Abstract
Interferons (IFN) are key cytokines with multifaceted antiviral and cell-modulatory properties. Three distinct types of IFN are recognized (I–III) based on structural features, receptor usage, cellular source and biological activities. The action of IFNs is mediated by a complex, partially overlapping, transcriptional program initiated by the interaction with specific receptors. Genetic diversity, with polymorphisms and mutations, can modulate the extent of IFN responses and the susceptibility to infections. Almost all viruses developed mechanisms to subvert the IFN response, involving both IFN induction and effector mechanisms. Interactions between IFN types may occur, for both antiviral and cell-modulatory effects, in a complex interplay, involving both synergistic and antagonistic effects. Interferon-associated diseases, not related to virus infections may occur, some of them frequently observed in IFN-treated patients. On the whole, IFNs are pleiotropic biologic response modifiers, that, upon activation of thousands genes, induce a broad spectrum of activities, regulating cell cycle, differentiation, plasma membrane molecules, release of mediators, etc., that can be relevant for cell proliferation, innate and adaptive immunity, hematopoiesis, angiogenesis and other body functions.
Abbreviations: ADAR1, adenosine deaminase acting on RNA 1; AP-1, activator protein 1; APCs, antigen presenting cells; ATF6, activating transcription factor 6; BCG, bacille Calmette–Guérin; cGAS, cyclic GMP-AMP synthase; CHIKV, Chickungunya virus; CRF2, class II cytokine receptor family; ds, double strand; EAE, experimental allergic encephalomyelitis; EM, environmental mycobacteria; EMCV, encephalomyocarditis virus; GAS, gamma interferon activation site; hCMV, human Cytomegalovirus; HCV, hepatitis C virus; HSE, herpes simplex encephalitis; HSV-1, herpes simplex virus-1; IFI16, gamma-interferon-inducible protein 16; IFN, interferon; IFNAR, interferon alpha receptor; IFNGR, interferon gamma receptor; IL, interleukin; IPS-1, interferon promoter-stimulating factor 1; IRAK-4, interleukin-1 receptor-associated kinase 4; IRF, interferon regulatory factor; ISGF3, interferon-stimulated gene factor 3; ISRE, interferon stimulated response element; JAK, Janus kinase; LASV, Lassa virus; MAPK, mitogen-activated protein kinases; MDA5, melanoma differentiation-associated protein 5; MHC, major histocompatibility complex; MS, multiple sclerosis; MSMD, Mendelian susceptibility to mycobacterial disease; MxA, Myxovirus Resistance Gene A; NEMO, nuclear factor (NF)-kB essential modulator; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NK, natural killer cells; OAS, 2′,5′-oligo adenylate synthetase A; p300/CBP, chromatin remodeling associated p300/CREB-binding protein; PD-L1, programmed death ligand 1; PKR, dsRNA-dependent protein kinase; PRRs, pattern-recognition receptors; RIG-1, retinoic-acid-inducible gene 1; SAMHD1, SAM domain and HD domain-containing protein 1; SARS, severe acute respiratory syndrome; SARS-CoV, severe acute respiratory syndrome-associated coronavirus; SNPs, single nucleotide polymorphisms; SOCS, suppressor of cytokine signaling; ss, single strand; STAT, Signal Transducer and Activator of Transcription; Th, lymphocytes T helper; TLR, Toll-like receptor; TREX1, three prime repair exonuclease 1; TYK2, tyrosine kinase 2; VSV, vesicular stomatitis virus; VZV, Varicella-Zoster virus; WNV, West Nile virus
Keywords: Interferon types I–III, Interferon genes and receptors, Interferon–interferon interactions, Anti-interferon strategies
1. Introduction
Interferon (IFN) was first discovered as an antiviral agent during studies on virus interference [1], [2]. Isaacs and Lindenmann reported in 1957 that influenza virus-infected chick cells produced a secreted factor that mediated the transfer of a virus-resistant state active against both homologous and heterologous viruses [1]. This seminal observation, along with similar findings described by Nagano and Kojima in 1958 [2], set the stage for subsequent studies that led to the elucidation of the IFN system in exquisite detail [3].
Almost 60 years after the discovery of Isaacs and Lindenmann, considerable progress has been made toward answering multiple questions about IFN family members and their effects on the virus–host interaction. Advances made while elucidating the IFN system contributed significantly to our understanding in multiple areas of mammalian cell biology and biochemistry, ranging from the pathways of signal transduction to the biochemical mechanisms of transcriptional and translational control to the molecular basis of viral pathogenesis [3], and led to the development of the first “cytokine-based” therapy in the 70s, now licensed worldwide for viral disease, malignant and even immune disorders [4].
2. Unraveling the complexity of the interferon system
After the first discovery of Type I IFN during 1950s [1] and Type II IFN in 1965 [5], in 2003 two groups independently reported the discovery of a trio of novel interferon-like cytokines that are referred to as either IFNλ1, λ2, and λ3 or interleukin-29 (IL-29), IL-28A, and IL-28B, respectively [6], [7], and constitute type III IFN. Both groups also identified and characterized the novel receptor, IFNλR1 (also known as IL-28RA), through which these cytokines mediate their biological activities [8].
2.1. Interferon genes and proteins
IFNs are key cytokines in the establishment of a multifaceted antiviral response. Three distinct types of IFNs are now recognized (types I, II, and III) based on their structural features, receptor usage and biological activities [8]. A synopsis of the major features of IFN family member components is reported in Table 1 . Type I IFN family in humans consists of 14 IFNα species [9] and a single species of IFNβ, IFNκ, IFNω, and IFNɛ, while there is only one type II IFN known as IFNγ. All the type I IFN components lack introns and are clustered on the short arm of chromosome 9 in human and chromosome 4 in mouse genome. The single IFNγ gene possesses three introns and maps to the long arm of chromosome 12 in human, and chromosome 10 in mouse genome. Although some IFNs are modified post-translationally by N- and O-glycosylation, the major human IFNα subspecies are not glycosylated [3]. IFNα and IFNβ gene products appear to function as monomers, whereas IFNγ appear to function as homodimers [10]. On the other hand, IFNλ genes are clustered together on human 19 or murine 7 chromosome. The IFNλ3 gene (IL28B) is transcribed in the opposite direction of the IFN-λ1 (IL29) and IFNλ2 (IL28A) genes [8].
Table 1.
Major features of IFN family components.
| IFN type | Name (no. of genes) | Location in human chromosomes | Receptors |
|---|---|---|---|
| Type I | IFNα (14) IFNβ (1) IFNκ (1) IFNω (1) IFNɛ (1) |
Chromosome 9 | IFNαR1 and IFNαR2 (also known as IFNAR1 and IFNAR2) |
| Type II | IFNγ (1) | Chromosome 12 | IFNγR1 and IFNγR2 (also known as IFNGR1 and IFNGR2) |
| Type III | IFNλ (3) | Chromosome 19 | IFNλR1 (also known as IL-28RA) and IL-10R2 |
Although all IFNs are important mediators of antiviral protection, their roles in antiviral defense vary. Type I IFNs have four major functions. First, they induce cell-intrinsic antimicrobial states in infected and neighboring cells that limit the spread of infectious agents, particularly viral pathogens. Second, they modulate innate immune responses in a balanced manner that promotes antigen presentation and natural killer cell functions while restraining pro-inflammatory pathways and cytokine production. Third, they activate the adaptive immune system, thus promoting the development of high-affinity antigen-specific T and B cell responses and immunological memory [9]. Fourth, they present an antiproliferative activity: in fact, several recent studies showed type I IFN-induced autophagy in multiple cancer cell lines highlighting a new function of type I IFN as an inducer of autophagy. This new function of type I IFN may play an important role in viral clearance, antigen presentation, inhibition of proliferation, as well as a positive feedback loop for the production of type I IFN [11]. Type I IFNs are protective in acute viral infections but can have either protective or deleterious roles in bacterial infections and autoimmune diseases. Most cell types produce IFNβ, whereas haematopoietic cells, particularly plasmacytoid dendritic cells, are the predominant producers of IFNα. Type I IFN production is induced after the sensing of microbial products by pattern-recognition receptors (PRRs)4–6 and by cytokines [9]. IFNγ is mainly secreted by T lymphocytes, natural killer cells and antigen presenting cells (APCs) such as monocytes, macrophages and dendritic cells. IFNγ secretion by natural killer (NK) cells and professional antigen presenting cells is likely to be important in early host defense against infection, whereas T lymphocytes become the major source of IFNγ in the adaptive immune response. IFNγ production is controlled by cytokines secreted by antigen presenting cells, most notably IL12 and IL18. Macrophage recognition of many pathogens induces the secretion of IL-12 and several chemokines. These chemokines attract natural killer cells to the site of inflammation, and IL-12 promotes IFNγ synthesis in these cells [12]. IFNγ also plays a central role in the development of antitumor immune responses, and it can amplify the induction of antiviral activity by IFNα or IFNβ. Therefore, type I and type II IFN often work together to activate a variety of innate and adaptive immune responses that result in the induction of effective antitumor immunity and the elimination of viral infections [10], [13]. On the other hand, the IFNλ signal transduction cascade is very similar to that induced by type I IFNs (IFNα or IFNβ). Therefore, it is not surprising that type I and type III IFNs exert similar biological activities. Both types of IFN possess the intrinsic ability to induce antiviral activity in cells [8]. In fact, the IFNλs are usually coexpressed together with type I IFNs by virus infected cells [6], [7].
2.2. IFN receptors
IFNs are part of the larger family of class II cytokines that also includes 6 IL-10-related cytokines: IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26 [14], [15], [16] as well as several viral IL-10-related cytokines [17]. Class II cytokines all signal via receptors that share common motifs in their extracellular domains, and are indicated as class II cytokine receptor family (CRF2). Consequently, IFNs and the IL-10-related cytokines are sometimes referred to as “CRF2 cytokines” [8]. However, although the tertiary structure of IFNγ resembles that of IL-10, its primary structure significantly diverges from all of the CRF2 ligands. The most recent addition to the CRF2 family members, type III IFN or IFNλ, demonstrates structural features of the IL-10-related cytokines but also induces antiviral activity in a variety of target cells, which supports their functional classification as a new IFN type [6], [7]. Phylogenetically, the IFNλ genes reside somewhere between the type I IFN and IL-10 gene families. Amino acid sequence comparison shows that the type III IFN members exhibit about 5–18% identity with either type I IFN members or the IL-10-related cytokines. The IFNλ proteins bind and signal through a receptor complex composed of the unique IFNλR1 chain (also known as IL-28RA) and the shared IL-10R2 chain which is also a part of the receptor complexes for IL-10, IL-22, and IL-26. In contrast, all type I IFN members exert their biological activities through a heterodimeric receptor complex composed of the IFNαR1 (IFNAR1) and IFNαR2 (IFNAR2) chains, and type II IFN (IFNγ) engages the IFNγR1 (IFNGR1) and IFNγR2 (IFNGR2) chains to assemble its functional receptor complex [8].
In the canonical type I IFN-induced signaling pathway described over 25 years ago, IFNAR engagement was shown to activate the receptor-associated protein tyrosine kinases Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which phosphorylate the latent cytoplasmic transcription factors Signal Transducer and Activator of Transcription 1 (STAT1) and STAT2. Tyrosine-phosphorylated STAT1 and STAT2 dimerize and translocate to the nucleus, where they assemble with IFN-regulatory factor 9 (IRF9) to form a trimolecular complex called IFN-stimulated gene factor 3 (ISGF3). ISGF3 binds to its cognate DNA sequences, which are known as IFN-stimulated response elements (ISREs), thereby directly activating the transcription of ISGs, many of which establish a cellular antiviral state. ISG-encoded proteins restrain pathogens by several mechanisms, including the inhibition of viral transcription, translation and replication, the degradation of viral nucleic acids and the alteration of cellular lipid metabolism. Cellular responses to IFNAR ligation are cell type- and context-dependent and vary during the course of an immune response [9]. Therefore, as said above, although the IFNλ members do not use the IFNα receptor complex for signaling, signaling through either IFNλ or IFNα receptor complexes results in the activation of the same Jak-STAT signal transduction cascade [8].
2.3. Genetic variations affecting IFN response
The relevance of genetic and epigenetic variations in determining the range of susceptibility and of clinical severity of several diseases, including those caused by infectious agents, in the human population is well known. Therefore, considering the importance of IFN system in antiviral defense mechanisms and, more broadly, in immune response, it is not surprising that genetic and epigenetic variations within the IFN genes and in those associated with sensing/signaling/response are associated with a range of diseases. In recent years, inborn errors affecting the production of IFN family members have been reported in human patients. Insights into the specific role of human IFNα/β and IFN-λ-induction pathways in anti-viral immunity have been provided by the study of herpes simplex encephalitis (HSE), the most common sporadic viral encephalitis in Western countries caused by the almost ubiquitous and almost innocuous herpes simplex virus-1 (HSV-1). HSE has been shown to result from a new group of primary immunodeficiencies; the first two genetic etiologies of this disease have been identified as UNC-93B and Toll-like receptor 3 (TLR3) deficiencies. These discoveries demonstrated the critical role of the UNC-93B-dependent, TLR3-IFNα, IFNβ, and IFNλ pathway in immunity to HSV-1 in children. The wide redundancy of other TLR-mediated IFN induction pathways is probably responsible for the lack of increased susceptibility to common viral infection and HSE that is reported for IL-1 receptor-associated kinase 4 (IRAK-4)-deficient patients, who display impaired production of IFNα, IFNβ, and IFNλ following the activation of TLR7, TLR8, and TLR9 [18]. Increased susceptibility to multiple viruses, including HSV-1, has been reported also for inborn errors affecting the response to IFN family members, such as STAT1 or TYK2 deficiency [19]. In addition, IFNγ deficiencies confer Mendelian susceptibility to mycobacterial disease (MSMD) that is a rare congenital syndrome characterized by the occurrence of severe, often disseminated, clinical disease, caused by weakly virulent mycobacterial species, such as bacille Calmette–Guérin (BCG) vaccines and non-tuberculous environmental mycobacteria (EM), in otherwise healthy individuals [18]. IFNγ production is controlled by cytokines secreted by myeloid cells, including IL-12 and IL23. Disorders of six genes of the IL-12/23–IFNγ circuit were discovered since 1996, demonstrating the critical role of this circuit in protective immunity to mycobacteria [20], [21]. The role of IL-12-dependent IFNγ production has been confirmed by recent studies, showing that defects in IL-12p40 or IL-12Rβ1 and specific mutations in NEMO [nuclear factor (NF)-kB essential modulator] are specifically associated with impaired IFNγ-mediated immunity and clinical MSMD [18]. Furthermore, over 347 IFNγ gene variants and several single nucleotide polymorphisms (SNPs) in either IFNγ promoter region or in NFκB binding regions, have been described in multiple ethnic populations, often affecting IFNγ gene expression. Many of these variants appear to modulate susceptibility not only to infectious diseases, especially tuberculosis and viral hepatitis, but also to some non-infectious conditions such as aplastic anemia and psoriasis; susceptibility to cervical cancer, that is in a position to bridge viral and non viral mechanisms, is also influenced by host genetic factors targeting IFNγ reviewed in [22]. Besides genetic variants, several epigenetic modifications are also described, increasing IFNγ expression in Th1 lymphocytes and reducing IFNγ expression in Th2 lymphocytes Increased methylation of IFNγ promoter and consequent decreased expression of this gene is associated with asthma; on the contrary, decreased methylation is associated with increased diastolic blood pressure, biliary atresia, dental pulp inflammation and chronic periodontitis [22]. Overall, human inborn errors affecting IFN system have provided conclusive evidence that the members of IFN family are crucial for protective immunity and innate defense against viral, bacterial and non-infectious diseases. However non-redundant role of these members seems to arise, with a certain polarization of protection, as IFNγ seems to be essential for anti-mycobacterial immunity, whereas IFNα/β and IFNλ are essential for anti-viral immunity [18].
3. Viral strategies to counteract the IFN system
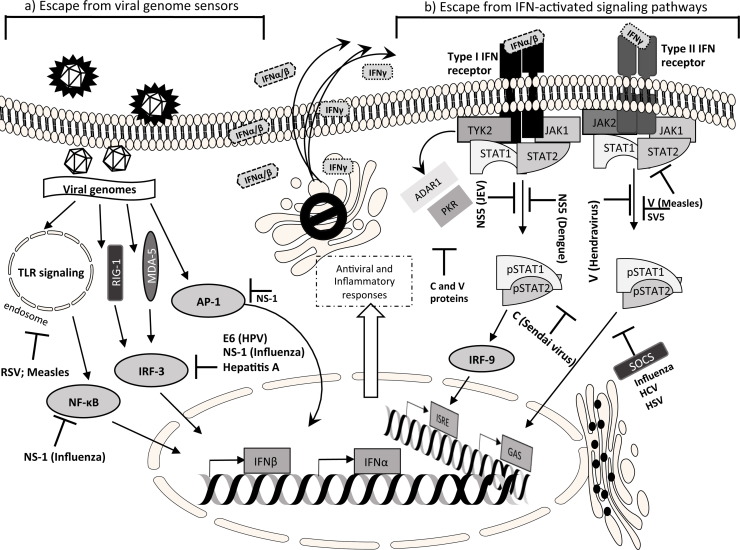
Virus survival and spread into the host depend on the ability to avoid recognition and control by the host defense. Therefore viruses use immunoevasion strategies to counteract the host defenses, particularly against effectors of the innate immunity, as the interferon system and the interferon-induced molecules (Fig. 1 ). Virtually all viruses of mammals have developed multiple mechanisms of immune evasion. Viruses with wide genomes and well adapted to the host (Poxviruses, Herpesviruses) have multiple tools to interfere with the IFN system. In general, differences in the strategies lie on the host cell machinery, rather than on the IFN type. Viruses can evade host defenses through inhibition of IFN induction and/or by inhibition of their signaling pathways, through activation of the IFN regulatory factors (IRFs), the Signal Transducer and Activator of Transcription (STAT) factors, or the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [23], [24], [25]. Viruses developed multiple escape strategies. The IFN-induced Mx pathway is active against a wide range of viruses by blocking primary transcription of the incoming viral genome, by its virionic polymerase, but so far there is no evidence for a virus-coded inhibitor of Mx. Since in the presence of Mx, the virus cannot replicate, the generation of Mx escape mutants/strategies is virtually impossible. Nevertheless, viruses subvert the Mx system, simply by suppressing IFN production in the host (see below), thereby avoiding Mx expression in potential target cells [26].
Fig. 1.
A simplified overview of the IFN signaling pathways counteracted by viruses. (a) After the viral entry and uncoating, the detection of viral genome by cytoplasmic or endosomal sensors is prevented by many viral strategies which, on the whole, target the nuclear translocation of transcription factors (NF-κB, IRF-3, AP-1); other strategies include the block of upstream mediators such as MDA-5 or TLRs. (b) Mechanisms that target the IFN cellular response through the block of JAK-STAT signaling pathway: degradation or cytoplasmic sequestration of STAT proteins by reduction of phosphorylation. See text for details. Abbreviations: ADAR1, adenosine deaminase acting on RNA 1; AP-1, activator protein 1; E6 early protein 6 (HPV); GAS, interferonγ-activated sequence; ISRE, interferon stimulated response element; IRF, interferon regulatory factor; JAK, janus kinase; MDA-5, melanoma differentiation-associated protein 5; NF-κB, NF-κB, nuclear factor κ-light-chain-enhancer of activated B cells; NS-1, non structural protein 1 (Influenza types A, B, C); PKR, protein kinase R; RIG-1, retinoic acid inducible gene 1; RSV, respiratory syncytial virus; SOCS, suppressor of cytokine signaling; STAT, signal transducers and activators of transcription; TYK-2, tyrosine kinase 2, TLR, toll-like receptors.
3.1. Virus inhibition of upstream mediators of IFN production
The cell can detect an ongoing viral invasion, through sensors located in the cytoplasm or on the membrane of endosomes. These molecules include the cytosolic retinoic-acid-inducible gene 1 (RIG-1) and the melanoma differentiation-associated gene 5 (MDA5) dsRNA helicases, through downstream interaction with the interferon promoter-stimulating factor 1 (IPS-1) adaptor protein, and some Toll-like receptors (TLR), as TLR3, 7, 8, and 9 [23], [24], [27]. The vast majority of human infections is due to RNA viruses; those with a dsRNA genome (as rotaviruses and other reoviruses) are detected by TLR3 or by MDA5. During replication of positive-sense ssRNA viruses (picornaviruses like hepatitis A virus and poliovirus, flaviviruses like hepatitis C virus and West Nile virus, coronaviruses like severe acute respiratory syndrome (SARS) virus), a dsRNA intermediate form is produced in the cytoplasm, which is recognized by MDA5. The latter two families may also be sensed by RIG-I. The viruses with a negative-sense ssRNA genome (orthomyxoviruses as influenza A virus, paramyxoviruses like measle virus, mumps virus, respiratory syncytial virus, parainfluenza virus, and Newcastle disease virus, rhabdoviruses like rabies virus and vesicular stomatitis virus, filoviruses like Ebola virus, hantaviruses, Borna disease virus, etc.) activate RIG-I with their ssRNA genome. Double-stranded DNA viruses (such as α, β and γ herpesviruses, adenoviruses, poxviruses) are sensed by TLR9; by convergent transcription they can also produce dsRNA and thus activate TLR3, IFN-inducible dsRNA-dependent protein kinase (PKR), and also MDA5 and/or RIG-I. Retroviruses (such as HIV-1 and human T-lymphotropic virus I and II) have a diploid positive-sense ssRNA genome in the virion, that is reverse transcribed into dsDNA provirus, which is then integrated into the host genome. Retroviral ssRNA is sensed by TLR7 and TLR8. The reverse transcription intermediates are recognized by the cytosolic IFI16 and cGAS DNA sensors, and this leads to the initiation of the type I IFN antiviral response, pyroptosis and apoptosis, under the regulation by host restriction factors such as SAMHD1 and TREX1.
3.2. Viral escape of signaling leading to IFN production
After virus sensing, a series of transcription factors are activated within the cell, leading to IFN production, i.e. IRF-3, NF-κB, the activator protein 1 (AP-1), and the chromatin remodeling associated p300/CREB-binding protein (p300/CBP). These activated molecules translocate to the nucleus and interact with the IFN promoter sequences, to upregulate the expression of interferon genes. IRF-3 activity is hampered by human papillomavirus (HPV) through the E6 protein, by the Influenza A virus through the NS1 protein, that is thought to bind and sequester viral dsRNA, to protect it from cell intrinsic viral RNA detection, and in Filoviruses there is an inverse correlation between the magnitude of IFNα/IFNβ responses and virulence in humans [23], [25], [28], [29].
3.3. Viral inhibition of the JAK-STAT signaling pathway
This signaling pathway (that acts through the JAK and TYK2 kinases, IRF-9, and STAT proteins) varies with the IFN type, since the IFNα/β receptor on the plasma membrane activates the JAK1 and TYK2 molecules, while the IFNγ receptor activates JAK1 and JAK2; by protein phosphorylation the signal is transmitted to the nucleus [30], [31]. Many viruses counteract the JAK-STAT pathway by their V and C proteins, as Paramyxoviruses, rubiviruses, etc., whose V and C proteins block the induction of IFNβ and can degrade the STAT proteins or prevent their phosphorylation or limit their nuclear translocation [23]. This can make the cells unresponsive to both type I and type II interferons, as for the Sendai virus, or the cells do not respond to IFNα but still remain responsive to IFNγ, as it occurs for measles virus [32]. The VP24 protein of the lethal Ebola virus binds the karyopherin alpha nuclear transporter, to inhibit STAT1 nuclear transport and render cells refractory to IFNs [33].
3.4. Inhibition of IFN-induced antiviral effectors
Viruses (influenza virus, HCV, herpes simplex virus) can also induce the suppressor of cytokine signaling (SOCS) proteins, negative regulators of the JAK-STAT signaling [23], [34]. The already mentioned virus-coded C and V proteins can interfere with the IFN-induced PKR (protein kinase R) and ADAR1 (adenosine deaminase acting on RNA 1) enzymes, thus preventing the inhibition of translation and the induction of apoptosis, thus helping virus presence and replication within the cell [35], [36]. Complex viruses have multiple mechanisms to interfere with the IFN system; the various poxviruses are active against type I and type III interferons through a secreted protein, antagonize NF-κB and have a specific interferon-gamma-binding protein that interfere with the binding to the cell receptor [37], [38], [39], [40]. Almost every step of the IFN response can be interfered by the vaccinia poxvirus: induction of IFNs, activation of the NF-κB and IRF-3 and of the JAK-STAT pathways, as well as the activity of the effectors of the antiviral state, such as those involving PKR- and the 2′-5′-Oligoadenylate synthetase [37], [41]. Recent studies have shown that the Acanthamoeba polyphaga mimivirus (APMV) may be pathogen for humans, causing pneumonia. In human PBMCs APMV replicates, inducing type I IFNs, but inhibiting IFN stimulated genes (ISG) induction by viroceptor and STAT-1 and STAT-2 dephosphorylation independent mechanisms; it is resistant to IFNα2, but sensitive to IFNβ [42].
4. Synergistic and antagonistic interactions
The ability of IFN family members to protect against various viral infections descends from the complex transcriptional programs they initiate. Each type of IFN can induce the expression of hundreds of genes to mediate various biological responses. Although IFNs are effective inhibitors of viruses such as vesicular stomatitis virus (VSV) and encephalomyocarditis virus (EMCV), almost all RNA and DNA viruses have evolved mechanisms to subvert the host IFN response, and are able to resist, at least in part, to the IFN-mediate inhibition. However, several studies have shown that viruses normally resistant to the effects of type I or type II IFN separately, are susceptible to IFNs when used in combination. The reciprocal potentiation of type I or type II IFN is a well known phenomenon described since 1979 [43], and has been observed for both antiviral and antitumor effects [44]. Synergistic antiviral activity resulting from the contemporary presence of both IFN types has also been described in HIV-infected patients and in some autoimmune diseases [45]. It has been demonstrated that IFNα/β and IFNγ synergistically inhibit the replication of HSV-1 both in vitro and in vivo [46], [47]. In addition, several reports have indicated that IFN family members used in combination have a synergistic antiviral activity against other viruses, such as hepatitis C virus (HCV) [48], Lassa virus (LASV) [49], Varicella-Zoster virus (VZV) [50], human Cytomegalovirus (hCMV) [47] and severe acute respiratory syndrome-associated coronavirus (SARS-CoV) [51], [52].
Peng and collaborators tried to better understand the mechanisms underlying the synergism between type I and type II IFN in the inhibition of HSV-1 replication. They showed that IFNβ1 and IFNγ interaction is exploited through either an independent, i.e. inducing distinct gene categories, or a cooperative mechanism, i.e. inducting a common subset of genes at a level that is much higher than with either IFN alone. The genes cooperatively induced by IFNβ1 and IFNγ included those involved in apoptosis, RNA degradation and inflammatory response. As result, the combination of IFNβ1 and IFNγ induced more apoptosis and inhibited HSV-1 gene expression and DNA replication significantly more than treatment with either IFN alone [53].
While synergism between type I and type II IFN is well established, little is known about the effect of type I and type III IFNs combination, as available studies report contrasting information, with synergism reported for some viruses (i.e. HCV) [54], [55], but not for other viruses (i.e. EMCV and HSV-2) [56]. In addition, antagonism for the intracellular pathways involved in antiviral activity has been reported [57]. In a recent study [58] possible synergism/antagonism between IFNα and/or IFNλ in the inhibition of virus replication (EMCV, WNV lineage 1 and 2, CHIKV and HSV-1) has been investigated in different cell lines. The results showed that IFNλ is less effective than IFNα for both the inhibition of virus replication, and that IFNλ antagonized the IFNα-driven inhibition of virus replication. The findings on antiviral activity are paralleled by analogous trends in the induction of the molecular mediators of antiviral activity (MxA and 2′-5′OAS). An overview of the interactions between IFN family members is reported in Table 2 .
Table 2.
Overview of synergistic and antagonistic interactions among IFN family members. See text for details and references.
| IFNs combination | Type of interaction | Outcome |
|---|---|---|
| IFNα or β + IFNγ | Synergy | Inhibition of HSV-1 replication |
| IFNα or β + IFNγ | Synergy | Inhibition of HCV replication |
| IFNα + IFNγ | Synergy | Inhibition of LASV replication |
| IFNβ + IFNγ | Synergy | Inhibition of VZV replication |
| IFNα or β + IFNγ | Synergy | Inhibition of hCMV replication |
| IFNα + IFNγ | Synergy | Inhibition of SARS-Cov replication |
| IFNα or β + IFNγ | Synergy | Inhibition of SARS-Cov replication |
| IFNβ + IFNγ | Synergy | Inhibition of HSV-1 replication |
| IFNα + IFNλ | Synergy | Inhibition of HCV replication |
| IFNα + IFNλ | Synergy | Inhibition of HCV replication |
| IFNα or β + IFNλ | Antagonism | Inhibition of EMCV and HSV-2 replication |
| IFNα + IFNλ | Antagonism | Inhibition of EMCV, WNV, CHIKV and HSV-1 replication |
5. Activities not related to viruses
Studies of the late Sixties revealed that the antiviral agent IFN was able also to suppress the growth of tumors [59]. Since then, IFNs were shown to be pleiotropic biologic response modifiers, that, upon activation of thousands genes [60] induce a broad spectrum of activities, regulating cell cycle, differentiation, expression of pivotal molecules on the plasma membrane, release of mediators, etc., that can be relevant for cell proliferation, innate and adaptive immunity, hematopoiesis, angiogenesis, and with important outcomes also for cancer, autoimmunity, and therapy [61], [62]. A survey of the spectrum of biological activities of the interferons is reported in Table 3 . Importantly, because of the multitude of IFN-induced genes, opposite effects can be determined by the same IFN, depending on timing and dosages, both in vitro and in vivo, as shown since the Seventies in the mouse Friend erythroleukemia cells [63], [64]; IFN effects on patients were defined “Janus-like”, as IFNs may have both beneficial and detrimental effects, depending on the disease context [65].
Table 3.
Spectrum of biological activities of the interferons. See text for details and references.
| Activity | Type I IFN | Type II IFN | Type III IFN |
|---|---|---|---|
| IFN-responsive genesa | 2169 | 2339 | 159 |
| Inducibility by virus infections | ++++ | +++ | ++ |
| Induction of antiviral effectors | ++++ | ++ | ++ |
| Cell growth inhibition | ++ | ++++ | + |
| Induction of autophagy | ++ | ++ | nsb |
| Stimulation/inhibition of cell differentiation | ++ | ++++ | ns |
| Stimulation of MHC class I antigens | ++ | ++++ | ++ |
| Stimulation of MHC class II antigens | ++ | ++++ | ns |
| Stimulation/inhibition of phagocytosis | ++ | ++++ | ns |
| Stimulation/inhibition of antibody production | ++ | ++++ | ns |
| Inhibition of intracellular parasites | +, indirect | ++, direct | ns |
| Induction of IFN, cytokines and chemokines | + | ++ | + |
| Induction of neurologic symptoms | ++ | ++ | − |
| Specific receptor distribution in the body | Ubiquitous | Ubiquitous | Tissue-restricted |
Cellular genes with p-value <0.05 and fold change ≥2, according to http://www.interferome.org[60].
ns: not studied, so far.
5.1. Anti-proliferative activity
This property of all IFNs can help the host by depletion of infected cells, restrain excess cell-mediated immunity and inhibit angiogenesis, with consequent reduction of tumor growth; it may occur both in cancer and normal cells, in vitro and in vivo. Type II IFN is more effective than type I IFN (being IFNβ more potent than IFNα), while type III IFN is restrained by the low or limited presence of the specific receptor on the membrane of the target cells [66], [67], [68]. IFNβ has a greater receptor-binding affinity than IFNα, and this finding may account for the more potent and different antiproliferative and perhaps immunoregulatory actions of IFNβ [69]. The anti-growth activity of IFNs is mediated by arrest of the cell cycle and induction of caspases and apoptosis; it occurs in all the somatic cells tested, with differences depending on the IFN type and dosage [66], [70]. The anti-proliferative activity of type III IFNs does not occur in all the receptor-positive cells but depends on the cell type, at variance with the antiviral activity and up-regulation of MHC class I molecules [66]. Because of possible therapy of melanoma, type I IFNs were extensively studied in keratinocytes and melanocytes [67]; of note, the IFNβ expression in vitro by undifferentiated, growth-arrested murine keratinocytes suggested that its production by terminally differentiated cells was associated with arrest of proliferation. Recently it was reported the induction of autophagy by type I and type II IFNs in cancer cells, a mechanism that may counteract the pro-apoptotic functions of IFNs [11], [71], [72], through the p38 MAPK pathway and ATF6 phosphorylation [72].
5.2. Cell differentiation
It can be modulated by IFNs, as shown by an endless list of reports. Type I and type II IFNs were shown to affect the erythropoiesis pathways [63], [64], [73]. Chronic IFNγ production may cause anemia and bone marrow failure, hence it may be involved in malignant hematopoietic malignancies [73]. Another IFN-regulated process is adypogenesis, that is inhibited by type I and II IFNs [74], [75]. In normal thyrocytes both IFNα and IFNγ regulate the expression of integrins (target for infiltrating lymphocytes) and the signaling induced by cell adhesion to fibronectin [76]. Moreover, IFNγ induces CXCR3-binding chemokines, which in turn recruit Th1 lymphocytes, with an important role in the initiation of autoimmune thyroiditis.
5.3. Immunomodulation
IFNs are potent immunomodulators, especially IFNγ; among the IFNα subtypes, each has its own immunomodulatory pattern [77]. In general, IFNs stimulate already differentiated cells in the short term, while, by growth inhibition, in the long term or with higher IFN concentrations, they may reduce the replacement of exhausted cells with their precursors. Immunomodulatory effects include enhancement of surface molecules such as MHC (major histocompatibility complex) antigens, several receptors, and modulation of the activation and differentiation of effector cells, as monocyte/macrophages, dendritic cells (DC), B, T and NK cells. Type I and III IFNs and, with higher efficiency, type II IFN upregulate the expression of class I MHC and co-stimulatory molecules by tissue cells and immune cells, as the DCs, that differentiate into efficient antigen- and self-antigen-presenting cells, leading to activation of quiescent reactive T helper cells [66], [78], [79], [80]. The expression of class II MHC molecules is upregulated by all three IFN types, being the type II the most active [81], [82]. The effect of the latter IFN can be interfered by type I IFNs [83]. The activation of DR and of chemokine receptors may modify the phenotype (and recognition of progeny virions), and virus binding on the membrane [84], [85]. Type I IFNs thus function to elevate both innate and adaptive immune responses, with induction of cytokines and chemokines, activation and differentiation of CD4+ T cells and CTLs, the activation and differentiation of B cells, the production of antibodies and the switching of immunoglobulin (Ig)isotypes [61], [66], [77]. Type III IFNs were not demonstrated to increase antibody formation, and were shown to modulate Th1/Th2 cells balance through a different mechanism from type I IFNs [66]. IFNγ, but not IFNα or IFNβ can induce neutrophils to suppress lymphocyte proliferation through the expression of programmed death ligand 1 (PD-L1) [86]. Type II IFN is essential also in immunity intracellular non-viral parasites such as Toxoplasma gondii, Cryptosporidium parvum, Leishmania major, Trypanosoma cruzi, etc., and elicits strong cell-autonomous killing of intracellular parasites; also type I IFN, however is induced during these infections: it does not exert an intrinsic control of parasites, but its signaling recruits NK cells, which then mediate parasite killing by producing type II IFN [87].
5.4. Interferon-induced diseases
Dysregulation in production/signaling of all IFN types are associated to a variety of pathologies. Activation of IFNα pathways were related to several autoimmune diseases, such as systemic lupus erythematosus, Addison's disease, etc. [88]. Miscarriage of humans and animals has been associated with higher levels of IFNγ, that, on the other hand, is involved in maintaining the decidual layer and vascular remodeling in the uterus [89]. Abnormality in IFN-stimulated gene patterns, causing failures in immunotolerance in early childhood were proposed to be contribute to the development of type 1 diabetes and cases of type 1 diabetes were also related to treatments with pegylated IFN and ribavirin for chronic hepatitis C [90]. Cardiovascular complications can be observed in IFN-treated subjects (arrhythmia, myocarditis, reversible hypertension, ischemic heart disease, pericarditis and pericardial effusion, cardiomyopathy) and are generally reversible [88]. Sarcoidosis, a systemic chronic granulomatous disease of unknown cause, is a rare adverse effect from the use of type I IFNα and IFNβ, that reflects an exaggerated cell-mediated immune response to an unknown persistent antigen, with higher levels of circulating IFNγ [91]. Thyroid dysfunctions in IFN-treated patients were observed since 1985 [92], and are still a significant problem for patients receiving either IFNα or IFNβ; in vitro and animal studies showed that thyroid inflammation and autoimmunity involve direct effects (increased expression of thyroid differentiation) as well as activation of destructive bystander immune responses [93]. Significant exacerbations were caused by treatment of multiple sclerosis patients with IFNγ, while IFNβ is currently given as therapy, and seems to be more potent than IFNα [94], [95]. In the EAE animal model of multiple sclerosis, a regulatory role of Type III IFN-producing CD4+ T cells conferring protection against EAE was proposed [96]. Depression and other cognitive and psychiatric disturbances are reported by approximately half of the patients treated with IFNα for chronic hepatitis C and B, Behçet's syndrome, melanoma and lymphoma [97]. IFNα determines abnormalities of the hypothalamo-pituitary-adrenal axis and disturbances of brain metabolism, and interferes with neurotrophic signaling, thus hindering neurite outgrowth, synaptic plasticity, endogenous neurogenesis and neuronal survival [97]. Comparing patients receiving pegylated IFNα plus ribavirin with those receiving IFNβ plus ribavirin, it was found that depressive symptoms were more prevalent following IFNα than IFNβ treatment [98]. In mice, IFNα was reported either to promote antidepressant-like effect associated with an increase in brain serotonin turn over, or to promote a depression-like phenotype, mediated by suppression of hippocampal neurogenesis and induction of depression [99], while the lack of IFNγ increased anxiety-like and depressive-like behaviors of IFNγ-knocked out animals, suggesting that IFNγ is involved in hippocampal neurogenesis and CNS plasticity [100].
6. Concluding remarks
Interferons have extraordinary antiviral efficacy; therefore viruses have developed multiple strategies of immune evasion to counteract IFN induction and signaling and the interferon-induced effector molecules. During the last decades, IFNs were shown to display a multifaceted, Janus-like congeries of activities, with synergisms and antagonisms between IFN types. Because of the multitude of IFN-induced genes, also opposite effects can be determined by the same IFN, depending on timing and dosages, and IFNs may have both beneficial and detrimental effects, depending on the disease context. Genetic diversity, with polymorphisms and mutations, can modulate the extent of IFN responses and the susceptibility to infections, and genetic and epigenetic variations within the IFN genes and in those associated with sensing/signaling/response can be linked to diseases. On the whole, IFNs are pleiotropic biologic response modifiers, that, upon activation of thousands genes, induce a broad spectrum of activities, regulating cell cycle, differentiation, plasma membrane molecules, release of mediators, etc., that can be relevant for cell proliferation, innate and adaptive immunity, hematopoiesis, angiogenesis and other body functions.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
This work was partly supported by grants from Italian Ministry of Health, Ricerca Corrente and Ricerca Finalizzata (M.R.C.), and from CARITRO 2013 (A.D.). E.U. was funded by a grant from Regione Autonoma Sardegna 2013 (Master & Back Program).
Biographies

Dr. Maria R. Capobianchi is PhD in Microbiology and Virology. She has been involved in research activity on viruses since 1977, when she joined the Institute of Virology at the University of Rome Medical School as fellow, and in 1981 got permanent position. Since 2000, Dr. Capobianchi is the Director of the Laboratory of Virology at the National Institute for Infectious Diseases, Rome, where she is in charge of diagnostic and research activity on viral diseases. Main scientific interests: Innate immunity and defence mechanisms against viruses; Interferon induction and action; Pathogenesis of persistent viral infections; Biosafety; Laboratory alert and preparedness to emerging viral diseases. As of October 2014, Dr. Capobianchi is listed as Author or co-author of more than 300 indexed scientific articles, with more than 5000 citations, H index 38.

Elena Uleri, Ph.D., is a Postdoctoral fellow at the Virology Section of the Department of Biomedical Sciences, University of Sassari (Italy). She earned her doctorate in Fundamental and Clinical Virology at University of Sassari in consortium with University of Pisa (Italy), where she studied the role of human endogenous retroviruses HERV-Ws in the pathogenesis of Multiple Sclerosis (MS). As a Visiting research scholar at Department of Neurovirology of Temple University, Philadelphia (USA), she studied the interplay between host restriction factors and JC virus life cycle in PML and animal tumors JCV-induced. She is currently focused on the role of JCV in colon cancer and on the modulation of host factors by MS drugs that could be involved in JCV reactivation, on the mechanisms by which HIV-tat reactivates HERV-Ws expression and on interactions between viruses and HERV-W relevant in neurodegenerative diseases, on the interferon and cytokines role in the modulation of HERV-Ws. She has a broad background in molecular biology. She is member of the Italian Society of Virology (SIV).

Claudia Caglioti graduated in Biology in 2012 at Roma Tre University (Italy). From October 2010 to January 2013 she attended the Laboratory of Virology at National Institute for Infectious Diseases “L. Spallanzani” in Rome, for developing her experimental thesis. Since 2013 she is attending the Postgraduate School in Microbiology and Virology. Currently she is research fellows at the National Institute for Infectious Diseases “L. Spallanzani”, where she is developing her PHD thesis focused on “Molecular basis of severe influenza A virus respiratory syndromes (pandemic and seasonal): study of viral evolution and identification of prognostic genetic markers”.

Antonina (Ninella) Dolei received her specialization/doctoral degree in Microbiology from the University of Rome in 1975 and in Virology from the University of Bologna in 1977. She became assistant professor of Virology, Faculty of Medicine at La Sapienza University, Rome in 1981. After serving as associate professor of Pathology, Faculty of Sciences at the University of Camerino, Italy until 1985, she joined the Faculty of Medicine at the University of Sassari as full professor of Virology. Since 1975, she studies INTERFERONS (antiretroviral and cell-modulating aspects, protective and immunopathogenic activities) and RETROVIRUSES (HIV pathogenesis, since 1984, and human endogenous retroviruses (HERV) in the pathogenesis of multiple sclerosis and of neuroAIDS, since 1998). ONGOING STUDIES: activation of HERV-Ws in pathogenic mechanisms linked to neurodegeneration and cancer, and as prognostic biomarker of multiple sclerosis progression and therapy outcome; immunosubversion of host defenses by retroviruses. She was co-founder of the Italian Society for Virology (2001, now serving as vice-President), and co-founder of the European Society for Virology (2009); she is in the board of directors of the International Society of Neurovirology, and in the Nomenclature Committee of the International Cytokine and Interferon Society, being member of the Interferon society from the beginnings (ISIR, ISICR and now ICIS).
References
- 1.Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Nagano Y., Kojima Y. Inhibition of vaccinia infection by a liquid factor in tissues infected by homologous virus. C R Seances Soc Biol Fil. 1958;152:1627–1629. [PubMed] [Google Scholar]
- 3.Samuel C.E. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touzot M., Soumelis V., Asselah T. A dive into the complexity of type I interferon antiviral functions. J Hepatol. 2012;56:726–728. doi: 10.1016/j.jhep.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Wheelock E.F. Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science. 1965;149:310–311. [PubMed] [Google Scholar]
- 6.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 7.Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly R.P., Kotenko S.V. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 11.Schmeisser H., Bekisz J., Zoon K.C. New function of type I IFN: induction of autophagy. J Interferon Cytokine Res. 2014;34:71–78. doi: 10.1089/jir.2013.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopušná K., Režuchová I., Betáková T., Skovranová L., Tomašková J., Lukáčiková L. Interferons lambda, new cytokines with antiviral activity. Acta Virol. 2013;57:171–179. doi: 10.4149/av_2013_02_171. [DOI] [PubMed] [Google Scholar]
- 13.Le Bon A., Tough D.F. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 14.Kotenko S.V. The family of IL-10-related cytokines and their receptors: related, but to what extent. Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 15.Renauld J.C. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat Rev Immunol. 2003;3:667–676. doi: 10.1038/nri1153. [DOI] [PubMed] [Google Scholar]
- 16.Pestka S., Krause C.D., Sarkar D., Walter M.R., Shi Y., Fisher P.B. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 17.Kotenko S.V., Langer J.A. Full house: 12 receptors for 27 cytokines. Int Immunopharmacol. 2004;4:593–608. doi: 10.1016/j.intimp.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S.Y., Boisson-Dupuis S., Chapgier A., Yang K., Bustamante J., Puel A. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 19.Jouanguy E., Zhang S.Y., Chapgier A., Sancho-Shimizu V., Puel A. Human primary immunodeficiencies of type I interferons. Biochimie. 2007;89:878–883. doi: 10.1016/j.biochi.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Newport M.J., Huxley C.M., Huston S., Hawrylowicz C.M., Oostra B.A., Williamson R. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 21.Jouanguy E., Altare F., Lamhamedi S., Revy P., Emile J.F., Newport M. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guérin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 22.Smith N.L., Denning D.W. Clinical implications of interferon gamma genetic and epigenetic variants. Immunology. 2014 doi: 10.1111/imm.12362. Accepted Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devasthanam A.S. Mechanisms underlying the inhibition of interferon signaling by viruses. Virulence. 2014;5:270–277. doi: 10.4161/viru.27902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Montfoort N., Olagnier D., Hiscott J. Unmasking immune sensing of retroviruses: interplay between innate sensors and host effectors. Cytokine Growth Factor Rev. 2014 doi: 10.1016/j.cytogfr.2014.08.006. pii:S1359-6101(14)00089-6. [DOI] [PubMed] [Google Scholar]
- 25.Orzalli M.H., Knipe D.M. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haller O., Kochs G., Weber F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007;18:425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang J.J., Davis M.E., Gack M.U. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev. 2014 doi: 10.1016/j.cytogfr.2014.06.005. pii:S1359-6101(14)00059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronco L.V., Karpova A.Y., Vidal M., Howley P.M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong G., Kobinger G.P., Qiu X. Characterization of host immune responses in Ebola virus infections. Expert Rev Clin Immunol. 2014;10:781–790. doi: 10.1586/1744666X.2014.908705. [DOI] [PubMed] [Google Scholar]
- 30.Horvath C.M. The Jak-STAT pathway stimulated by interferon alpha or interferon beta. Sci STKE. 2004;2004:tr10. doi: 10.1126/stke.2602004tr10. [DOI] [PubMed] [Google Scholar]
- 31.Stark G.R., Kerr I.M. Interferon-dependent signaling pathways: DNA elements, transcription factors, mutations, and effects of viral proteins. J Interferon Res. 1992;12:147–151. doi: 10.1089/jir.1992.12.147. [DOI] [PubMed] [Google Scholar]
- 32.Yokota S., Saito H., Kubota T., Yokosawa N., Amano K., Fujii N. Measles virus suppresses interferon-alpha signaling pathway: suppression of Jak1 phosphorylation and association of viral accessory proteins, C and V, with interferon-alpha receptor complex. Virology. 2003;306:135–146. doi: 10.1016/s0042-6822(02)00026-0. [DOI] [PubMed] [Google Scholar]
- 33.Xu W., Edwards M.R., Borek D.M., Feagins A.R., Mittal A., Alinger J.B. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014;16:187–200. doi: 10.1016/j.chom.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bode J.G., Ludwig S., Ehrhardt C., Albrecht U., Erhardt A., Schaper F. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 35.Li Z., Okonski K.M., Samuel C.E. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J Virol. 2012;86:3787–3794. doi: 10.1128/JVI.06307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith G.L., Benfield C.T., Maluquer de Motes C., Mazzon M., Ember S.W., Ferguson B.J. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 38.Huang J., Smirnov S.V., Lewis-Antes A., Balan M., Li W., Tang S. Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc Natl Acad Sci U S A. 2007;104:9822–9827. doi: 10.1073/pnas.0610352104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brady G., Bowie A.G. Innate immune activation of NFκB and its antagonism by poxviruses. Cytokine Growth Factor Rev. 2014 doi: 10.1016/j.cytogfr.2014.07.004. pii:S1359-6101(14)00066-5. [DOI] [PubMed] [Google Scholar]
- 40.Puehler F., Schwarz H., Waidner B., Kalinowski J., Kaspers B., Bereswill S. An interferon-gamma-binding protein of novel structure encoded by the fowlpox virus. J Biol Chem. 2003;278:6905–6911. doi: 10.1074/jbc.M207336200. [DOI] [PubMed] [Google Scholar]
- 41.Perdiguero B., Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res. 2009;29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 42.Silva L.C., Almeida G.M., Oliveira D.B., Dornas F.P., Campos R.K., La Scola B. A resourceful giant: APMV is able to interfere with the human type I interferon system. Microbes Infect. 2014;16:187–195. doi: 10.1016/j.micinf.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Fleischmann W.R, Jr., Georgiades J.A., Osborne L.C., Johnson H.M. Potentiation of interferon activity by mixed preparations of fibroblast and immune interferon. Infect Immun. 1979;26:248–253. doi: 10.1128/iai.26.1.248-253.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Clercq E., Zhang Z.X., Huygen K. Synergism in the antitumor effects of type 1 and type II interferon in mice inoculated with leukemia L1210 cells. Cancer Lett. 1982;15:223–228. doi: 10.1016/0304-3835(82)90122-7. [DOI] [PubMed] [Google Scholar]
- 45.Capobianchi M.R., Mattana P., Mercuri F., Conciatori G., Ameglio F., Ankel H. Acid lability is not an intrinsic property of interferon-alpha induced by HIV-infected cells. J Interferon Res. 1992;12:431–438. doi: 10.1089/jir.1992.12.431. [DOI] [PubMed] [Google Scholar]
- 46.Sainz B., Jr., Halford W.P. Alpha/beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J Virol. 2002;76:11541–11550. doi: 10.1128/JVI.76.22.11541-11550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sainz B., Jr., LaMarca H.L., Garry R.F., Morris C.A. Synergistic inhibition of human cytomegalovirus replication by interferon-alpha/beta and interferon-gamma. Virol J. 2005;2:14. doi: 10.1186/1743-422X-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larkin J., Jin L., Farmen M., Venable D., Huang Y., Tan S.L. Synergistic antiviral activity of human interferon combinations in the hepatitis C virus replicon system. J Interferon Cytokine Res. 2003;23:247–257. doi: 10.1089/107999003321829962. [DOI] [PubMed] [Google Scholar]
- 49.Asper M., Sternsdorf T., Hass M., Drosten C., Rhode A., Schmitz H. Inhibition of different Lassa virus strains by alpha and gamma interferons and comparison with a less pathogenic arenavirus. J Virol. 2004;78:3162–3169. doi: 10.1128/JVI.78.6.3162-3169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desloges N., Rahaus M., Wolff M.H. Role of the protein kinase PKR in the inhibition of varicella-zoster virus replication by beta interferon and gamma interferon. J Gen Virol. 2005;86:1–6. doi: 10.1099/vir.0.80466-0. [DOI] [PubMed] [Google Scholar]
- 51.Castilletti C., Bordi L., Lalle E., Rozera G., Poccia F., Agrati C. Coordinate induction of IFN-alpha and -gamma by SARS-CoV also in the absence of virus replication. Virology. 2005;341:163–169. doi: 10.1016/j.virol.2005.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scagnolari C., Trombetti S., Alberelli A., Cicetti S., Bellarosa D., Longo R. The synergistic interaction of interferon types I and II leads to marked reduction in severe acute respiratory syndrome-associated coronavirus replication and increase in the expression of mRNAs for interferon-induced proteins. Intervirology. 2007;50:156–160. doi: 10.1159/000098242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng T., Zhu J., Hwangbo Y., Corey L., Bumgarner R.E. Independent and cooperative antiviral actions of beta interferon and gamma interferon against herpes simplex virus replication in primary human fibroblasts. J Virol. 2008;82:1934–1945. doi: 10.1128/JVI.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcello T., Grakoui A., Barba-Spaeth G., Machlin E.S., Kotenko S.V., MacDonald M.R. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 55.Shindo H., Maekawa S., Komase K., Miura M., Kadokura M., Sueki R. IL-28B (IFN-λ3) and IFN-α synergistically inhibit HCV replication. J Viral Hepat. 2013;20:281–289. doi: 10.1111/j.1365-2893.2012.01649.x. [DOI] [PubMed] [Google Scholar]
- 56.Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.François-Newton V., Magno de Freitas Almeida G., Payelle-Brogard B., Monneron D., Pichard-Garcia L., Piehler J. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bordi L., Lalle E., Lapa D., Caglioti C., Quartu S., Capobianchi M.R. Type III interferon (IFN-lambda) antagonizes the antiviral activity of interferon-alpha in vitro. J Biol Regul Homeost Agents. 2013;27:1001–1009. [PubMed] [Google Scholar]
- 59.Gresser I. Antitumor effects of interferon. Adv Cancer Res. 1972;16:97–140. doi: 10.1016/s0065-230x(08)60339-5. [DOI] [PubMed] [Google Scholar]
- 60.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(database issue):D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wijesundara D.K., Xi Y., Ranasinghe C. Unraveling the convoluted biological roles of type I interferons in infection and immunity: a way forward for therapeutics and vaccine design. Front Immunol. 2014;5:412. doi: 10.3389/fimmu.2014.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.El-Baky N.A., Redwan E.M. Therapeutic alpha-interferons protein: structure production, and biosimilar. Prep Biochem Biotechnol. 2015;45:109–127. doi: 10.1080/10826068.2014.907175. [DOI] [PubMed] [Google Scholar]
- 63.Swetly P., Ostertag W. Friend virus release and induction of haemoglobin synthesis in erythroleukaemic cells respond differently to interferon. Nature. 1974;251:642–644. doi: 10.1038/251642a0. [DOI] [PubMed] [Google Scholar]
- 64.Rossi G.B., Dolei A., Cioé L., Benedetto A., Matarese G.P., Belardelli F. Inhibition of transcription and translation of globin messenger RNA in dimethyl sulfoxide-stimulated Friend erythroleukemic cells treated with interferon. Proc Natl Acad Sci USA. 1977;74:2036–2040. doi: 10.1073/pnas.74.5.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Axtell R.C., Raman C. Janus-like effects of type I interferon in autoimmune diseases. Immunol Rev. 2012;248:23–35. doi: 10.1111/j.1600-065X.2012.01131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q., Kawamura K., Tada Y., Shimada H., Hiroshima K., Tagawa M. Novel type III interferons produce anti-tumor effects through multiple functions. Front Biosci (Landmark Ed) 2013;18:909–918. doi: 10.2741/4152. [DOI] [PubMed] [Google Scholar]
- 67.Ismail A., Yusuf N. Type I interferons: key players in normal skin and select cutaneous malignancies. Dermatol Res Pract. 2014;2014:847545. doi: 10.1155/2014/847545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balachandran S., Adams G.P. Interferon-γ-induced necrosis: an antitumor biotherapeutic perspective. J Interferon Cytokine Res. 2013;33:171–180. doi: 10.1089/jir.2012.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaitin D.A., Roisman L.C., Jaks E., Gavutis M., Piehler J., Van der Heyden J. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levin D., Schneider W.M., Hoffmann H.H., Yarden G., Busetto A.G., Manor O. Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci Signal. 2014;7:ra50. doi: 10.1126/scisignal.2004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ambjørn M., Ejlerskov P., Liu Y., Lees M., Jaatela M., Issazadeh-Navikas S. IFNbeta1/interferon-beta-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy. 2013;9:287–302. doi: 10.4161/auto.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gade P., Manjegowda S.B., Nallar S.C., Maachani U.B., Cross A.S., Kalvakolanu D.V. Regulation of the death-associated protein kinase 1 expression and autophagy via ATF6 requires apoptosis signal-regulating kinase 1. Mol Cell Biol. 2014;34:4033–4048. doi: 10.1128/MCB.00397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Bruin A.M., Voermans C., Nolte M.A. Impact of interferon-γ on hematopoiesis. Blood. 2014 doi: 10.1182/blood-2014-04-568451. pii:blood-2014-04-568451. [DOI] [PubMed] [Google Scholar]
- 74.Cioé L., O’Brien T.G., Diamond L. Inhibition of adipose conversion of BALB/c 3T3 cells by interferon and 12-O-tetradecanoylphorbol-13-acetate. Cell Biol Int Rep. 1980;4:255–264. doi: 10.1016/0309-1651(80)90057-0. [DOI] [PubMed] [Google Scholar]
- 75.Vidal C., Bermeo S., Li W., Huang D., Kremer R., Duque G. Interferon gamma inhibits adipogenesis in vitro and prevents marrow fat infiltration in oophorectomized mice. Stem Cells. 2012;30:1042–1048. doi: 10.1002/stem.1063. [DOI] [PubMed] [Google Scholar]
- 76.Russo E., Salzano M., Postiglione L., Guerra A., Marotta V., Vitale M. Interferon-γ inhibits integrin-mediated extracellular signal-regulated kinase activation stimulated by fibronectin binding in thyroid cells. J Endocrinol Invest. 2013;36:375–378. doi: 10.3275/8649. [DOI] [PubMed] [Google Scholar]
- 77.Bekisz J., Sato Y., Johnson C., Husain S.R., Puri R.K., Zoon K.C. Immunomodulatory effects of interferons in malignancies. J Interferon Cytokine Res. 2013;33:154–161. doi: 10.1089/jir.2012.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcin G., Bordat Y., Chuchana P., Monneron D., Law H.K., Piehler J. Differential activity of type I interferon subtypes for dendritic cell differentiation. PLOS ONE. 2013;8:e58465. doi: 10.1371/journal.pone.0058465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heron I., Hokland M., Berg K. Enhanced expression of β2-microglobulin and HLA antigens on human lymphoid cells by interferon. Proc Natl Acad Sci USA. 1978;75:6215–6219. doi: 10.1073/pnas.75.12.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dolei A., Ameglio F., Capobianchi M.R., Tosi R. Human β-type interferon enhances the expression and shedding of Ia-like antigens. Comparison to HLA-A, B, C and β2-microglobulin. Antiviral Res. 1981;1:367–381. [Google Scholar]
- 81.Mennechet F.J., Uzé G. Interferon-lambda-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood. 2006;107:4417–4423. doi: 10.1182/blood-2005-10-4129. [DOI] [PubMed] [Google Scholar]
- 82.Dolei A., Capobianchi M.R., Ameglio F. Human interferon-gamma enhances the expression of class I and class II major histocompatibility complex products in neoplastic cells more effectively than interferon-alpha and interferon-beta. Infect Immun. 1983;40:172–176. doi: 10.1128/iai.40.1.172-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barna B.P., Chou S.M., Jacobs B., Yen-Lieberman B., Ransohoff R.M. Interferon-beta impairs induction of HLA-DR antigen expression in cultured adult human astrocytes. J Neuroimmunol. 1989;23:45–53. doi: 10.1016/0165-5728(89)90072-6. [DOI] [PubMed] [Google Scholar]
- 84.Capobianchi M.R., Serra C., Abbate I., Marongiu P., Castilletti C., Tilocca F. Treatment of HIV-infected fibroblasts with human leukocyte antigen (HLA)-DR-inductive cytokines leads to infectious virions with newly acquired HLA-DR. AIDS. 1994;8:1731–1733. doi: 10.1097/00002030-199412000-00017. [DOI] [PubMed] [Google Scholar]
- 85.Serra C., Biolchini A., Mei A., Kotenko S., Dolei A. Type III and I interferons increase HIV uptake and replication in human cells that overexpress CD4, CCR5, and CXCR4. AIDS Res Hum Retroviruses. 2008;24:173–180. doi: 10.1089/aid.2007.0198. [DOI] [PubMed] [Google Scholar]
- 86.de Kleijn S., Langereis J.D., Leentjens J., Kox M., Netea M.G., Koenderman L. IFN-γ-stimulated neutrophils suppress lymphocyte proliferation through expression of PD-L1. PLOS ONE. 2013;8:e72249. doi: 10.1371/journal.pone.0072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beiting D.P. Protozoan parasites and type I interferons: a cold case reopened. Trends Parasitol. 2014;30:491–498. doi: 10.1016/j.pt.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Hellesen A., Edvardsen K., Breivik L., Husebye E.S., Bratland E. The effect of types I and III interferons on adrenocortical cells and its possible implications for autoimmune Addison's disease. Clin Exp Immunol. 2014;176:351–362. doi: 10.1111/cei.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Micallef A., Grech N., Farrugia F., Schembri-Wismayer P., Calleja-Agius J. The role of interferons in early pregnancy. Gynecol Endocrinol. 2014;30:1–6. doi: 10.3109/09513590.2012.743011. [DOI] [PubMed] [Google Scholar]
- 90.Panarina M., Kisand K., Alnek K., Heilman K., Peet A., Uibo R. Interferon and interferon-inducible gene activation in patients with type 1 diabetes. Scand J Immunol. 2014;80:283–292. doi: 10.1111/sji.12204. [DOI] [PubMed] [Google Scholar]
- 91.Buss G., Cattin V., Spring P., Malinverni R., Gilliet M. Two cases of interferon-alpha-induced sarcoidosis Koebnerized along venous drainage lines: new pathogenic insights and review of the literature of interferon-induced sarcoidosis. Dermatology. 2013;226:289–297. doi: 10.1159/000346244. [DOI] [PubMed] [Google Scholar]
- 92.Burman P., Karlsson F.A., Öberg K., Alm G., Orava M., Vihko R. Autoimmune thyroid disease in interferon-treated patients. Lancet. 1985;2:100–101. [PubMed] [Google Scholar]
- 93.Akeno N., Smith E.P., Stefan M., Huber A.K., Zhang W., Keddache M. IFN-α mediates the development of autoimmunity both by direct tissue toxicity and through immune cell recruitment mechanisms. J Immunol. 2011;186:4693–4706. doi: 10.4049/jimmunol.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reder A.T., Feng X. How type I interferons work in multiple sclerosis and other diseases: some unexpected mechanisms. J Interferon Cytokine Res. 2014;34:589–599. doi: 10.1089/jir.2013.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Panitch H.S., Hirsch R.L., Haley A.S., Johnson K.P. Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet. 1987;1:893–895. doi: 10.1016/s0140-6736(87)92863-7. [DOI] [PubMed] [Google Scholar]
- 96.Rynda A., Maddaloni M., Ochoa-Repáraz J., Callis G., Pascual D.W. IL-28 supplants requirement for T(reg) cells in protein sigma1-mediated protection against murine experimental autoimmune encephalomyelitis (EAE) PLoS ONE. 2010;5:e8720. doi: 10.1371/journal.pone.0008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoyo-Becerra C., Schlaak J.F., Hermann D.M. Insights from interferon-α-related depression for the pathogenesis of depression associated with inflammation. Brain Behav Immun. 2014 doi: 10.1016/j.bbi.2014.06.200. pii:S0889-1591(14)00365-1. [DOI] [PubMed] [Google Scholar]
- 98.Nomura H., Miyagi Y., Tanimoto H., Yamashita N., Oohashi S., Nishiura S. Occurrence of clinical depression during combination therapy with pegylated interferon alpha or natural human interferon beta plus ribavirin. Hepatol Res. 2012;42:241–247. doi: 10.1111/j.1872-034X.2011.00930.x. [DOI] [PubMed] [Google Scholar]
- 99.Zheng L.S., Hitoshi S., Kaneko N., Takao K., Miyakawa T., Tanaka Y. Mechanisms for interferon-α-induced depression and neural stem cell dysfunction. Stem Cell Rep. 2014;3:73–84. doi: 10.1016/j.stemcr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campos A.C., Vaz G.N., Saito V.M., Teixeira A.L. Further evidence for the role of interferon-gamma on anxiety- and depressive-like behaviors: involvement of hippocampal neurogenesis and NGF production. Neurosci Lett. 2014;578:100–105. doi: 10.1016/j.neulet.2014.06.039. [DOI] [PubMed] [Google Scholar]