Abstract
Infectious diarrhea is the most frequent cause of morbidity and mortality in neonatal calves. Cryptosporidium parvum is one of the main pathogens associated with calf diarrhea. Although diarrhea is a symptom of infection with various pathogens, investigations to detect the types of pathogens have never been performed in Japan. This study investigated the prevalence of four major diarrhea-causing pathogens in calves: C. parvum, rotavirus, coronavirus, and enterotoxigenic Escherichia coli (E. coli K99). Commercial immunochromatography testing of all four pathogens and molecular analysis of C. parvum with diarrhea in calves from southernmost Okinawa and northernmost Hokkaido, Japan, were conducted. The frequencies of C. parvum, rotavirus, coronavirus, and E. coli (K99) in Okinawa were 50%, 28%, 2.3%, and 4.7%, respectively. Watery fecal stools were significantly correlated with C. parvum (p < 0.05). In oocyst calculations for C. parvum, no significant difference was observed between the single-infection cases and the mixed-infection cases with rotavirus. Interestingly, molecular analyses targeting small subunit ribosomal RNA as well as glycoprotein 60 (GP60) genes revealed that the C. parvum nucleotide sequences from the two prefectures were identical, indicating that C. parvum with a uniform characteristic is distributed throughout Japan. GP60 subtyping analysis identified C. parvum from Okinawa and Hokkaido as belonging to the IIaA15G2R1 subtype, a known zoonotic subtype. Hence, control of cryptosporidiosis is important not only for pre-weaned calves, but also for human health.
Keywords: Cryptosporidium parvum, Immunochromatography test, SSUrRNA, GP60, Japan
Graphical abstract
Highlights
-
•
ICT strips were used for calf diarrhea to detect four major enteric pathogens.
-
•
C. parvum showed the highest frequency in the southernmost Okinawa prefecture, Japan.
-
•
C. parvum from the northernmost Hokkaido prefecture was used for a comparative study.
-
•
C. parvum from the two prefectures had a uniform character in SSUrRNA and GP60 genes.
-
•
GP60 subtyping revealed that IIaA15G2R1, a known zoonotic subtype, was predominant.
1. Introduction
Cryptosporidium parvum, a well-known protozoan intestinal parasite of the phylum Apicomplexa, causes enteric diseases in domestic ruminants and humans. C. parvum is strongly associated with diarrhea in neonatal calves [1]. A report mentioned that the prevalence of this parasite reached 75% in pre-weaned calves in Japan [2]. Diarrhea is a complex and multifactorial disease symptom influenced by the nutritional and immunological status of a calf, the management of the herd, the environment, and various infectious agents [3]. Co-infection with multiple pathogens may increase diarrhea severity and mortality rates. In addition to C. parvum, rotavirus, bovine coronavirus and enterotoxigenic Escherichia coli (E. coli K99) are well-known enteropathogens associated with neonatal diarrhea in calves throughout the world [4], [5], [6]. However, investigations to identify the multiple causative agents of diarrhea in calves have not been performed yet in Japan. Therefore, one of the aims of the present study was to determine the prevalence of the four major causative agents associated with clinical diarrheal cases in pre-weaned calves through use of commercial immunochromatography test (ICT) strips.
Traditionally, C. parvum has been diagnosed by microscopy of fecal samples following sample concentration by the sugar floatation method. However, C. parvum cannot be differentiated from C. bovis and C. ryanae using microscopy because their oocysts are of similar size and morphology [7]. C. bovis and C. ryanae have been detected exclusively from cattle, and infections with the two species are most likely asymptomatic [8]. It is therefore important to accurately distinguish those species from C. parvum. Recently, the nucleotide sequences obtained from nested polymerase chain reactions (PCRs) of the small subunit ribosomal RNA (SSUrRNA) and glycoprotein 60 (GP60) genes have been employed as tools for accurate species identification and subtyping analysis, respectively [9], [10], [11], [12], [13]. Although some epidemiological studies on C. parvum in Japan have been performed using these DNA markers [14], [15], [16], [17], they did not characterize the C. parvum populations from more than one region of the country. Therefore, the aim of the present study was not only to obtain epidemiological information about C. parvum based on SSUrRNA and GP60 gene sequences, but also to compare the characteristics of this parasite in two different Japanese prefectures, southernmost Okinawa and northernmost Hokkaido.
2. Materials and methods
2.1. Diarrhea samples from the two locations
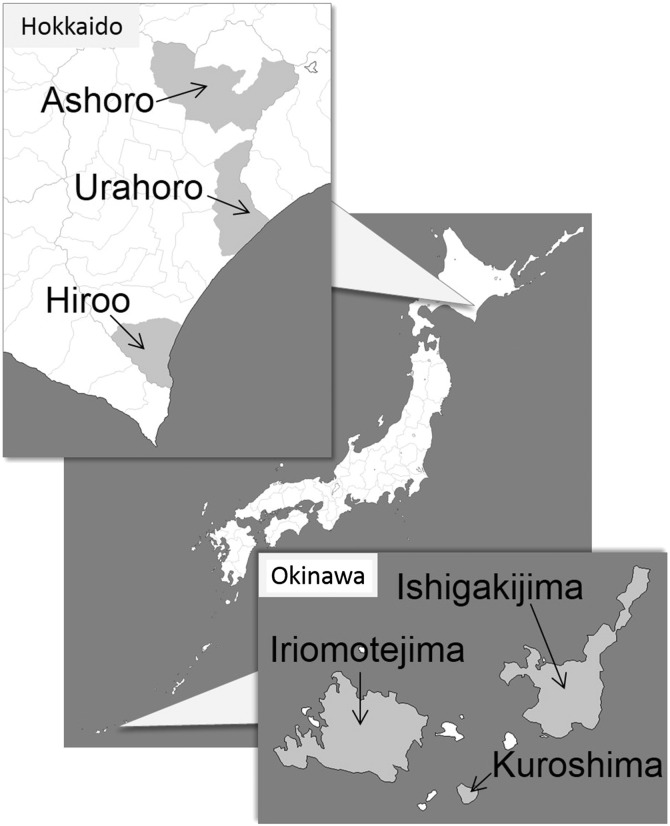
Sampling location information is summarized in Fig. 1 . We collected 50 diarrheic samples from calves aged from 6 to 88 days, from 29 farms located on three islands in Okinawa prefecture, Japan, from December 2012 to January 2013. One to seven samples were collected from each farm and the fecal properties of the samples were recorded. Commercial ICT strips (Bio-X Diagnostics SPRL, Jemelle, Belgium) were used to detect C. parvum, rotavirus, coronavirus, and E. coli K99 antigens in the diarrhea samples (Table 1 ). Twenty-five diarrheic samples were also collected from calves aged from 4 to 21 days in Hokkaido prefecture, Japan, from December 2012 to May 2013. One to five samples were collected from 11 farms (Table 2 ). These samples tested positive for C. parvum antigen using ICT strips, and molecular characterizations were performed to compare those with C. parvum from Okinawa prefecture. Rotavirus, coronavirus, and E. coli K99 were not investigated for the samples from Hokkaido prefecture. All the fecal samples were stored at 4 °C and transported to laboratories for further analyses.
Fig. 1.
Geographical origins of the calf fecal samples. We compared the characteristics of C. parvum from the northernmost prefecture (Hokkaido) with those from the southernmost prefecture (Okinawa) of Japan. Sample collection sites in Hokkaido were Ashoro, Urahoro, and Hiroo. Those in Okinawa were Ishigakijima, Iriomotejima, and Kuroshima Islands.
Table 1.
Origin of diarrheal samples from Okinawa and the number of positive samples obtained.
| Location | Number of farms (breed of calf) | Number of diarrhea samples | ICT positive |
Oocyst positive |
Nested PCR-positive |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| C. parvum | Rotavirus | Coronavirus | E. coli (K99) | C. parvum | Eimeria spp. | SSUrRNA | GP60 | |||
| Ishigakijima | 21 (Japanese black) | 42 | 23 | 14 | 1 | 2 | 16 | 1 | 24 | 20 |
| Iriomotejima | 4 (Japanese black) | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 |
| Kuroshima | 2 (Japanese black) | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| No datab | 2 (Japanese black) | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| Total | 29 | 50 | 25 (50.0%) | 14 (28.0%) | 1 (2.3%)a | 2 (4.7%)a | 18 (36.0%) | 3 (6.0%) | 29 (58.0%) | 20 (40.0%) |
Immunochromatography test (ICT) for coronavirus and E. coli antigens were not performed on 7 samples from Ishigakijima.
One of the three locations.
Table 2.
Origin of diarrheal samples from Hokkaido and the number of positive samples obtained.
| Location | Number of farms (breed of calf) | Number of diarrheal samples | Oocyst positive |
Nested PCR-positive |
||
|---|---|---|---|---|---|---|
| C. parvum | Eimeria spp. | SSUrRNA | GP60 | |||
| Ashoro | 8 (Japanese black), 1 (Holstein) | 20 | 20 | 0 | 20 | 20 |
| Hiroo | 1 (Japanese black) | 4 | 4 | 0 | 4 | 4 |
| Urahoro | 1 (Japanese black) | 1 | 1 | 0 | 1 | 1 |
| total | 11 | 25 | 25 (100%) | 0 | 25 (100%) | 25 (100%) |
2.2. Oocysts per gram calculations
The centrifugal sugar flotation method was used to detect Cryptosporidium and other parasitic oocysts and/or eggs that might cause diarrhea. Floatation was performed after suspending 1 g of a diarrheal sample in sucrose solution (1.2 g/ml), thereby allowing the approximate number of oocysts per gram (OPG) to be calculated. The average number of oocysts for 10 fields at a magnification of 400 was calculated, and the average number was converted into OPG by using the total number of fields under the microscope.
2.3. Statistical analyses
OPG values were compared between C. parvum single infections and mixed infections with rotavirus using a Student's t-test. Correlations between the fecal properties and the pathogens were evaluated for C. parvum-associated and non-associated diarrhea using Fisher's exact test.
2.4. DNA analysis
Total DNA was extracted using a QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Before DNA extraction, 0.2 g of each fecal sample was transferred to a 1.5 ml plastic tube and three cycles of freezing/thawing were performed; quick freezing was performed in a − 80 °C freezer for 15 min, while quick thawing was conducted in a 37 °C water bath for 15 min. Thereafter, SSUrRNA and GP60 genes were amplified by nested PCR. A previously described primer set [9] was used for nested PCR to amplify the SSUrRNA gene. For the GP60 gene, gp60 nested 1st F (5′-ATG CAA AAA TAC GTG GAC TGG GT-3′) and gp60 nested 1st R (5′-TCG CAC GAA AGA TTT CCA TTG-3′) primers were used for the first round of PCR, followed by gp60 nested 2nd orfF (5′-ATG AGA TTG TCG CTC ATT ATC GTA T-3′) and gp60 nested 2nd orfR (5′-TTA CAA CAC GAA TAA GGC TGC AAA G-3′) primers for the second PCR round. Outer PCR was performed in a 25 μl volume with 2 μl of template DNA, 12.5 μl of 2 × Gflex PCR buffer (TaKaRa, Shiga, Japan; Mg2 +, dNTP plus), 0.13 μl of each primer (10 μM), and 1.25 U of Tks Gflex DNA polymerase (TaKaRa) using the GeneAmp® PCR system 2700 (Applied Biosystems, Foster City, USA). Thermal cycling consisted of an initial denaturation step at 94 °C for 1 min, followed by 35 cycles at 98 °C for 10 s, 55 °C for 15 s, and 68 °C for 30 s, with a final extension at 68 °C for 7 min. Inner PCRs were performed using the same conditions with 2 μl of the first PCR amplification mixture as the template after 10-fold dilution in double-distilled water. Inner PCR amplicons were purified with the NucleoSpin® Gel and PCR Clean-up Kit (MACHEREY-NAGEL, Düren, Germany) according to the manufacturer's instructions, followed by direct sequencing from both directions using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The resultant sequence ladders were determined using a 3500-Genetic Analyzer (Applied Biosystems). The SSUrRNA nucleotide sequences were compared with C. parvum GenBank reference sequences for accurate species identification. A neighbor-joining phylogram inferred from the GP60 sequences was constructed by PAUP version 4.0b10 [18]. In the phylogram construction, the GP60 sequences obtained in this study along with the C. parvum reference sequences were used. Bootstrap analyses were conducted using 1000 replicates.
3. Results
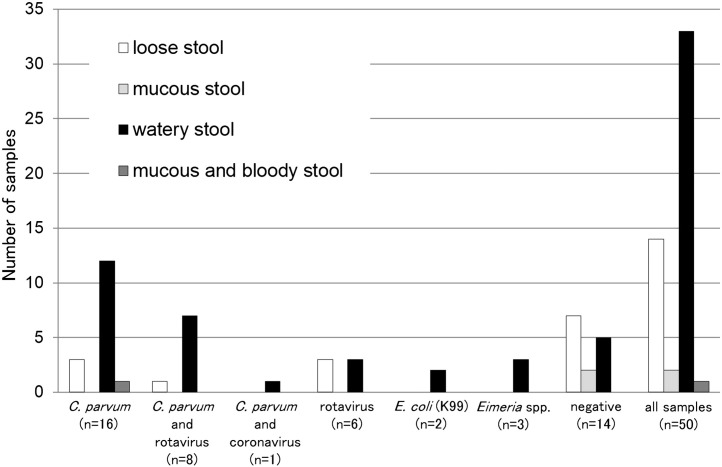
For the 50 diarrheal samples from Okinawa, 25 (50.0%) were C. parvum antigen-positive, 14 (28.0%) were rotavirus antigen-positive, one (2.3%) was coronavirus-positive, and two (4.7%) were E. coli K99 antigen-positive. Eight samples were positive for both C. parvum and rotavirus, and one was positive for both C. parvum and coronavirus. The properties of the diarrheal samples are summarized in Fig. 2 , and watery stools were significantly correlated (p < 0.05) with C. parvum infections (Table 3 ).
Fig. 2.
Okinawa prefecture samples, feces type, and pathogen identification. C. parvum, rotavirus, coronavirus, and E. coli (K99) were detected using commercial immunochromatography test strips. Eimeria spp. was diagnosed by the sugar floatation method.
Table 3.
Correlation between watery stool samples and C. parvum in the samples from Okinawa.
| C. parvum associated | C. parvum non-associated | Total | |
|---|---|---|---|
| Watery stool | 20 | 13 | 33 |
| Other fecal propertiesa | 5 | 12 | 17 |
| Total | 25 | 25 | 50 |
C. parvum and watery stool were statistically correlated. (p < 0.05).
Loose stool, mucous stool, and mucous and bloody stool are included.
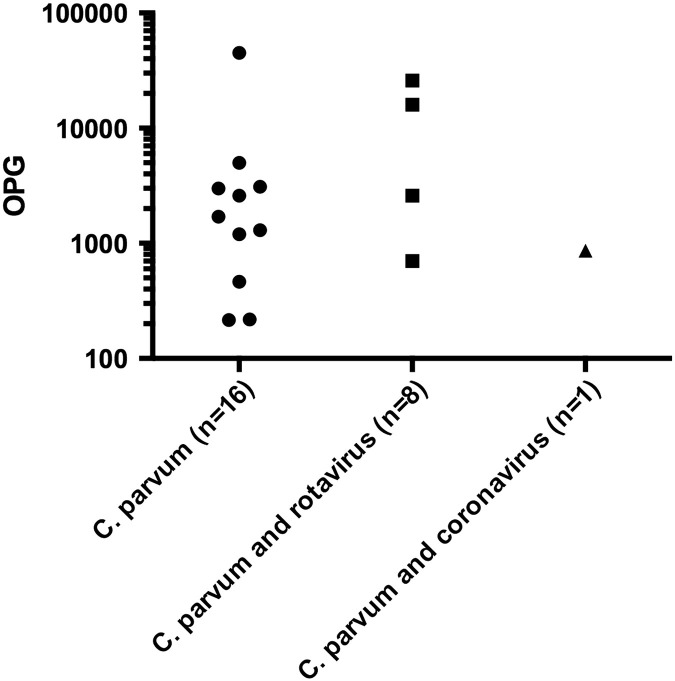
Among the samples from Okinawa, the OPG values ranged from 12 to 4.5 × 104. Cryptosporidium oocysts were not found in seven of the samples although they were positive for C. parvum antigen (Ishigakijima Island; Table 1). No significant difference was observed in OPG values between C. parvum single infections and mixed infections with rotavirus (Fig. 3 ). Eimeria spp. oocysts were detected in three calves from Okinawa (Table 1).
Fig. 3.
Oocysts per gram (OPG) counts on a logarithmic scale for C. parvum single infections, mixed infections of C. parvum and rotavirus, and C. parvum and coronavirus. Pathogen detection in the fecal samples from Okinawa prefecture was based on commercial immunochromatography test strips. No significant difference was observed between C. parvum single infections and mixed infections with rotavirus.
SSUrRNA and GP60 genes were analyzed for accurate species identification and for subtyping analysis, respectively, and PCR amplicons were obtained from 29 and 20 samples in the both targeting regions (Table 2). The SSUrRNA gene was amplified in one sample from Ishigakijima Island, two samples from Iriomotejima Island, and one sample with no location data, even though all were negative for C. parvum antigen. In contrast, GP60 gene was not amplifiable from three Ishigakijima Island samples despite being positive for C. parvum antigen. Surprisingly, we could not obtain a GP60 amplicon from one sample from Kuroshima Island and another sample with no location data, although both were C. parvum antigen- and oocyst-positive. However, the two DNA target sequences were successfully amplified from the 25 samples from Hokkaido (Table 2).
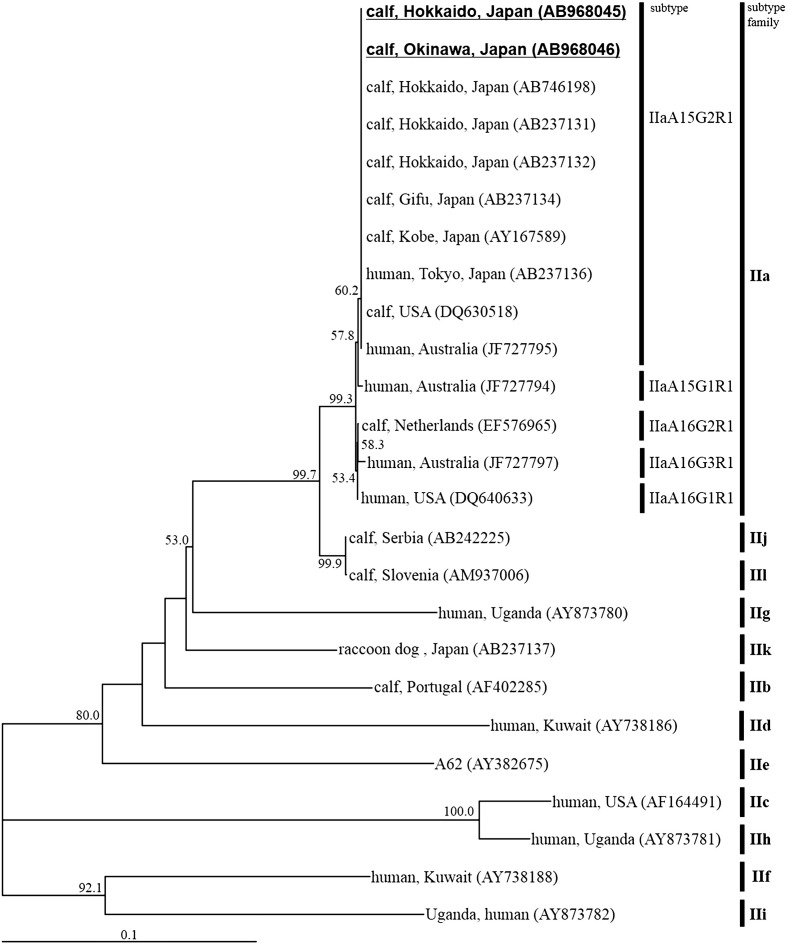
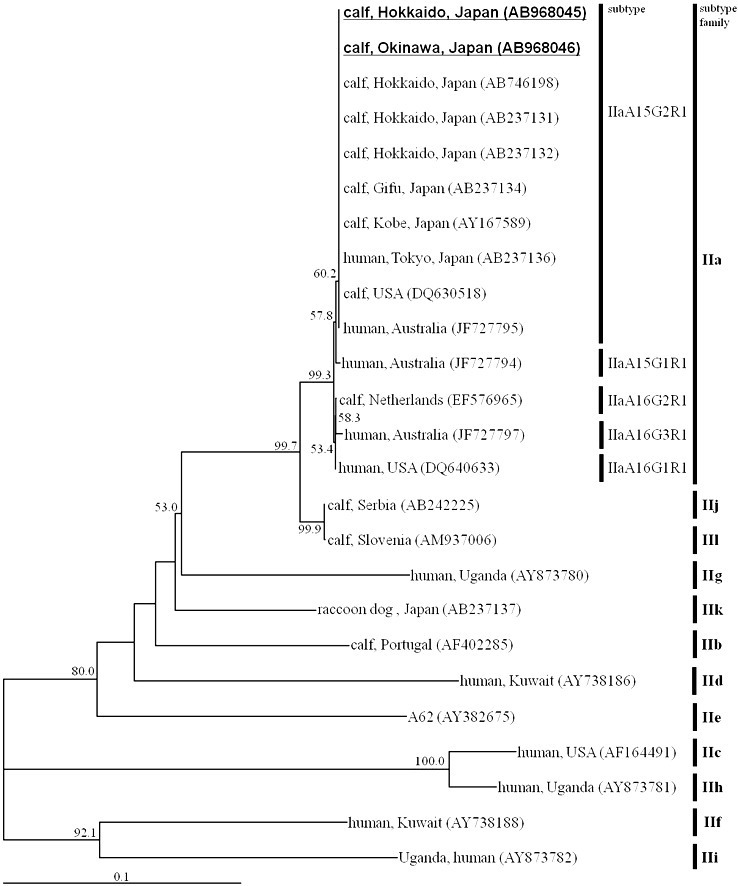
The nucleotide sequences of all the 786-bp SSUrRNA PCR amplicons from the two prefectures were identical to that of the C. parvum sequence deposited in GenBank (accession no. AB746195). The nucleotide sequences of all the GP60 PCR amplicons (925bp) were also identical to each other, and shared 100% similarity values with the IIaA15G2R1 subtype sequence deposited in GenBank (Fig. 4 ). Nucleotide sequences representative of the SSUrRNA and GP60 genes have been deposited in GenBank under accession nos. AB968045 to AB968048AB968045AB968046AB968047AB968048.
Fig. 4.
Neighbor-joining phylogram inferred from the glycoprotein 60 sequences of C. parvum. The nucleotide sequences obtained in this study are highlighted in bold with underlining. The host species, geographical origin, and the accession no. of each sequence are shown in the phylogram. Bootstrap values over 50% are labeled on the nodes. The subtype and subtype family of each sequence are displayed on the right-hand side.
4. Discussion
This study is the first attempt in Japan to detect the causative agents of diarrhea in pre-weaned calves using commercial ICT strips. Among the diarrhea samples from Okinawa, the C. parvum antigen showed the highest frequency (50%). A previous study performed in Hokkaido reported that the prevalence of C. parvum reached 75% in pre-weaned calves [2]. In contrast, the previously reported prevalence rates for rotavirus in Japan were 9.6% [19] and 5.2% [20]. Further investigations on diarrhea cases from different locations in Japan may reveal the major pathogens involved in calf diarrhea more clearly. The present study revealed that the diarrhea cases associated with C. parvum tended to have watery diarrhea (Table 3). Watery diarrhea causes severe dehydration in neonatal calves, which in turn may lead to a significant economic loss in cattle industry. Our study did not reveal any significant difference in the OPG values between single infections of C. parvum and mixed infections with another diarrhea-causing agent. However, the number of co-infections observed was insufficient to draw any conclusions about the effect that more than one pathogenic organism might have on the OPG values. Hence, continuous sampling from clinical cases of calf diarrhea is needed to obtain more accurate OPG calculations to evaluate whether any correlations between C. parvum oocyst numbers and the presence or absence of other pathogens exist.
In this study, the nested SSUrRNA gene appeared to be the more efficient of the two PCR tests at detecting C. parvum. The SSUrRNA gene was probably amplified efficiently because it is a multicopy gene whereas GP60 is a single locus gene [21]. The ICT strip readily detected C. parvum antigen; however, our results suggest that there is a risk of misdiagnosis when this test strip alone is used to detect C. parvum. The GP60 nested PCR did not seem to always be effective at diagnosing C. parvum because we were unable to obtain amplicons from two of the Okinawa samples, despite them being C. parvum antigen- and oocyst-positive according to microscopic observations. Therefore, the results of this study suggest that it is better to use more than one method to reliably detect C. parvum.
Many C. parvum subtypes are categorized as the IIa subtype family on the basis of their worldwide GP60 sequences [11], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]. Interestingly, the GP60 nucleotide sequences detected from the two different locations were identical to each other, and were classified as the IIaA15G2R1 subtype. Furthermore, previous analyses of the GP60 subtype of C. parvum from Gifu, Kobe, Hokkaido, and the most recent epidemiological study performed in the Ishikari district of Hokkaido also reported the occurrence of the IIaA15G2R1 subtype alone [14], [17], [37]. These findings suggest that C. parvum with the IIaA15G2R1 subtype is predominant in Japan. Because IIaA15G2R1 is a zoonotic subtype [10], cryptosporidiosis control in Japan is important for the health of domestic ruminants and humans alike.
Conflict of interest statement
The authors declare that there are no competing interests.
Acknowledgments
The authors would like to thank Noboru Inoue (Obihiro University of Agriculture and Veterinary Medicine) for his useful advices about this study. This study was supported in part by a Joint Research Grant from the National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine (*25-joint-10) and the Adaptable and Seamless Technology Transfer Program through target driven R & D (A-STEP) from the Japan Science and Technology Agency (AS242Z03137P).
References
- 1.Uga S., Matsuo J., Kono E., Kimura K., Inoue M., Rai S.K. Prevalence of Cryptosporidium parvum infection and pattern of oocyst shedding in calves in Japan. Vet Parasitol. 2000;94:27–32. doi: 10.1016/s0304-4017(00)00338-1. [DOI] [PubMed] [Google Scholar]
- 2.Karanis P., Eiji T., Palomino L., Boonrod K., Plutzer J., Ongerth J. First description of Cryptosporidium bovis in Japan and diagnosis and genotyping of Cryptosporidium spp. in diarrheic pre-weaned calves in Hokkaido. Vet Parasitol. 2010;169:387–390. doi: 10.1016/j.vetpar.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Bendali F., Saanaa M., Bbichet H., Scchelcher F. Risk factors associated with diarrhoea in newborn calves. Vet Res. 1999;30:509–522. [PubMed] [Google Scholar]
- 4.Uhde F.L., Kaufmann T., Sager H., Albini S., Zanoni R., Schelling E. Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland. Vet Rec. 2008;163:362–366. doi: 10.1136/vr.163.12.362. [DOI] [PubMed] [Google Scholar]
- 5.Izzo M.M., Kirkland P.D., Mohler V.L., Perkins N.R., Gunn A.A., House J.K. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust Vet J. 2011;89:167–173. doi: 10.1111/j.1751-0813.2011.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Y.I., Sun D., Cooper V., Dewell G., Schwartz K., Yoon K.J. Evaluation of a commercial rapid test kit for detecting bovine enteric pathogens in feces. J Vet Diagn Invest. 2012;24:559–562. doi: 10.1177/1040638712440997. [DOI] [PubMed] [Google Scholar]
- 7.Plutzer J., Karanis P. Genetic polymorphism in Cryptosporidium species: an update. Vet Parasitol. 2009;165:187–199. doi: 10.1016/j.vetpar.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Šlapeta J. Cryptosporidiosis and Cryptosporidium species in animals and humans: a thirty colour rainbow? Int J Parasitol. 2013;44:957–970. doi: 10.1016/j.ijpara.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Xiao L., Morgan U.M., Limor J., Escalante A., Arrowood M., Shulaw W. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Peng M.M., Matos O., Gatei W., Das P., Stantic-Pavlinic M., Bern C. A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol. 2001:28S–31S. doi: 10.1111/j.1550-7408.2001.tb00442.x. [Suppl.] [DOI] [PubMed] [Google Scholar]
- 12.Alves M., Xiao L., Sulaiman I., Lal A.A., Matos O., Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–2747. doi: 10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y., Ortega Y., He G., Das P., Xu M., Zhang X. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Parasitol. 2007;144:1–9. doi: 10.1016/j.vetpar.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Abe N., Matsubayashi M., Kimata I., Iseki M. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res. 2006;99:303–305. doi: 10.1007/s00436-006-0140-0. [DOI] [PubMed] [Google Scholar]
- 15.Amer S., Honma H., Ikarashi M., Oishi R., Endo M., Otawa K. The first detection of Cryptosporidium deer-like genotype in cattle in Japan. Parasitol Res. 2009;104:745–752. doi: 10.1007/s00436-008-1250-7. [DOI] [PubMed] [Google Scholar]
- 16.Murakoshi F., Xiao L., Matsubara R., Sato R., Kato Y., Sasaki T. Molecular characterization of Cryptosporidium spp. in grazing beef cattle in Japan. Vet Parasitol. 2012;187:123–128. doi: 10.1016/j.vetpar.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Murakoshi F., Tozawa Y., Inomata A., Horimoto T., Wada Y., Kato K. Molecular characterization of Cryptosporidium isolates from calves in Ishikari District, Hokkaido, Japan. J Vet Med Sci. 2013;75:837–840. doi: 10.1292/jvms.12-0435. [DOI] [PubMed] [Google Scholar]
- 18.Swofford D.L. Sinauer Associates; Sunderland, Massachussetts: 2001. PAUP*. Phylogenetic analysis using parasimony and other methods ver. 4.0beta. [Google Scholar]
- 19.Okada N., Matsumoto Y. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet Microbiol. 2002;84:297–305. doi: 10.1016/s0378-1135(01)00445-x. [DOI] [PubMed] [Google Scholar]
- 20.Abe M., Ito N., Morikawa S., Takasu M., Murase T., Kawashima T. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res. 2009;144:250–257. doi: 10.1016/j.virusres.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Strong W.B., Gut J., Nelson R.G. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feltus D.C., Giddings C.W., Schneck B.L., Monson T., Warshauer D., McEvoy J.M. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol. 2006;44:4303–4308. doi: 10.1128/JCM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L., Zhou L., Santin M., Yang W., Fayer R. Distribution of Cryptosporidium parvum subtypes in calves in eastern United States. Parasitol Res. 2007;100:701–706. doi: 10.1007/s00436-006-0337-2. [DOI] [PubMed] [Google Scholar]
- 24.Wielinga P.R., de Vries A., van der Goot T.H., Mank T., Mars M.H., Kortbeek L.M. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int J Parasitol. 2008;38:809–817. doi: 10.1016/j.ijpara.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Brook E.J., Anthony Hart C., French N.P., Christley R.M. Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Vet J. 2009;179:378–382. doi: 10.1016/j.tvjl.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Silverlås C., Näslund K., Björkman C., Mattsson J.G. Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet Parasitol. 2010;169:289–295. doi: 10.1016/j.vetpar.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Widmer G., Lee Y. Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl Environ Microbiol. 2010;76:6639–6644. doi: 10.1128/AEM.01268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pangasa A., Jex A.R., Nolan M.J., Campbell B.E., Haydon S.R., Stevens M.A. Highly sensitive non-isotopic restriction endonuclease fingerprinting of nucleotide variability in the gp60 gene within Cryptosporidium species, genotypes and subgenotypes infective to humans, and its implications. Electrophoresis. 2010;31:1637–1647. doi: 10.1002/elps.200900706. [DOI] [PubMed] [Google Scholar]
- 29.Chalmers R.M., Smith R.P., Hadfield S.J., Elwin K., Giles M. Zoonotic linkage and variation in Cryptosporidium parvum from patients in the United Kingdom. Parasitol Res. 2011;108:1321–1325. doi: 10.1007/s00436-010-2199-x. [DOI] [PubMed] [Google Scholar]
- 30.Waldron L.S., Dimeski B., Beggs P.J., Ferrari B.C., Power M.L. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Appl Environ Microbiol. 2011;77:7757–7765. doi: 10.1128/AEM.00615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imre K., Lobo L.M., Matos O., Popescu C., Genchi C., Dărăbuş G. Molecular characterisation of Cryptosporidium isolates from pre-weaned calves in Romania: is there an actual risk of zoonotic infections? Vet Parasitol. 2011;181:321–324. doi: 10.1016/j.vetpar.2011.04.042. [DOI] [PubMed] [Google Scholar]
- 32.Herges G.R., Widmer G., Clark M.E., Khan E., Giddings C.W., Brewer M. Evidence that Cryptosporidium parvum populations are panmictic and unstructured in the Upper Midwest of the United States. Appl Environ Microbiol. 2012;78:8096–8101. doi: 10.1128/AEM.02105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rzeżutka A., Kaupke A. Occurrence and molecular identification of Cryptosporidium species isolated from cattle in Poland. Vet Parasitol. 2013;196:301–306. doi: 10.1016/j.vetpar.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Rieux A., Paraud C., Pors I., Chartier C. Molecular characterization of Cryptosporidium isolates from pre-weaned calves in western France in relation to age. Vet Parasitol. 2013;197:7–12. doi: 10.1016/j.vetpar.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Silverlås C., Bosaeus-Reineck H., Näslund K., Björkman C. Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Int J Parasitol. 2013;43:155–161. doi: 10.1016/j.ijpara.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Coco V.F., Córdoba M.A., Bilbao G., de Almeida Castro A.P., Basualdo J.A., Fayer R. Cryptosporidium parvum GP60 subtypes in dairy cattle from Buenos Aires, Argentina. Res Vet Sci. 2014;96:311–314. doi: 10.1016/j.rvsc.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Wu Z., Nagano I., Boonmars T., Nakada T., Takahashi Y. Intraspecies polymorphism of Cryptosporidium parvum revealed by PCR-restriction fragment length polymorphism (RFLP) and RFLP-single-strand conformational polymorphism analyses. Appl Environ Microbiol. 2003;69:4720–4726. doi: 10.1128/AEM.69.8.4720-4726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]