Abstract
Background
Multiplex real time PCR is increasingly used to diagnose respiratory viruses and has shown to be superior to traditional methods, like culture and antigen detection. However, comprehensive data on sensitivity, specificity and performance of the multiplex PCR compared to the single target PCR's is limited for most published respiratory multiplex real time PCR assays.
Objectives
Development and extensive analysis of an internally controlled multiplex real time rt-PCR for detection of respiratory viruses.
Study design
The assay was validated in comparison to single-target PCR's using plasmid targets and prospectively collected nasopharyngeal aspirates.
Results
Using plasmid targets the multiplex format was found to be as least as sensitive and specific as the single-target PCR and no competition was observed when different targets were present at different amounts in one tube. Clinical validation showed high concordance for all viruses tested except for samples with low levels of enterovirus.
Conclusion
This multiplex showed excellent specificities for all 14 respiratory viruses and sensitivity was high except for clinical samples with low levels of enterovirus.
Keywords: Respiratory, Virus, Multiplex, PCR, Realtime
1. Background
Demonstrating the presence of a viral pathogen in patients with respiratory symptoms has long been challenging. Traditional methods like viral culture and antigen detection have moderate sensitivity and specificity. Moreover, the relative long turnaround time for some of these techniques makes them less useful for diagnosis of acute clinical problems. With the development of real time (RT) reverse transcriptase (rt) PCR, direct detection of amplified nucleic acids became possible, minimizing cross-contamination and reducing hands on time. In addition, RT rtPCR provides the possibility of easy quantitation of the amplified target. This progress has led to the development of a large number of rtPCR assays for the detection of individual viral pathogens. The introduction of multiplex RT rtPCR assay has increased the efficiency of routine molecular diagnostics of respiratory viruses and has been shown to be cost effective.1, 2, 3
Past studies have extensively proven superior sensitivity of (real time) multiplex PCR over traditional methods.4, 5, 6 However comprehensive analysis of analytical sensitivity and specificity of a multiplex PCR as compared to single-target assays has not been described for respiratory viruses. Puppe et al.7 compared a multiplex assay with corresponding single-target PCR assays and showed a mean loss of 1.3 log for the multiplex assay. However their PCR was qualitative and did not use a real time format. Gunson et al.8 reported similar median crossing-point (Cp) values when comparing duplex versus triplex real time assays.
2. Objective
In the study described here, a systematic analysis of multiplex RT rtPCR was performed. Sensitivity and specificity of the multiplex assay were compared to its single-target counterparts. This was done in serial dilutions of plasmids, samples with mixed plasmid input and in prospectively gathered nasopharyngeal aspirates (NPA).
3. Study design
3.1. Preparation of controls
DNA targets controls were constructed by elongation of oligonucleotide linkers followed by amplification with specific primers, as described before.9 The PCR products were ligated into PCRII-TOPO vector (Invitrogen, Paisley, UK) according to the instructions of the manufacturer. The sequences of all plasmids obtained were confirmed by sequencing using the BigDye sequencing kit (ABI, Nieuwerkerk a/d lJssel, The Netherlands).
Equine arteritis virus (EAV) was used as internal control (IC)10 and was measured during multiplex PCR in respiratory viral package (RVP) number one.
3.2. Primers and probes
Specific primers and probes for all targets (Table 1 ) were designed using published sequences from Gene Bank. Primers and 6FAM and HEX labeled probes were obtained from Biolegio (Nijmegen, the Netherlands), CYAN500, LCRED610, and LCRED670 labeled probes from TIB Molbiol (Berlin, Germany) and MGB probes from ABI (6FAM or VIC labeled).
Table 1.
Primers and probes.
| Virus | Sequence 5′–3′ | Target gene | Labels 5′/3′ | |
|---|---|---|---|---|
| Influenza A | F | GACAAGACCAATCCTGTCACYTCTG | ||
| R | AAGCGTCTACGCTGCAGTCC | M | ||
| P | TTCACGCTCACCGTGCCCAGTGAGC | LCRED610/BBQ | ||
| Influenza B | F | TCGCTGTTTGGAGACACAAT | ||
| R | TTCTTTCCCACCGAACCA | M | ||
| P | AGAAGATGGAGAAGGCAAAGCAGAACT | CYAN500/DB | ||
| Enterovirus | F | GGCCCTGAATGCGGCTAAT | ||
| R | GGGATTGTCACCATAAGCAGCC | 5′-UTR | ||
| P | GCGGAACCGACTACTTTGGGT | FAM/MGB-NFQ | ||
| Adenovirus | F | CAGGACGCCTCGGRGTAYCTSAG | ||
| R | GGAGCCACVGTGGGRTT | Hexon | LCRED670/BBQ | |
| P | CGGGTCTGGTGCAGTTTGCCCGC | |||
| RSV | F | ATGAACAGTTTAACATTACCAAGT | ||
| R | GTTTTGCCATAGCATGACAC | F | ||
| P1 | TGACTTCAAAAACAGATGTAAGCAGCTCC | LCRED610/BBQ | ||
| P2 | TTATGACATCAAAAACAGACATAAGCAGCTCAG | |||
| Rhinovirus | F | CAAGCACTTCTGTTTCCCC | ||
| R | GGCAGCCACGCAGGC | 5′-UTR | ||
| P1 | TAGACCTGGCAGATGA+G+G+CT | FAM/BBQ | ||
| P2 | TAGTTTGGTCGA+T+GA+GGCT | FAM/BBQ | ||
| P3 | CTAGTYTGGTCGAT+G+A+GGC | FAM/BBQ | ||
| Coronavirus | F1 | GGTGGYTGGGAYGATATGTTACG | ||
| F2 | GCTRAGCATGATTTCTTTACTTGG | Replicase | ||
| R1 | KRTTTGGCATAGCACGATCACA | |||
| R2 | CARTYTTKTTCATCAAAGTTACGCA | Replicase | ||
| P1 | ATGTTGACAAYCCTGTWCTTATGGGTTGGG | FAM/MGB-NFQ | ||
| P2 | CAGARTCATTTATGGTAATGTTAGTAGACA | FAM/MGB-NFQ | ||
| Metapneumovirus | F | AGCTTCAGTCAATTCAACAGAAG | ||
| R | CCTGCAGATGTYGGCATGT | F | ||
| P | TGTTGTGCGGCAGTTTTCAGACAATGC | LCRED670/BBQ | ||
| Parechovirus | F | CTGGGGCCAAAAGCCA | ||
| R | GGTACCTTCTGGGCATCCTTC | 5′-UTR | ||
| P | AAACACTAGTTGTAWGGCCC | FAM/MGB-NFQ | ||
| Parainfluenzavirus 1 | F | ATCTCATTATTACCYGGACCAAGTCTACT | ||
| R | CATCCTTGAGTGATTAAGTTTGATGAATA | HN | ||
| P | AGGATGTGTTAGAYTACCTTCATTATCAATTGGTGATG | CYAN500/DB | ||
| Parainfluenzavirus 2 | F | CTGCAGCTATGAGTAATC | ||
| R | TGATCGAGCATCTGGAAT | NP | ||
| P | AGCCATGCATTCACCAGAAGCCAGC | LCRED610/BBQ | ||
| Parainfluenzavirus 3 | F | ACTCTATCYACTCTCAGACC | ||
| R | TGGGATCTCTGAGGATAC | NP | ||
| P | AAGGGACCACGCGCTCCTTTCATC | LCRED670/BBQ | ||
| Parainfluenzavirus 4 | F | GATCCACAGCAAAGATTCAC | ||
| R | GCCTGTAAGGAAAGCAGAGA | NP | ||
| P | TATCATCATCTGCCAAATCGGCAA | HEX/BHQ | ||
| HBoV | F | CAAATCTCTTCTGGCTACACG | ||
| R | CTCTGCGATCTCTATATTGAAGG | NS1 | ||
| P | ATGTTGCCGCCAGTAACTCCACC | LCRED670/BBQ | ||
| IC (EAV) | F | CATCTCTTGCTTTGCTCCTTAG | ||
| R | AGCCGCACCTTCACATTG | Replicase | ||
| P | TGTGGGCAATAATGTTGTTCTGACAGCG | HEX/BHQ |
+ denotes a locked nucleic acid (LNA) nucleotide.
3.3. Nucleic acid extraction and real-time multiplex RT rtPCR
Extraction of nucleic acids (NA) from 200 μl of NPA was performed by MagNA Pure LC extraction using the total nucleic acid extraction kit (Roche Diagnostics, Penzberg, Germany). cDNA synthesis was performed as described earlier.11 All PCR (multiplex and single-target) reactions were performed on a Roche LC480 (Roche Diagnostics, Penzberg, Germany) using exactly the same conditions. Reactions contained 10 μl of 2× Probes Master (Roche Diagnostics, Penzberg, Germany), 900 nM of primer (each) 200 nM of probe (each), and 5 μl of a cDNA in a total volume of 20 μl. Cycling conditions were as follows: 2 min at 50 °C and 10 min at 95 °C, followed by 45 cycles each consisting of 15 s at 95 °C and 1 min at 60 °C. Data were analyzed using the LC480 software. Color compensation and calculation of Cp-values was done using LC480 software. The crossing point (Cp) value, which is comparable to a cycle threshold (CT) value, was used as an approximation of the amount of virus present. The Cp-value reflects the cycle at which a positive PCR signal is detected (the lower the Cp-value the more target DNA or RNA is present in the sample). The LC480 software obtains a Cp-value using the second derivative maximum of the amplification curve, which is more precise than cycle tresholds or crossing point algorithms with normalized, proportional, arithmetic or no background adjustment.12 However small differences between Cp value of mono and multiplex pcr seem inevitable, since PCR reaction mixture between single and multiplex intrinsically different and to some degree will influence outline of amplification curves used to determine Cp value.
A sample was considered positive if the results showed Cp-values ≤ 40. Samples were considered negative if Cp-values were above 40 and the Cp-value for the corresponding IC was ≤32.8 (i.e. two times the standard deviation) above the mean IC for the negative samples).
3.4. PCR design
We have recently described an internally controlled multiplex RT rtPCR for the detection of influenzavirus A (InfA) and B (InfB), enterovirus (EV), and adenovirus (AdV) which will be referred to as respiratory virus package 1 (RVP1).7 In the current study we have broadened our diagnostic repertoire by designing three additional RVP's: RVP2 for the detection of respiratory syncytial virus (RSV), rhinovirus (RV) and human-metapneumovirus (hMPV); RVP3 for the detection of parainfluenzavirus 1-4 (PIV1-4) and human-parechovirus (hPeV); RVP4 for the detection of human-coronavirus (hCoV: HKU1, NL63, 229E and OC43), and human-bocavirus (hBoV).
3.5. Assessment of sensitivity and specificity
Sensitivity and specificity of the multiplex assay was analyzed using the single target PCR's as the gold standard.
This was done in four ways. One; dynamic ranges of the multiplex and single-target PCR's were assessed by testing serial dilutions of plasmids. Two; testing of mixed plasmid solutions containing different viral DNA targets. Each target was tested by spiking one target in an end dilution of 105 copies/reaction (i.e. 107 copies/ml) in a background of 103 copies/reaction (i.e. 105 copies/ml) of all other targets for respectively RVP1, RVP2, RVP3 and RVP4. Samples were analyzed by multiplex PCR and single-target PCR and Cp-values were determined and compared. Three; testing of prospectively gathered clinical samples. A panel of 133 NPA's was taken from cohort of children who were admitted for a suspected respiratory tract infection to the Academical Medical Centre of Amsterdam, during the winter season of 2007–08. The study was approved by the local ethical committee and participants were included after written informed consent by one of the parents. A basic population description is presented in Table 2 . Four; for viruses species that were detected in less than 4 NPA samples, QCMD (Quality Control for Molecular Diagnostics, Glasgow Scotland) control panels were run (Table 3 ). These samples consisted of cultured virus in various concentrations.
Table 2.
Patients characteristics.
| Clinical characteristics from children (n = 133) admitted to the Academical Medical Centre for an acute respiratory tract infection during the winter of 2007–08. | |
|---|---|
| Age in months; median (inter quartile range) | 12.1 (4.8–31.5) |
| Sex | ♂ 82 (62%), ♀ 51 (38%) |
| Reason of admittance: | |
| Upper respiratory tract infection + imminent dehydration | 15 (11%) |
| Wheezing illness | 30 (23%) |
| Pneumonia | 12 (9%) |
| Impending respiratory failure | 17 (13%) |
| Dyspnoea not further classified | 13 (10%) |
| Fever without specific respiratory symptoms | 24 (18%) |
| Other (=not classifiable) | 22 (16%) |
Table 3.
Clinical evaluation.
| (A) Analysis of prospectively collected nasopharyngeal aspirates (n = 133) from children admitted to the Academical Medical Centre for an acute respiratory tract infection during the winter of 2007–08 | |||
|---|---|---|---|
| Single target PCR | Multiplex PCR | ||
| InfA | 9/133 | 9/133 | (p = 1.00a) |
| InfB | 3/133 | 3/133 | (p = 1.00b) |
| EV | 7/133 | 1/133 | (p = 0.01b) |
| ADV | 15/133 | 14/133 | (p = 0.86a) |
| RV | 36/133 | 36/133 | (p = 1.00a) |
| RSV | 23/133 | 22/133 | (p = 0.89a) |
| hMPV | 7/133 | 8/133 | (p = 0.80a) |
| PIV-1 | 2/133 | 2/133 | (p = 1.00b) |
| PIV-2 | 1/133 | 1/133 | (p = 1.00b) |
| PIV-3 | 1/133 | 2/133 | (p = 1.00b) |
| PIV-4 | 1/133 | 1/133 | (p = 1.00b) |
| hPeV | 4/133 | 3/133 | (p = 0.70b) |
| hCoV | 3/133 | 4/133 | (p = 0.70b) |
| hBoV | 8/133 | 7/133 | (p = 0.80a) |
| Total | |||
| Excluding EV | 113/133 | 112/133 | (p = 0.96a) |
| Including EV | 120/133 | 113/133 | (p = 0.73a) |
| (B) Quality control panel (2007) for PIV 1, 2, 3, 4, InfB, hPeV and hCoV (n = 10) | ||||
|---|---|---|---|---|
| Single target PCR | Multiplex PCR | QCMD | ||
| PIV-1 | 4/10 | 4/10 | 4/10 | (p = 1.00b) |
| PIV-2 | 1/10 | 1/10 | 1/10 | (p = 1.00b) |
| PIV-3 | 1/10 | 1/10 | 1/10 | (p = 1.00b) |
| PIV-4 | 3/10 | 3/10 | 3/10 | (p = 1.00b) |
| InfB | 1/14 | 1/14 | 1/14 | (p = 1.00b |
| hCoV | 5/14 | 5/14 | 5/14 | (p = 1.00a) |
Chi square
Fisher exact test.
Assessment of specificity was done by analyzing potential “crosstalk” (i.e. the fluorescence of a single target influencing detection of one of the others targets, generating a false positive signal) and potential false positivity due to unspecific primer–primer combinations (some multiplex packages contained up to 10 primers) in prospectively gathered NPA.
4. Results
4.1. Analyses of dynamic ranges and mixed input of plasmids
Dynamic ranges were determined to be linear between 500 and 108 copies/PCR of target plasmid for both multiplex and single target assay's. The lower limit of detection for the multiplex assay was between 40 and 50 copies/reaction for every target, which was similar to the individual single-target PCR's (data not shown).
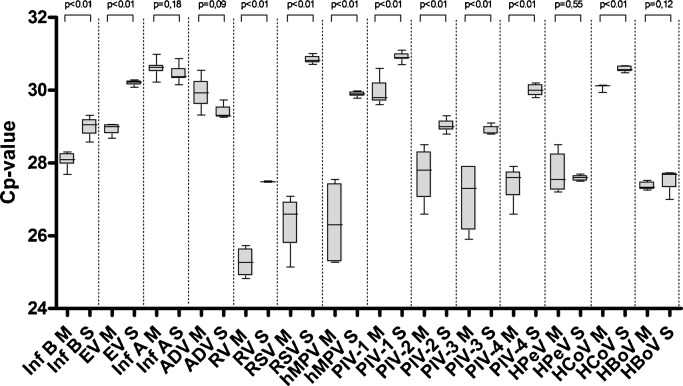
When analyzing mixed plasmid input, median Cp-values in the multiplex PCR were never higher than the single-target assay's, and in most cases the median Cp-value of multiplex analyses was even statistically lower than its single-target counterpart (Fig. 1 ). Variation was slightly higher for the multiplex analysis, with a coefficient of variation (CV) range for the multiplex assay between 0.4% (hCoV) and 3.3% (PIV3), compared to the CV range for single-target assay's between 0.3% (hPeV) and 1.0% (hBoV).
Fig. 1.
Analysis of sensitivity using mixed plasmids containing multiple viral DNA targets in different quantities (see method section for details). Samples were analyzed by multiplex PCR (M) and single-target PCR (S). Median CP values and inter quartile range are indicated by box plots.
4.2. Analyses of clinical samples and quality control panels
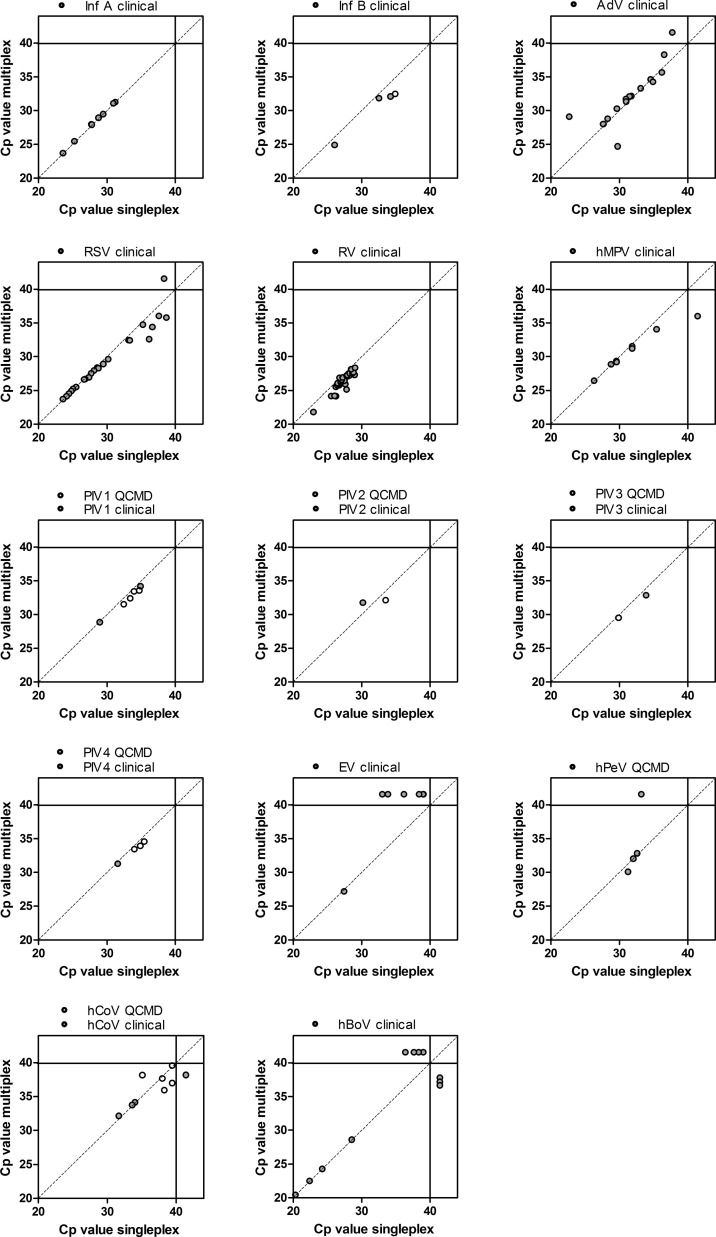
Overall, good concordance was observed between the multiplex PCR in comparison with the single-target PCR assays for clinical samples (Table 3 and Fig. 2 ). Of the 133 tested samples 70% (93/133) gave a positive result in one or more of the single-target assays (52% [70/133] mono-infections, 16% [22/133] double and <1% [1/133] triple infections). In 68% (90/133) the multiplex assay gave one or more positive results (50% [67/133] mono-infections, 17% [23/133] double-infections, no triple-infections). All discrepancies are described in Table 4 . Opposed to plasmid analysis, the multiplex assay failed to detect EV in clinical samples with low EV loads, since Cp-values ≥ 33 in the single-target PCR were not detected in the multiplex PCR. Moreover low viral loads of hBoV (Cp-values ≥ 36) seemed to be detected alternately by single-target PCR or by multiplex assay.
Fig. 2.
Analysis of sensitivity and specificity using prospective gathered nasopharyngeal samples (grey dots) and QCMD samples (white dots). Crossing point (Cp) values of single target assays (i.e. singleplex) are plotted against Cp values of multiplex analysis, for every individual virus species. Dots with a Cp > 40 represent discrepant PCR results (i.e. sole detection by one of the assays).
Table 4.
Discrepancies.
| Virus | Single target | Multiplex | Remarks |
|---|---|---|---|
| Cp value | Cp value | ||
| hBoV | 39.1 | neg | Co-infection with RV (Cp 27.76) |
| hBoV | 38.4 | neg | Co-infection with ADV (Cp 36.64) |
| hBoV | 37.35 | neg | No remarks |
| hBoV | 36.48 | neg | No remarks |
| hBoV | neg | 37.25 | Co-infection with RSV (Cp 28.83) |
| hBoV | neg | 37.1 | Co-infection with RV (Cp 31.68) |
| hBoV | neg | 36.94 | Co-infection with CoV (Cp 34.11) |
| EV | 39.11 | neg | Co-infection with RV (Cp 26.55) & ADV (Cp 37.83) |
| EV | 39.06 | neg | Co-infection with ADV (Cp 34.17) |
| EV | 38.47 | neg | Co-infection with RV (Cp 27.13) |
| EV | 36.25 | neg | Co-infection with RV (Cp 27.04) |
| EV | 33.92 | neg | Co-infection with RV (Cp 26.27) & ADV (Cp 33.23) |
| EV | 33.08 | neg | Co-infection with RV (Cp 26.33) |
| ADV | 37.83 | neg | Co-infection with RV (Cp 26.55) & EV (Cp 39.11) |
| hMPV | neg | 35.94 | Co-infection with Inf A (Cp 23.65) |
| PIV 3 | neg | 35.14 | No remarks |
| hPeV | 33.32 | neg | No remarks |
| CoV | neg | 38.11 | Co-infection with ADV (Cp 28.38) |
| RSV | 38.45 | neg | No remarks |
Median Cp-values in clinical samples of the single-target assay was 29.7 (Inter Quartile Range [IQR]; 27.5–33.3) which did not differ statistically from the median Cp-value of the multiplex assay 28.8 (IQR 26.9–32.0) (p = 0.2). Species specific analyses did also not show statistical differences for median Cp-values. However, somewhat lower Cp-values for the multiplex assay were observed for InfB, RSV and RV in samples with high Cp-values (Fig. 2).
Analyses of quality controls was performed to test viral targets that were less prevalent (i.e. PIV 1, 2, 3, 4, InfB and hCoV) in the clinical samples. Results are summarized in Table 3 and Fig. 2. Complete concordance was observed between the multiplex assay and the individual single-target assays for quality controls.
5. Discussion
Real time multiplex PCR is a technique that has been shown to be fast and cost effective for the diagnosis of respiratory viral infections. Most papers on respiratory multiplex PCR's use traditional methods like virus culture, antigen tests or direct immune fluoresce as comparison,8 hence questions can be raised about their diagnostic accuracy compared to sensitive single-target assays. We report the development and the extensive validation of an internally controlled four-tube RT rtPCR for the detection of 14 different viruses associated with respiratory infections.
5.1. Sensitivity
Combining primers and probes in a single reaction can influence the sensitivity of the assay as primer–primer interactions may lead to a lower availability of specific primers. In addition, the presence of multiple targets in one reaction may result in competition for enzymes and nucleotides. Analysis of dynamic ranges and mixed input of plasmids, revealed no loss of sensitivity. In fact, for most viruses Cp-values were slightly, but significantly, lower in the multiplex analyses. Most likely, this difference is due to the algorithm used by the LC480 software to calculate crossing point. The LC480 software calculates the second derivatives of the entire amplification curve and determines where this value is at its maximum. This value is the Cp-value and represents the cycle at which the increase of fluorescence is highest (i.e. the start logarithmic phase of the PCR). Using Cp values by the second derivative maximum of the amplification curve is known to be more precise than cycle thresholds values or other crossing point algorithms.12 We have observed that in our multiplex reactions the total fluorescence is slightly lower and the slope of the amplification curve is less steep as compared to the corresponding single-target PCR's (which is most likely due to inevitable minute differences in PCR reaction mixtures of singleplex versus multiplex pcr's). This difference could lead to slightly lower Cp-values for the multiplex reactions, but does not represent a difference in absolute sensitivity.
In prospectively collected clinical samples the multiplex PCR showed concordant results to the single-target PCR's. For InfA, hCoV, PIV1, PIV2, PIV3, PIV 4, hMPV, AdV and hPeV Cp-values were nearly identical for multiplex and single-target assays. However for certain viruses (InfB, PIV 1, 2, 3 & 4, hPeV, and hCoV) the number of clinical samples was relatively small and extensive comparisons could be merely performed in the mixed input of plasmids analysis, but not by clinical samples. For RSV, RV and InfB multiplex showed slightly lower Cp-values than the single target PCR, particularly when single-target Cp-values were ≥30. This is probably due to a comparable phenomenon as described for slightly lower multiplex Cp-values in the plasmid evaluation. Discrepancies in clinical samples were mostly seen for hBoV and EV. Samples with a very low hBoV load (Cp-value > 36) were variable positive in either multiplex or single-target assay. This seems to represent a stochastic process around the detection limit for both multiplex and single-target assays. Most of the EV positive respiratory samples, with Cp-values higher than 33 in single-target assays were negative in the multiplex assay, suggesting reduced sensitivity of EV detection of our multiplex assay. Additional analysis of dilution series of 4 different EV subtypes (representing group A, B, C and D EV's) showed concordant results for EV Cp-values in the multiplex and the singleplex assay (data not shown). However the fluorescence plateau's of the EV multiplex assay was notably lower compared to its singleplex equivalent. It might be possible that in samples with low abundance of multiple viral targets this lower fluorescence could lead to false-negativity. In this respect, it is noteworthy that all clinical EV samples that were “missed” by the multiplex PCR were samples positive two or three respiratory viruses (Table 4).
5.2. Specificity
Another explanation the missing EV in the multiplex PCR could be due to possible cross reactivity between EV PCR and RV. The EV primers and probes we used were previously developed in our department and described by Beld et al.9 EV specificity was assessed in this publication and did not show any cross-reactivity with a panel of culture typed rhinoviruses. Since then, we have also tested the EV PCR against a panel of RV type C virus isolates (N = 9) which showed absent of cross reactivity against these viruses as well. Morever extensive in silico analysis (data not shown) did not show cross reactivity between our EV primers and RV sequences. However, as RV comprises over a hundred subtypes we cannot completely rule out cross reactivity with less common HRV strains. In literature cross-reactivity of molecular test between rhinovirus and enterovirus is not uncommon13 and some authors even suggest that complete specificity for Rhinovirus and Enterovirus in impossible.14 Yet the ongoing discovery of new respiratory EV's implies that separate analyses of EV in respiratory samples is of clinical importance15 and optimization RV and EV pcr specificity is an important topic for future study.
In addition when addressing specificity issues in multiplex PCR: false positive results due to fluorescence cross-talk from one detection channel into other detection channels one was not seen in analysis with mixed plasmid input. Furthermore false positivity in the multiplex by other causes, like non-specific primer–target interactions, was studied by analyzing prospective clinical samples (including viral negative samples) of which the results were not known before testing. Some discrepancies were seen in which positive multiplex results were not confirmed by single-target testing. Most of these samples were hBoV positive samples with Cp-values > 36 and are probably due to a stochastic process at the detection limit, as mentioned earlier.
In summary, we present here an extensively evaluated multiplex PCR assay for the detection of 14 major causative agents of respiratory tract disease. Good concordance was observed between multiplex and single target analysis, except for clinical samples with low level of enterovirus. The assay is internally controlled, does not need post-PCR manipulations and can be used in routine diagnostic laboratories, providing a fast and proper validated way for the detection of respiratory viruses.
Conflicts of interest
None.
Acknowledgments
The authors want to thank Dr. E.C. Claas (Leiden University Medical Centre, The Netherlands) for kindly making available sequences of primers and probes for the detection of EAV and hBoV.
References
- 1.Brittain-Long R., Nord S., Olofsson S., Westin J., Anderson L.M., Lindh M. Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol. 2008;41(1):53–56. doi: 10.1016/j.jcv.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahony J.B., Blackhouse G., Babwah J., Smieja M., Buracond S., Chong S. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol. 2009;47(9):2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Ren P., Sheng J., Mardy S., Yan H., Zhang J. Simultaneous detection of respiratory viruses in children with acute respiratory infection using two different multiplex reverse transcription-PCR assays. J Virol Methods. 2009;162(1–2):40–45. doi: 10.1016/j.jviromet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freymuth F., Vabret A., Cuvillon-Nimal D., Simon S., Dina J., Legrand L. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006;78(11):1498–1504. doi: 10.1002/jmv.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Elden L.J., van Kraaij M.G., Nijhuis M., Hendriksen K.A., Dekker A.W., Rozenberg-Arska M. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis. 2002;34(2):177–183. doi: 10.1086/338238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Pol A.C., van Loon A.M., Wolfs T.F., Jansen N.J., Nijhuis M., Breteler E.K. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J Clin Microbiol. 2007;45(7):2260–2262. doi: 10.1128/JCM.00848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puppe W., Weigl J.A., Aron G., Grondahl B., Schmitt H.J., Niesters H.G. Evaluation of a multiplex reverse transcriptase PCR ELISA for the detection of nine respiratory tract pathogens. J Clin Virol. 2004;30(2):165–174. doi: 10.1016/j.jcv.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Gunson R.N., Collins T.C., Carman W.F. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J Clin Virol. 2005;33(4):341–344. doi: 10.1016/j.jcv.2004.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beld M., Minnaar R., Weel J., Sol C., Damen M., van der Avoort H. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J Clin Microbiol. 2004;42(7):3059–3064. doi: 10.1128/JCM.42.7.3059-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheltinga S.A., Templeton K.E., Beersma M.F., Claas E.C. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J Clin Virol. 2005;33(4):306–311. doi: 10.1016/j.jcv.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molenkamp R., van der H.A., Schinkel J., Beld M. Simultaneous detection of five different DNA targets by real-time Taqman PCR using the Roche LightCycler480: application in viral molecular diagnostics. J Virol Methods. 2007;141:205–211. doi: 10.1016/j.jviromet.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Wittner C.T., Kusukawa N. Real-Time PCR. In: Persing D.H., editor. Molecular microbiology: diagnostic principles and practice. ASM Press; Washington, DC: 2004. p. 71–4. [Google Scholar]
- 13.Lu X., Holloway B., Dare R.K., Kuypers J., Yagi S., Williams J.V. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46(2):533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tapparel C., Junier T., Gerlach D., Van-Belle S., Turin L., Cordey S. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis. 2009;15(5):719–726. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tapparel C., Cordey S., Van B.S., Turin L., Lee W.M., Regamey N. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J Clin Microbiol. 2009;47(6):1742–1749. doi: 10.1128/JCM.02339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]