Abstract
Background
Human rhinoviruses (HRVs) are associated with more acute respiratory tract infections than any other viral group yet we know little about viral diversity, epidemiology or clinical outcome resulting from infection by strains, in particular the recently identified HRVs.
Objectives
To determine whether HRVC-QCE was a distinct HRV-C strain, by determining its genome and prevalence, by cataloguing genomic features for strain discrimination and by observing clinical features in positive patients.
Study design
Novel real-time RT-PCRs and retrospective chart reviews were used to investigate a well-defined population of 1247 specimen extracts to observe the prevalence and the clinical features of each HRV-QCE positive case from an in- and out-patient pediatric, hospital-based population during 2003. An objective illness severity score was determined for each HRVC-QCE positive patient.
Results
Differences in overall polyprotein and VP1 binding pocket residues and the predicted presence of a cis-acting replication element in 1B defined HRVC-QCE as a novel HRV-C strain. Twelve additional HRVC-QCE detections (1.0% prevalence) occurred among infants and toddlers (1–24 months) suffering mild to moderate illness, including fever and cough, who were often hospitalized. HRVC-QCE was frequently detected in the absence of another virus and was the only virus detected in three (23% of HRVC-QCE positives) children with asthma exacerbation and in two (15%) toddlers with febrile convulsion.
Conclusions
HRVC-QCE is a newly identified, genetically distinct HRV strain detected in hospitalized children with a range of clinical features. HRV strains should be independently considered to ensure we do not overestimate the HRVs in asymptomatic illness.
Keywords: Rhinovirus, Acute respiratory tract infection, HRV C, Wheeze, Respiratory virus epidemiology, Virus discovery
1. Background
Acute respiratory tract infections (ARTIs), a major cause of human morbidity and hospitalization, are most frequently viral in origin. The human rhinoviruses (HRVs) have for over a decade been associated, more than any other individual factor, with asthma1, 2, 3 and chronic obstructive pulmonary disease exacerbations4 and the majority of cold and flu-like illnesses (CFLIs).5, 6 It has been shown that HRVs infect both upper and lower respiratory tract tissues.7, 8 Classical serotypes (HRV-1 to 100) or “strains”, the term we use to indicate a molecular equivalent, form two accepted species within the heavily populated genus Enterovirus, family Picornaviridae. Despite the considerable economic burden of HRV infections,9 deliberate and comprehensive investigation of strain-specific clinical data is lacking.
In 2006 we proposed that a clade of divergent sequences belonged to a novel viral subgroup, HRV-A2,10 which we later redefined11 as a putative third species, HRV-C, in agreement with Lau et al.12 To date, nine complete polyprotein sequences of HRV-C strains have been determined from Australia (HRVC-QPM10), the United States (HRVC-NAT001/X-1, HRVC-NAT045/X-213 and HRVC-NY07414), Hong Kong (HRVC-C024, HRVC-C025 and HRVC-C02612) and Shanghai (HRVC-N4 and HRVC-N1015).
Many HRV strains have distinct genetic, antigenic, immunogenic and epidemiologic profiles which are similar in nature to those of other respiratory virus species including human respiratory syncytial virus (HRSV). The prospect of variation in sensitivity to antivirals is also an important reason to investigate each novel strain comprehensively, as is the chance that infection by specific HRV species could be associated with more severe illness or particular pre-existing conditions. Disappointingly, the possibility that discrete HRV strains exist as distinct viral entities is not accounted for in asymptomatic control populations because strain identification is not routinely conducted. As a consequence elevated prominence is, by default, assigned to HRVs as if to a single virus.
2. Objectives
We sought to genomically characterize HRV-QCE, an apparently distinct HRV-C strain whose novel subgenomic sequences were first detected in a child with exacerbated asthma and to define distinct molecular features among the HRV-Cs that are useful in discriminating strains for future studies and observe the clinical features amongst HRVC-QCE positive patients.
3. Study design
Clinical specimens (n = 1247; three additions were made to the previously described HRVC-QPM population10) were predominantly nasopharyngeal aspirates (NPA; 93%) from patients (59% male) aged 1 day to 80 years (mean 9.2 years, median 1.3 years) presenting to Queensland hospitals with symptoms of ARTI during 2003. Extracts were stored and tested as previously described.10 A clinical illness severity scoring system was applied as previously described.11 This study was approved by the Royal Children's Hospital and University of Queensland Medical Research Ethics Committees (specific approvals #2006/097 and #2008/020 and #2007000004).
Primers for genome deduction were first designed using early HRVC-QCE sequences in 1A/1B (EU131891 and EU155152). All oligonucleotide sequences are available upon request. Additional primers supplemented the process.10, 12, 16 To confirm our primary sequence belonged to a distinct virus and was not the result of mixed HRV template, a second round of RT-PCR and sequencing of overlapping fragments was performed on RNA from the original patient specimen using newly designed HRVC-QCE-specific primers. Discrepant sequence results were confirmed by additional rounds of RT-PCR and sequencing. Two specific diagnostic real-time RT-PCRs (RT-rtPCRs) were performed on the Rotor-Gene™ 3000 (Corbett Research) for diagnostic screening.
Polyprotein protease cleavage sites were sought using the NetPicoRNA server.17 Cis-acting replication element (cre) sequences18 were sought using the RNAalifold server and structures were predicted using MFold.19
4. Results
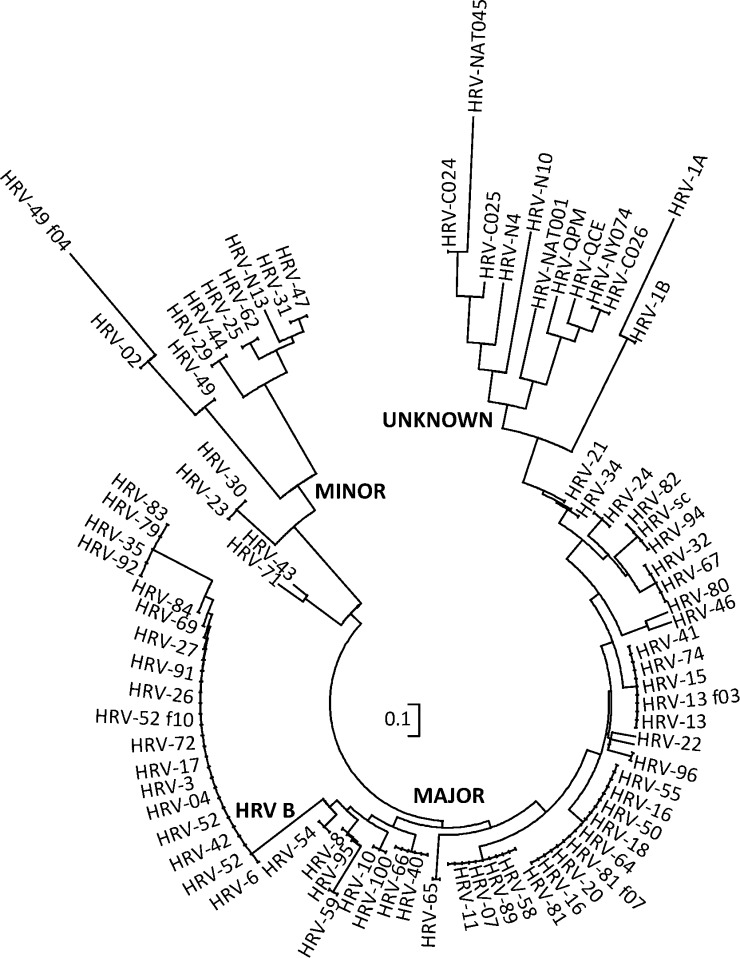
The polyprotein encoding sequence (GenBank #GQ323774) of HRVC-QCE, a new member of the species Human rhinovirus C, spanned 6423 nt; shorter than all HEVs and HRVs except HRV-NY074.14, 20, 21 It had a G + C content of 43% and 2140 predicted amino acids terminating in an HRV-C-specific isoleucine.11 The HRVC-QCE polyprotein shared 54% average amino acid identity with HRV-A strains, 50% with HRV-Bs and 79% with other HRV-Cs. Ten protease cleavage sites were predicted to reduce the HRVC-QCE polyprotein into typical picornavirus structural (VP1-4) and non-structural proteins (2A-C and 3A-D17, 22, 23). HRVC-QCE shared a translation initiation site (MGAQVS) with most other HRVs and three motifs crucial for RNA polymerase binding (YGDD, TFLKR and SIRWT) shared by HRVs, and HEVs24, 25 but lacked two of three exposed VP1 motifs previously conserved among HRV polyproteins26 were absent in HRV-QCE. Phylogeny inferred from a discontinuous alignment of 10 amino acids in VP3 and VP1 which comprise the predicted intercellular adhesion molecule (ICAM)-1 footprint27 in major group HRV strains, placed HRVC-QCE amongst other HRV-Cs on a branch with HRV-1 (a minor group HRV which employs a different receptor) but distinct from the known major (employing the ICAM-1 molecule as receptor) or remaining minor group HRV strains (Fig. 1 ). Key residues on the VP1 BC loop involved in receptor contact with the VLDLR19 are missing in HRVC-QCE and there are differences in the HI loop compared to the classical strains. Antigenic site A28 is absent in HRVC-QCE and the sequence at site B is unique to HRVC-QCE. As with other HRV-C strains identified to date,18 the conserved loop sequence motif constituting a cre, R 1NNNA1A2 R 2NNNNNNR 3 was identified (GCUCAAGCAAAUCA) in 1B of HRVC-QCE in the context of an appropriate predicted RNA stem-loop structure (not shown).
Fig. 1.
Alignment of HRV amino acids in the regions that are known to interact with ICAM-1. A circular representation of HRV strains (some strain titles removed for clarity) created from a set of discontinuous VP3 and VP1 amino acids reflecting the ICAM-1 footprint. Nucleotide and amino acid alignments using Muscle (64 iterations) were created in Geneious Pro 4.7.35 Neighbour-joining consensus trees constructed in Mega 4.1 were used to infer phylogeny.
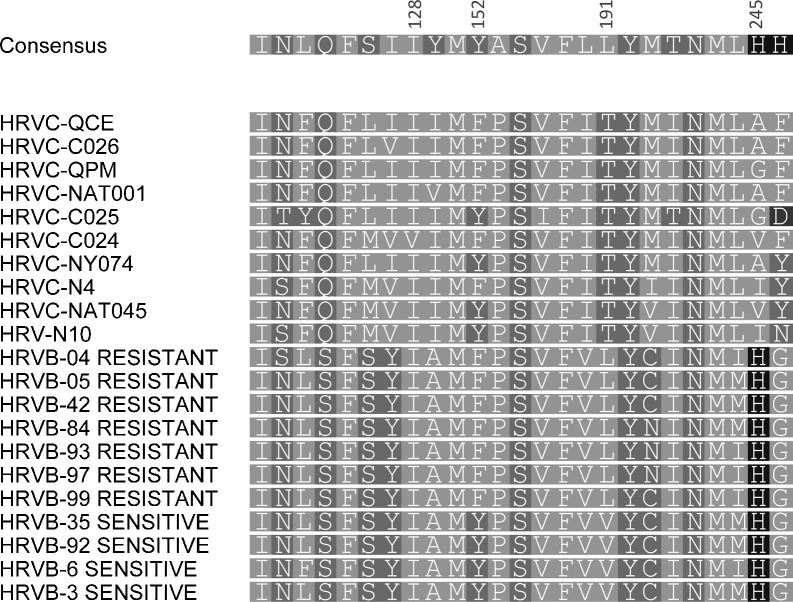
All classical HRV-A strains and 18/25 HRV-B strains are susceptible to the antiviral compound, pleconaril. Among the 25 key contact residues lining the antiviral binding pocket, there were 11 or 12 HRVC-QCE-specific differences compared to HRV-A or HRV-B consensus sequences,29 respectively (Fig. 2 ); 11 differences exist between HRV-As and Bs and two differences between the consensus sequence of HRV-B sensitive versus resistant strains. Changes at two HRVC-QCE residues historically important for identifying naturally resistant strains were identified as Tyr152 (in resistant strains) to Phe152 and Val191 to Thr191. His245, conserved in nearly all other classical HRV strains, varied in HRVC-QCE and among other HRV-Cs. Other conserved HRV residues including Ile/Leu106 and Tyr/Phe/Ala150, differed in HRVC-QCE (Fig. 2).
Fig. 2.
Alignment of the 25 key VP1 amino acids making contact with pleconaril. Representative HRV-A, -B and -C strains were included. The consensus amino acid sequence and a graphical representation of the contribution of residues occupying each position are shown above the alignment. Consensus residue numbering is from Ref. 30.
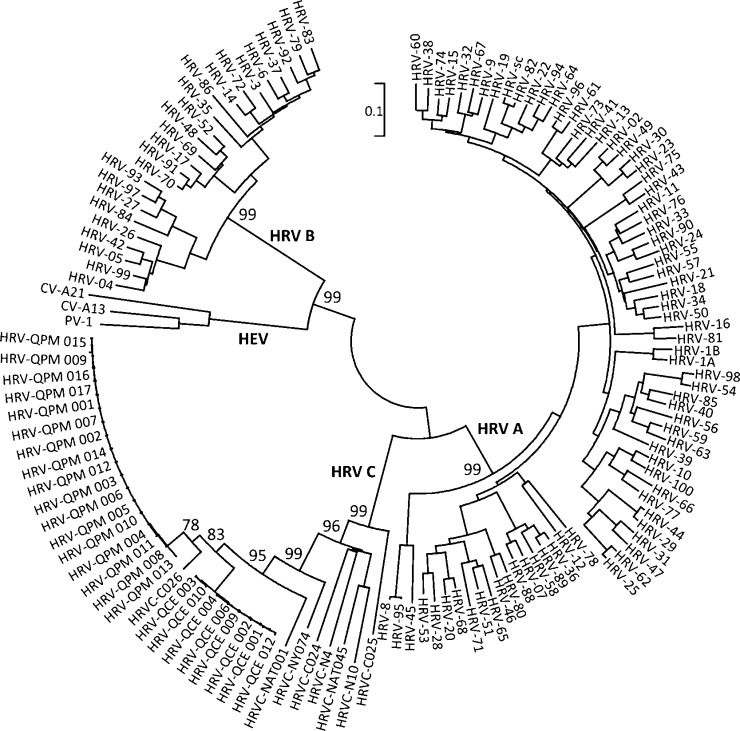
To examine whether HRVC-QCE was a variant of a previously identified HRV strain, we first compared the full polyprotein against public sequence databases. The nearest matches were HRVC-C026 (93.5% identity, 2003 identical amino acids) and HRVC-QPM (92.9%, 1991 amino acids). The inferred amino acid sequences from complete 1D regions of eight HRVC-QCE-positive extracts (variants 001–004, 006, 009, 010, 012; intra-strain amino acid identity, 99.5%; GenBank accession number GQ323774 and bankit numbers, 1304337, 1304336, 1304343, 1304341, 1304344, 1304335, 1304320) were compared to those from known HRV and HEV serotypes and to previously described HRVC-QPM variants (n = 17; intra-strain identity of 99.8%; Fig. 3 ). The amino acid identity between variants of the HRVC-QPM and HRVC-QCE strains (91.2%) was lower than among variants of the two strains (≥99.5%). At least 93% polyprotein-length pairwise identity is also shared by 80% (n = 60) of antigenically distinct and officially recognised HRV-A strains (60 strains occur in at least one pairing in which sequence identity is ≥93%; HRV-8 and 95, HRV-1A and 1B and HRV-82 and sc were not considered discrete viruses). The HRVC-QCE VP1 (272 aa) is 9–24 amino acids shorter than that of other HRV serotypes. The HRVC-QCE residues aligning with the 10 residue ICAM-1 footprint differed by one from those of the most similar strain (HRVC-C026). In VP1, two of 6 amino acids from antigenic site B, four of 20 from the EF loop, one of 3 from the FG loop, two of 32 from the GH loop, three of 8 from the HI loop, one of 24 constituting the binding pocket and two of 48 nt in the 1B cre differed from the most similar HRV-C strain.
Fig. 3.
Alignment of complete VP1 coding sequences. Complete VP1 sequences of known HRV-A, -B, -C and representative HEV strains (those included in Palmenberg et al.; f-field strain). The HRVC-QCE (GenBank accession number GQ323774 and bankit numbers, 1304337, 1304336, 1304343, 1304341, 1304344, 1304335, 1304320) and HRVC-QPM strain variants are indicated with three-digit numbers. Phylogeny was inferred by using MEGA 4.1. Evolutionary distances are in the units of the number of amino acid substitutions per site (243 positions in the final dataset). Rooted on the midpoint. Only early bootstrap (n = 500) support values are shown. PV = Human Poliovirus, CV-A = Coxsackievirus A.
Since our description of HRVC-QCE 1A/1B sequences in 2006 (EU131891 = QPID03-0076/EU155152 = QPID03-0035, representing HRVC-QCE strain variants 006/002, respectively) we herein note that detections of a similar viral strain, based on our comparisons of the highly conserved VP4 sequences (100% amino acid identity), have been made in China (EU687526 = 112807/EU687524 = 111007 and EU822837 = hRVLz124), Finland (EU590049 = PNC40135), South Africa (ACE77006 = SA365412), Thailand (ACR44015 = HRVCU156) and the United States (ABQ16562 = HRVC-NAT001; DQ875928 = NY60). The complete polyprotein of most of these strains has not been described and for the one that has (HRVC-NAT001), no additional variants were sought and only preliminary clinical studies were performed.
HRVC-QCE RNA was detected in 1.0% (n = 13) of 1247 specimen extracts collected from January to December 2003 using a 1B targeted RT-rtPCR and confirmed by an assay targeting the 2C region. No control patients were sampled in 2003. HRVC-QCE prevalence was bimodal with the major peak in summer when 3.5% of specimens were positive (7.6% of 119 specimens from February were positive representing 76.9% of all HRVC-QCE positives and 16.1% of all virus detections (n = 56) that month). HRVC-QCE was the only HRV detected in January. The remaining 23.1% of HRVC-QCE positives were detected in winter (0.8% of specimens, peaking at 3.8% of all viral detections in June). No other viruses were detected in 61.5% (n = 8) of HRVC-QCE positives (Table 1 ) leaving five extracts in which another viral sequence was identified. Respiratory picornaviruses were the most frequently detected virus at 40.9% of detections (n = 351) followed by HBoV (11.8%, n = 101), HRSV (8.3%, n = 71) and HMPV (7.7%, n = 66). No viruses were found in 46.0% (n = 574) of extracts. Isolation of HRVC-QCE in culture was not attempted because of the age of the clinical specimens.
Table 1.
HRVC-QCE detections, severity of respiratory illness and diagnosis.
| Diagnosis and HRVC-QCE variant | Age in months (number and proportion of HRVC-QCE detections)/sex and pre-existing conditions | Other viruses co-detected | Hospital admission | Cough/fever | Severity score |
|---|---|---|---|---|---|
| Expiratory wheezing (n = 5; 38.5%) | |||||
| HRVC-QCE 001 | 42/Ma | Y | Y/N | 2 | |
| HRVC-QCE 004 | 24/Fa,b | Y | Y/Y | 1 | |
| HRVC-QCE 005 | 10/Ma | HBoV | Y | Y/N | 3 |
| HRVC-QCE 006 | 18/Ma | HBoV; KIPyV | Y | Y/N | 1 |
| HRVC-QCE 007 | 12/Ma | Y | Y/N | 2 | |
| Transplant or underlying conditions (n = 5; 38.5%) | |||||
| HRVC-QCE 003 | 10/F ALL. Deceasedb | Y | Y/N | 3 | |
| HRVC-QCE 010 | 9/F AMLb | Y | N/N | 1 | |
| HRVC-QCE 009 | 5/M CFb | Y | Y/N | 2 | |
| HRVC-QCE 013 | 26/M CFb | HRSV | N | Y/NR | 0 |
| HRVC-QCE 012 | 7/M Lung disease of prematurity; bronchiolitisa | HBoV | Y | NR/N | 1 |
| Febrile convulsion (n = 2; 15.4%) | |||||
| HRVC-QCE 008 | 20/Fb | Y | Y/Y | 2 | |
| HRVC-QCE 011 | 15/M | HBoV | Y | N/Y | 1 |
| Upper respiratory tract infection (n = 1; 7.7%) | |||||
| HRVC-QCE 002 | 9/F | N | Y/Y | 0 | |
ALL = acute lymphoblastic leukemia; AML = acute myeloblastic leukemia; CF = cystic fibrosis; HBoV = human bocavirus; HRSV = human respiratory syncytial virus; KIPyV = KI polyomavirus; M = male; F = female; NR = not recorded in chart.
Bronchodilators.
Antibiotics.
Chart review of the 13 HRVC-QCE positive cases revealed presentation with lower respiratory tract illnesses (6/13; 46.2%), most commonly involving wheeze and administration of bronchodilators (Table 1). Antibiotics were not usually given to wheezers compared to non-wheezers. Eleven of the 13 cases (84.6%) were admitted to hospital and HRVC-QCE was the sole detection in 61.5% (8/13) of patients. The average severity score among single HRVC-QCE detections was higher (1.6) than that for co-detections (1.2) and was higher in summer (1.6) than in winter (1.0). Average scores were highest among lower respiratory tract presentations (1.8) and scores of 0 were obtained from an upper respiratory tract illness and a cystic fibrosis patient with an HRSV co-detection. There were no adult cases, despite adults comprising 15.8% of the screened population. Most (84.6%) patients were 2 years of age or younger.
5. Discussion
We have described the detection, polyprotein coding region and distinguishing genetic features of a newly identified and evidently genetically distinct HRV-C strain. We have summarised the clinical features of a predominantly pediatric population from whom HRVC-QCE variants were detected. The relatively shortened genome with truncations and sequence diversity in the 1D capsid region may affect receptor usage, antigenicity and sensitivity to antivirals. Phe152 in VP1 is a distinctive feature of pleconaril resistant clinical HRV-B strains,30 and was present in the VP1 of HRVC-QCE as was Thr191, which is found in both resistant and sensitive strains. The predicted HRV-C-specific cre motif in 1B, the terminal isoleucine, the comparatively elevated G + C content and other features we have previously defined,11 identified HRVC-QCE as a candidate for the proposed new species HRV-C.
HRVC-QCE appears to be a distinct global pathogen however no consensus molecular criteria exist which define strains or provide identity thresholds below which a clinical detection may be defined as a novel strain nor above which it may best be called a variant of a previously described strain.11 The HRV literature notes infections which apparently have not elicited a cross-protective immune response as demonstrated by patients infected by distinct strains of the same or different species such over a defined period which share 62–93% amino acid identity in VP4.12, 31, 32 These biological data together with strain-determining genetic criteria permit the proposal of a practical molecular threshold, for example ≤93% amino acid identity, that could be used as a surrogate to define the biological distinctiveness of HRVs that cannot be cultured. Strain-defining criteria are especially useful to determine whether it is more accurate to consider “the HRVs” as a single group or as a collection of related but mostly antigenically discrete viruses, as is the norm for HRSV or HMPV for example. Applying strain-defining criteria to control populations may very likely redefine our concept of the HRVs in asymptomatic illness, since the proportion of specimens positive for any distinct HRV strain in controls will be less than that which is positive for the HRV super-group when considered as a whole.
Approximately half of HRVC-QCE detections were from patients with lower respiratory tract symptoms and 85% of HRVC-QCE positives were detected from hospitalized children. HRVC-QCE was the sole virus detected in over half of all cases.
The paucity of sequence data, strain-defining criteria and clinical investigations of the novel HRVs or of individual classical strains parallel our limited knowledge of the extent and clinical impact of HRV diversity. The most recent significant healthcare impact attributed to HRVs was the 2009 H1N1v influenza pandemic when HRVs were the most common viruses detected among patients qualifying, but laboratory-negative for, pandemic virus testing.33 Similar findings were described during the SARS-CoV outbreak.34 A lack of viral sequence data weakens our differential diagnostic and overall detection capabilities which limits our ability to define epidemiological features as well as the occurrence of strain-, or species-specific clinical outcomes and potentially differentiating genetic features. Due to the retrospective nature of our study, no temporally comparable control population was available for testing. This is a limitation among most other HRV studies as is the current inability to culture the newly identified HRVs. When a control population is included however, strain typing is usually not conducted and so a higher proportion of asymptomatic illness is attributed to all HRVs as a total group. Additionally, because we and others cannot fulfil Koch's postulates for the currently unculturable HRV-Cs, the clinical observations cannot be specifically associated with the PCR-determined presence of HRVC-QCE RNA. Our findings highlight the need to better understand the genomics, taxonomy, epidemiology and clinical impact of the newly identified HRV strains to accurately quantify the scope of their clinical impact as distinct respiratory viruses.
Competing interests
None.
Acknowledgments
We are grateful to Patrick Woo and Susanna Lau for primer and HRV strain sequences and to Daniel Gerlach for bioinformatics support. We thank Pathology Queensland Central for the provision of clinical specimens.
Funding. This work was supported by an Australian NHMRC Project grant 455905.
References
- 1.Chauhan A.J., Inskip H.M., Linaker C.H., Smith S., Schreiber J., Johnston S.L. Personal exposure to nitrogen dioxide (NO2) and the severity of virus-induced asthma in children. Lancet. 2003;361(9373):1939–1944. doi: 10.1016/S0140-6736(03)13582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston S.L., Pattemore P.K., Sanderson G., Smith S., Lampe F., Josephs L. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. Br Med J. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakes G.P., Arruda E., Ingram J.M., Hoover G.E., Zambrano J.C., Hayden F.G. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care. Am J Resp Crit Care Med. 1999;159:785–790. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- 4.Johnston S.L. Overview of virus-induced airway disease. Proc Am Thorac Soc. 2005;2(2):150–156. doi: 10.1513/pats.200502-018AW. [DOI] [PubMed] [Google Scholar]
- 5.Rotbart H.A., Hayden F.G. Picornavirus infections: a primer for the practitioner. Arch Fam Med. 2000;9:913–920. doi: 10.1001/archfami.9.9.913. [DOI] [PubMed] [Google Scholar]
- 6.Ruohola A., Waris M., Allander T., Ziegler T., Heikkinen T., Ruuskanen O. Viral etiology of common cold in children, Finland. Emerg Infect Dis. 2009;15(2):344–346. doi: 10.3201/eid1502.081468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arruda E., Boyle T.R., Winther B., Pevear D.C., Gwaltney J.M., Jr., Hayden F.G. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J Infect Dis. 1995;171(5):1329–1333. doi: 10.1093/infdis/171.5.1329. [DOI] [PubMed] [Google Scholar]
- 8.Jakiela B., Brockman-Schneider R., Amineva S., Lee W.-M., Gern J.E. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am J Respir Cell Mol Biol. 2008;38(5):517–523. doi: 10.1165/rcmb.2007-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert S.B., Allen K.M., Carter R.C., Nolan T.M. The cost of community-managed viral respiratory illnesses in a cohort of healthy preschool-aged children. Resp Res. 2008;9(1):1–11. doi: 10.1186/1465-9921-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McErlean P., Shackleton L.A., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McErlean P., Shackelton L.A., Andrewes E., Webster D.R., Lambert S.B., Nissen M.D. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS One. 2008;3(4):e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau S.K.P., Yip C.C.Y., Tsoi H.-W., Lee R.A., So L.-Y., Lau Y.-L. Clinical features and complete genome characterization of a distinct human rhinovirus genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45(11):3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmenberg A.C., Spiro D., Kuzmickas R., Wang S., Djikeng A., Rathe J.A. Sequencing and analyses of all known human rhinovirus genomes reveals structure and evolution. Science. 2009;324(5923):55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang T., Wang W., Bessaud M., Ren P., Sheng J., Yan H. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4(7):e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmonds P., Welch J. Frequency and dynamics of recombination within different species of human enteroviruses. J Virol. 2006;80(1):483–493. doi: 10.1128/JVI.80.1.483-493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blom N., Hansen J., Blaas D., Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Prot Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordey S., Gerlach D., Junier T., Zdobnov E.M., Kaiser L., Tapparel C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA. 2008;14(8):1568–1578. doi: 10.1261/rna.1031408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol. 2004;11(5):429–434. doi: 10.1038/nsmb753. [DOI] [PubMed] [Google Scholar]
- 20.Lukashev A.N., Lashkevich V.A., Ivanova O.E., Koroleva G.A., Hinkkanen A.E., Ilonen J. Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J Gen Virol. 2005;86:3281–3290. doi: 10.1099/vir.0.81264-0. [DOI] [PubMed] [Google Scholar]
- 21.Oberste M.S., Maher K., Schnurr D., Flemister M.R., Lovchik J.C., Peters H. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol. 2004;85:2577–2584. doi: 10.1099/vir.0.79925-0. [DOI] [PubMed] [Google Scholar]
- 22.Skern T., Sommergruber W., Blaas D., Gruendler P., Fraundorfer F., Pieler C. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13(6):2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanway G., Hughes P.J., Mountford R.C., Minor P.D., Almond J.W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12(20):7859–7877. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savolainen C., Laine P., Mulders M.N., Hovi T. Sequence analysis of human rhinoviruses in the RNA-dependent RNA polymerase coding region reveals within-species variation. J Gen Virol. 2004;85(8):2271–2277. doi: 10.1099/vir.0.79897-0. [DOI] [PubMed] [Google Scholar]
- 25.Brown B., Oberste M.S., Maher K., Pallansch M.A. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J Virol. 2003;77:8973–8984. doi: 10.1128/JVI.77.16.8973-8984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laine P., Blomqvist S., Savolainen C., Andries K., Hovi T. Alignment of capsid protein VP1 sequences of all human rhinovirus prototype strains: conserved motifs and functional domains. J Gen Virol. 2006;87(1):129–138. doi: 10.1099/vir.0.81137-0. [DOI] [PubMed] [Google Scholar]
- 27.Blomqvist S., Savolainen-Kopra C., Paananen A., Hovi T., Roivainen M. Molecular characterization of human rhinovirus field strains isolated during surveillance of enteroviruses. J Gen Virol. 2009;90(Pt 6):1371–1381. doi: 10.1099/vir.0.008508-0. [DOI] [PubMed] [Google Scholar]
- 28.Verdaguer N., Blaas D., Fita I. Structure of human rhinovirus serotype 2 (HRV2) J Mol Biol. 2000;300:1179–1194. doi: 10.1006/jmbi.2000.3943. [DOI] [PubMed] [Google Scholar]
- 29.Ledford R.M., Patel N.R., Demenczuk T.M., Watanyar A., Herbertz T., Collett M.S. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78(7):3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ledford R.M., Collett M.S., Pevear D.C. Insights into the genetic basis for natural phenotypic resistance of human rhinoviruses to pleconaril. Antiviral Res. 2005;68(3):135–138. doi: 10.1016/j.antiviral.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Linsuwanon P., Payungporn S., Samransamruajkit R., Theamboonlers A., Poovorawan Y. Recurrent human rhinovirus infections in infants with refractory wheezing. Emerg Infect Dis. 2009;15(6):978–980. doi: 10.3201/eid1506.081558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau S.K., Yip C.C., Lin A.W., Lee R.A., So L.Y., Lau Y.L. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis. 2009;200(7):1096–1103. doi: 10.1086/605697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Follin P., Lindqvist A., Nystrom K., Lindh M. A variety of respiratory viruses found in symptomatic travellers returning from countries with ongoing spread of the new influenza A(H1N1)v virus strain. Euro Surveill. 2009;14(24) doi: 10.2807/ese.14.24.19242-en. [DOI] [PubMed] [Google Scholar]
- 34.Schrag S.J., Brooks J.T., Van B.C., Parashar U.D., Griffin P.M., Anderson L.J. SARS surveillance during emergency public health response, United States, March–July 2003. Emerg Infect Dis. 2004;10(2):185–194. doi: 10.3201/eid1002.030752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetic analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]