translocation and assembly of MITA containing signalosomes, which is essential for maximal activation of the innate immune response triggered by cytosolic DNA. First, Brefeldin A, known to cause disassembly of the Golgi complex [66], blocked MITA trafficking, indicating that Golgi apparatuses are involved in the translocation of MITA. Second, in the presence of dsDNA, MITA co-localized with the early endosome marker early endosome antigen 1 (EEA1) and the recycling endosome marker transferrin receptor (TFR) [13]. Third, upon ISD stimulation, MITA associates with Sec5, a component of exocysts that is involved in vesicle trafficking. Depletion of Sec5 impairs the function of MITA, suggesting that exocysts are involved in MITA-mediated signal transduction [9], [13]. Finally, HSV-1 infection causes MITA to predominantly associate with microsomes, complexes of continuous membranes that include the ER, Golgi and transport vesicles [13]. In summary, DNA stimulation causes MITA to translocate from the ER via the Golgi apparatuses to vesicles in the perinuclear region, where it forms punctate structures.
Coincidently, it has been demonstrated that, in response to intracellular DNA, TBK1 also aggregates in perinuclear punctate structures in a MITA-dependent manner [13]. Additionally, co-localization of a phosphorylation defective mutant of IRF3 with MITA in punctate structures was also detected [67], suggesting that IRF3 might be activated in these punctate structures. Together, these studies demonstrate that DNA-stimulated translocation of MITA to perinuclear regions where MITA-TBK1-IRF3 complex assemblies is essential for IRF3 activation.
Although MITA is necessary for the localization of TBK1 to perinuclear regions, some details still need to be revealed. First, where does the MITA-TBK1interaction happen, does it occur before or after translocation to the perinuclear region? Specific inhibitors of such transportation pathways may be helpful to answer this question. Another important question is how do the puncta form and what are they? MITA was found to co-localize with several autophagy-related proteins after DNA stimulation, including LC3 and Atg9a, which are components of the autophagosomes [65]. However, electron microscopy analyses revealed that MITA-containing puncta induced by dsDNA stimulation did not exhibit the morphological characteristics of autophagosomes, suggesting that the puncta may represent a unique membrane structure. Another study proposed that this puncta is an endo/lysosome [13]. However, it has also been shown that MITA does not localize to endosomes or lysosomes after dsDNA stimulation [65]. Further work will be required to clarify the characteristics of the puncta, which will contribute to understanding the molecular mechanisms of DNA-stimulated signal transduction.
Overall, these observations clearly suggest that membrane-associated protein trafficking is closely related to MITA-mediated signal transduction in innate immunity. In light of these observations, it has also been reported that translocon-mediated RIG-I redistribution from the cytosol to MAM is essential for downstream innate immune signaling [68].
4. Regulation of MITA-mediated signaling
Because MITA exerts critical roles in cytosolic nucleic acids-triggered innate immune responses, the regulation of MITA-mediated signal transduction has been extensively investigated. Many host factors have been implicated in modulating MITA-mediated signal transduction to generate an appropriate immune response. Furthermore, MITA-mediated signaling has also been targeted by viral proteins for immune evasion.
4.1. Ubiquitination-mediated regulation of MITA
Ubiquitination has emerged as a central posttranslational regulatory mechanism in the positive and negative control of antiviral signaling [69]. Two typical linkages of polyubiquitin chains, K48 and K63 (polyubiquitin chains that are linked through lysine at position 48 or 63 of ubiquitin, respectively), have been extensively characterized. In most cases, K48-linked polyubiquitin chains target substrate proteins for proteasome-dependent degradation, whereas K63-linked polyubiquitin chains usually enhance substrate protein functions by regulating cellular localization or protein–protein interactions.
It was reported that TRIM56 is a positive regulator of MITA-mediated signaling. Knockdown of TRIM56 impaired poly(I:C)- and poly(dA:dT)-stimulated type I IFN production in transformed cell lines and normal human lung fibroblasts, indicating that TRIM56 is required for both dsRNA- and dsDNA-induced responses [70]. TRIM56 interacted with MITA and preferentially mediated the K63-linked ubiquitination of MITA on K150, which is required for MITA dimerization and subsequent recruitment of TBK1 [70]. However, a structural study of the MITA dimer suggested that K150 may not play a major role in the dimerization of MITA, and a K150 mutation had no effect on MITA dimerization, but did impair its association with TBK1 [54].
In addition to TRIM56, TRIM32 was also identified as a positive regulator of MITA-mediated signaling in response to cytosolic poly(I:C) and poly(dA:dT), as well as SeV and HSV-1 infection [71]. TRIM32 targeted MITA for K63-linked ubiquitination at K20, K150, K224, and K236 through its E3 ubiquitin ligase activity, and promoted the interaction of MITA with TBK1. These findings suggested that TRIM32 is an important regulatory protein for innate immunity against both RNA and DNA viruses.
Both TRIM32 and TRIM56 are IFN-induced genes that act as positive feedback regulators of cytosolic RNA- and DNA-triggered signaling. The relationship between these two E3 ligases remains unclear. It seems reasonable that they have complementary functions because their underlying mechanisms are somewhat different. TRIM56 interacts with the C-terminal region of MITA and partially colocalizes with MITA at punctate structures after poly(dA:dT) stimulation. By contrast, TRIM32 interacts with the N-terminal transmembrane domain-containing fragment of MITA and colocalizes with MITA at the ER and mitochondria. To better clarify these potentially distinct roles of TRIM32 and TRIM56, additional studies using knockout mice and cells will be needed.
In addition to the positive regulation of MITA by ubiquitination, it was also reported that MITA is negatively regulated by RNF5-mediated ubiquitination [61]. RNF5 targets MITA for K48-linked ubiquitination at the mitochondria, leading to its degradation and inhibition of virus-induced IRF3 activation, IFN-β expression, and cellular antiviral response [61].
MITA has also been reported to undergo degradation after DNA virus infection [72]. It has been proposed that CDNs activate ULK1(ATG1) to phosphorylate MITA on S366, which causes the degradation of MITA, thus triggering a negative feedback control of MITA activity. Chloroquine, which inhibits the lysosomal degradation pathway, could only partially block the degradation of MITA [72], suggesting MITA is mostly degraded by a non-lysosomal degradation pathway. The degradation of MITA remains incompletely characterized, as the major degradation pathways need to be determined and the role of ubiquitination in DNA-induced degradation of MITA needs to be clarified in future studies.
4.2. Phosphorylation-mediated regulation of MITA
The phosphorylation of MITA has been reported by different groups, and several phosphorylation sites have been identified in different cell types and in response to different stimuli [10], [11], [62], [72]. Two groups identified distinct sites of MITA that are phosphorylated after stimulation by cytosolic dsDNA. One group found that recombinant human MITA in L929 cells was phosphorylated on S353, S358 and S379 after ISD stimulation [62], while another group found that endogenous human MITA was phosphorylated on S345, S358, S366 and S379 after dsDNA stimulation [72]. Contrary to DNA virus infection, upon SeV infection, MITA is mainly phosphorylated on Ser358 by TBK1, which is critical for the activation of IRF3. Furthermore, the mutation of Ser358 to alanine impaired the ability of MITA to interact with TBK1 [10]. However, the function of S358, which was identified by three groups, is obscure as both S358A and S358D mutants partially impair their abilities to activate IRF3 [10], [62], [72]. It is possible that the phosphorylation of MITA is dynamic and different sites are phosphorylated at different stages following stimulation.
Depletion of TBK1 prevented dsDNA-induced phosphorylation of MITA, indicating that TBK1 is essential for MITA phosphorylation [54]. MITA forms puncta in TBK1- and IKKɛ- double-deficient MEFs following dsDNA stimulation, indicating that the translocation of MITA is independent of its phosphorylation [55]. Additionally, MITA phosphorylation likely occurs after trafficking from the ER to the Golgi apparatus [59]. Recently, one study reported that ULK1(ATG1), an autophagy-related kinase, could phosphorylate MITA at S366 and cause MITA to be degraded. This finding suggests that phosphorylation at S366 is important for negative regulation of MITA-mediated signaling [72]. Surprisingly, both S366A and S366D mutants were found to be inactive [62], [72], suggesting that S366 is not only a phosphorylation site. Indeed, another study found that S366 is important for IRF3 binding [62].
4.3. Regulation of MITA-mediated signaling by viral proteins
Viruses have evolved elaborate mechanisms to antagonize the innate immune system. For example, Hepatitis C virus (HCV) can evade innate immunity and establish chronic infection by cleaving VISA via HCV-NS3/4A serine protease [73]. As a critical component in the antiviral innate immune response, MITA is also targeted by various viruses for immune evasion.
Recently, two groups reported that the HCV non-structural protein NS4B could abrogate RIG-I-mediated type I IFN induction by targeting MITA [74], [75]. MITA shares a structurally homologous domain with flavivirus NS4B, which suggests a direct protein–protein interaction. NS4B colocalizes with MITA in the ER and MAM, and impairs the interaction between MITA and VISA, which is required for a robust IFN-β induction [74]. These studies suggest that HCV NS3/4A and NS4B may cooperate to block IFN-β induction [74]. Independently, another group found that NS4B can suppress dsRNA- or RNA virus-induced IFN production by disrupting the MITA-TBK1 interaction [75]. Additionally, NS4B of Yellow fever virus (YFV) was found to inhibit MITA activity, most likely by a similar mechanism [13].
Dengue virus (DENV) can evade the innate immune system through cleavage of human MITA by its NS2B3 proteinase, thereby inhibiting type I IFN production [76], [77]. In MITA-deficient cells, the replication of DENV was enhanced, indicating an antiviral role for MITA against DENV infection [76]. The cleavage site of MITA was mapped to LRR96/G, which is not conserved in mouse MITA and render it resistant to NS2B3 cleavage [77]. The absence of this cleavage site explains why the replication of DENV in mouse cells is severely restricted, and this is consistent with the notion that MITA plays a key role in inhibiting DENV infection and propagation in mice.
Human coronavirus HCoV-NL63 and severe acute respiratory syndrome (SARS) papain-like proteases (PLPs) antagonize innate immune signaling by inhibiting MITA-mediated IRF3 activation [78]. PLPs from human HCoV-NL63 or SARS-CoV interact with MITA, block MITA dimerization and negatively regulate the assembly of VISA-MITA-TBK1/IKKɛ complexes that are required for activation of IRF-3. Furthermore, PLPs reduce the levels of ubiquitinated forms of MITA. Thus, the HCoV PLPs seem to disrupt MITA-mediated IFN induction by distinct strategies.
Compared to the extensive targeting of MITA by RNA viruses, the regulation of MITA by DNA virus-encoded proteins has not been reported. As an essential component in cytosolic DNA-induced signaling, it is reasonable that MITA may also be targeted by DNA viruses, bacteria and parasites.
4.4. Other regulatory mechanisms
Recently, NLRC3 was reported to diminish MITA-dependent innate immune responses to cytosolic DNA, c-di-GMP and DNA viruses [79]. NLRC3 is associated with both MITA and TBK1, which hampers the MITA-TBK1 interaction and thus impairs type I IFN production. Nlrc3-deficient mice exhibit enhanced innate immunity and reduced morbidity and viral loads after HSV-1 infection. This study demonstrates crosstalks exist between two key pathways of innate immune regulation, NLR- and MITA-mediated signaling pathways. In addition to NLRC3, MITA was reported to be regulated by its alternatively spliced isoform [80]. It has also been reported that treatment of cells with type I IFNs decreases the mRNA levels of MITA in an IFI16-dependent manner by an unknown mechanism, indicating that MITA is regulated by a negative feedback mechanism at the transcriptional level [81].
5. Concluding remarks
This review summarizes our current knowledge of the molecular mechanisms of MITA-mediated innate immune responses to cytosolic nucleic acids, including the recognition of cytosolic nucleic acids, subsequent signaling to induce type I IFNs, and regulation of the signaling pathways. Many studies have confirmed that MITA is important in RNA virus-triggered signaling and MITA is targeted by various viruses for immune evasion. It has also been established that MITA is essential for innate immune responses against DNA-producing pathogens, including DNA viruses, retroviruses, bacteria, and parasites. Thus, additional studies of differential regulation of MITA-mediated signaling in response to RNA and DNA, and the crosstalks between these pathways will benefit our understanding of mechanisms of innate immune responses. The identification of cGAS as a critical DNA sensor and CDNs (including c-di-AMP, c-di-GMP and cGAMP) as direct ligands of MITA significantly advanced our understanding of DNA sensing and signaling. Nevertheless, it will also be necessary to clarify and confirm the role of other DNA sensors using more rigorous strategies, including a determination of the relationships of other sensors with cGAS. Additionally, the mechanisms of MITA activation remain obscure, as the crystallography studies did not provide evidence of ligand-induced conformational changes coupled to downstream signaling. The crystallization of full-length MITA associated with ligands or downstream components could be critical for further uncovering the mechanisms of MITA-mediated signal transduction. Another important question is how MITA mediates NF-κB activation, a critical event in innate immunity. It will be interesting to investigate whether NF-κB activation is also associated with formation of punctate structures. Furthermore, given the different responses in mouse and human cells, the functions of murine and human MITA might not be completely similar; discrepancies could result from critical amino acid changes, differential regulation by viruses, or other mechanisms [9], [10], [13], [45], [52]. Resolving these issues will contribute to our understanding of innate immune responses, and provide clues for drug and vaccine development against infectious and autoimmune diseases.
Acknowledgements
Work in the authors’ laboratories is supported by Chinese Ministry of Science and Technology Grants 2014CB542603 and 2012CB910201; National Natural Science Foundation of China Grants 31170792, 31270932, 31130020 and 31321001.
Biographies

Dr. Yong Ran graduated with a B.S. degree in biology from Wuhan University in 2007. He received his Ph.D. degree in cell biology from Wuhan University in 2012. He has been an assistant investigator in Wuhan Institute of Virology, Chinese Academy of Sciences since 2012. He has been working the molecular mechanisms of innate antiviral immunity in the past years.

Dr. Hong-Bing Shu graduated from Lanzhou University with a B.S. degree in biology (1987) and Peking Union Medical College with a M.S. degree in cell biology (1990) in China. He received his Ph.D. degree in cell and developmental biology from Emory University (1995) and finished his post-doctoral training with Dr. David Goeddel at Tularik Inc. (1995–1997) in the US. He was recruited as an assistant professor in 1999 and promoted to associate professor in 2003 in the Department of Immunology at National Jewish Medical and Research Center at Denver in the US. He served as a Changjiang professor at Peking University (2000–2004) and later on the Dean at College of Life Sciences at Wuhan University in China (2005–2013). Currently, he is the vice president for research and graduate studies at Wuhan University. He is also the vice president of the Chinese Society for Cell Biology and an elected member of the Chinese Academy of Sciences. For ∼20 years, he has been investigating the molecular mechanisms of signaling triggered by the tumor necrosis factor family members as well as innate antiviral immune response. He has published 100 peer-reviewed papers in internationally recognized journals and some of these papers have been highly cited.

Dr. Yan-Yi Wang received her B.S., M.S., and Ph.D. degrees from Peking University, University of Colorado School of Medicine, and Wuhan University, respectively. In 2006, she started to work in Wuhan University as an instructor and later on promoted to associate professor. She joined the Institute of Virology, Chinese Academy of Sciences in 2012 as a principle investigator. Her research focuses on molecular mechanisms of antiviral innate immunity and inflammation.
References
- 1.Yoneyama M., Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 2.Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 5.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Ahn J., Gutman D., Saijo S., Barber G.N. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paludan S.R., Bowie A.G. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdette D.L., Vance R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Sun W., Li Y., Chen L., Chen H., You F., Zhou X. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin L., Hill K.K., Filak H., Mogan J., Knowles H., Zhang B. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer J.D., Sotelo-Troha K., von Moltke J., Monroe K.M., Rae C.S., Brubaker S.W. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H., Sun H., You F., Sun W., Zhou X., Chen L. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Nazmi A., Mukhopadhyay R., Dutta K., Basu A. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci Rep. 2012;2:347. doi: 10.1038/srep00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sze A., Belgnaoui S.M., Olagnier D., Lin R., Hiscott J., van Grevenynghe J. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe. 2013;14:422–434. doi: 10.1016/j.chom.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Koppe U., Hogner K., Doehn J.M., Muller H.C., Witzenrath M., Gutbier B. Streptococcus pneumoniae stimulates a STING- and IFN regulatory factor 3-dependent type I IFN production in macrophages, which regulates RANTES production in macrophages, cocultured alveolar epithelial cells, and mouse lungs. J Immunol. 2012;188:811–817. doi: 10.4049/jimmunol.1004143. [DOI] [PubMed] [Google Scholar]
- 21.Watson R.O., Manzanillo P.S., Cox J.S., Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Almeida L.A., Carvalho N.B., Oliveira F.S., Lacerda T.L., Vasconcelos A.C., Nogueira L. MyD88 and STING signaling pathways are required for IRF3-mediated IFN-beta induction in response to Brucella abortus infection. PLoS ONE. 2011;6:e23135. doi: 10.1371/journal.pone.0023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma S., DeOliveira R.B., Kalantari P., Parroche P., Goutagny N., Jiang Z. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo T., Kobayashi J., Saitoh T., Maruyama K., Ishii K.J., Barber G.N. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber G.N. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moeller H.B. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun F., Zhang Y.B., Liu T.K., Shi J., Wang B., Gui J.F. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J Immunol. 2011;187:2531–2539. doi: 10.4049/jimmunol.1100642. [DOI] [PubMed] [Google Scholar]
- 28.Biacchesi S., Merour E., Lamoureux A., Bernard J., Bremont M. Both STING and MAVS fish orthologs contribute to the induction of interferon mediated by RIG-I. PLoS ONE. 2012;7:e47737. doi: 10.1371/journal.pone.0047737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Z., Chen X., Yu B., Chen D. Cloning and functional characterization of rat stimulator of interferon genes (STING) regulated by miR-24. Dev Comp Immunol. 2012;37:414–420. doi: 10.1016/j.dci.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Wu J., Sun L., Chen X., Du F., Shi H., Chen C. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burckstummer T., Baumann C., Bluml S., Dixit E., Durnberger G., Jahn H. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., Alnemri E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muruve D.A., Petrilli V., Zaiss A.K., White L.R., Clark S.A., Ross P.J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 36.Bauer S., Kirschning C.J., Hacker H., Redecke V., Hausmann S., Akira S. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 38.Ishii K.J., Kawagoe T., Koyama S., Matsui K., Kumar H., Kawai T. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 39.Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Chen R., Zhou Q., Xu Z., Li C., Wang S. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc Natl Acad Sci U S A. 2012;109:11770–11775. doi: 10.1073/pnas.1203405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe T., Harashima A., Xia T., Konno H., Konno K., Morales A. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diner E.J., Burdette D.L., Wilson S.C., Monroe K.M., Kellenberger C.A., Hyodo M. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoggins J.W., Macduff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam E., Stein S., Falck-Pedersen E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J Virol. 2014;88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McWhirter S.M., Barbalat R., Monroe K.M., Fontana M.F., Hyodo M., Joncker N.T. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodward J.J., Iavarone A.T., Portnoy D.A. c-di-AMP secreted by intracellular listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao P., Ascano M., Zillinger T., Wang W., Dai P., Serganov A.A. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y.H., Liu X.Y., Du X.X., Jiang Z.F., Su X.D. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 54.Ouyang S., Song X., Wang Y., Ru H., Shaw N., Jiang Y. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang G., Zhu D., Li N., Zhang J., Zhu C., Lu D. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 56.Shu C., Yi G., Watts T., Kao C.C., Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin Q., Tian Y., Kabaleeswaran V., Jiang X., Tu D., Eck M.J. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 61.Zhong B., Zhang L., Lei C., Li Y., Mao A.P., Yang Y. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai X., Chen J., Xu H., Liu S., Jiang Q.X., Halfmann R. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R. Unified polymerization mechanism for the assembly of asc-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujiwara T., Oda K., Yokota S., Takatsuki A., Ikehara Y., Brefeldin A. causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- 67.Bowie A. The STING in the tail for cytosolic DNA-dependent activation of IRF3. Sci Signal. 2012;5:pe9. doi: 10.1126/scisignal.2002919. [DOI] [PubMed] [Google Scholar]
- 68.Liu H.M., Loo Y.M., Horner S.M., Zornetzer G.A., Katze M.G., Gale M., Jr. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhoj V.G., Chen Z.J. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 70.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J., Hu M.M., Wang Y.Y., Shu H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Konno H., Konno K., Barber G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of sting to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nitta S., Sakamoto N., Nakagawa M., Kakinuma S., Mishima K., Kusano-Kitazume A. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 75.Ding Q., Cao X., Lu J., Huang B., Liu Y.J., Kato N. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 76.Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu C.Y., Chang T.H., Liang J.J., Chiang R.L., Lee Y.L., Liao C.L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L., Mo J., Swanson K.V., Wen H., Petrucelli A., Gregory S.M. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen H., Pei R., Zhu W., Zeng R., Wang Y., Wang Y. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J Immunol. 2014;192:1162–1170. doi: 10.4049/jimmunol.1300798. [DOI] [PubMed] [Google Scholar]
- 81.Panchanathan R., Liu H., Xin D., Choubey D. Identification of a negative feedback loop between cyclic di-GMP-induced levels of IFI16 and p202 cytosolic DNA sensors and STING. Innate Immun. 2013;0:1–9. doi: 10.1177/1753425913507097. [DOI] [PubMed] [Google Scholar]