Highlights
-
•
Overview on arthropod-associated virus discovery.
-
•
Description of newly characterized virus species.
-
•
Projections for further research.
Abstract
Recent studies on virus discovery have focused mainly on mammalian and avian viruses. Arbovirology with its long tradition of ecologically oriented investigation is now catching up, with important novel insights into the diversity of arthropod-associated viruses. Recent discoveries include taxonomically outlying viruses within the families Flaviviridae, Togaviridae, and Bunyaviridae, and even novel virus families within the order Nidovirales. However, the current focusing of studies on blood-feeding arthropods has restricted the range of arthropod hosts analyzed for viruses so far. Future investigations should include species from other arthropod taxa than Ixodita, Culicidae and Phlebotominae in order to shed light on the true diversity of arthropod viruses.
Current Opinion in Microbiology 2013, 16:507–513
This review comes from a themed issue on Host–microbe interactions: viruses
Edited by Carlos F Arias
For a complete overview see the Issue and the Editorial
Available online 11th July 2013
1369-5274/$ – see front matter, © 2013 Elsevier Ltd. All rights reserved.
Introduction
In recent years, the systematic discovery of novel viruses has undergone a renaissance as a field of research [1, 2, 3]. Virus discovery has yielded important and in part spectacular new insights into virus diversity. Starting from improved approaches to clone and screen cDNA libraries, the availability of novel amplification-based and sequencing-based techniques has enabled the characterization of viral isolates and even uncultured viruses. Some of the milestones in this field included the untargeted sequencing of the Severe Acute Respiratory Syndrome (SARS) agent [4], discoveries of relevant human respiratory viruses including human metapneumovirus and human coronavirus NL63 [5, 6, 7], as well as applications of technology for the clarification of mysterious outbreaks, for instance, one caused by a novel old world arenavirus involving haemorrhagic fever [8].
Out of the necessity to find diagnoses and etiologies in medicine, the field has so far placed a lot of focus on human viruses. Even in veterinary medicine, the paramount discovery of Schmallenberg virus as a novel pathogen in livestock ungulates was driven by clinical necessity, rather than pathogen surveillance [9]. Nevertheless, the growing interest in epidemic preparedness and pathogen ecology has triggered an expansion of fields of application for virus discovery. Efforts to investigate viral animal reservoirs have begun to yield fundamental insight into virus diversity and evolution, exemplified by the clarification of the reservoir of Ebola virus [10], the recent discovery of mammalian viruses co-ancestral with influenza A virus [11], as well as the analysis of the roles of different mammalian orders in paramyxovirus evolution [12]. According to the orientation of pathogen surveillance programmes, those investigations have focused on mammalian and avian viruses. In terms of arthropod-associated viruses, the harvest from virus discovery studies has been thinner [13] but there is now a number of remarkable arthropod virus findings that deserve attention and discussion.
Virus discovery in vertebrates and arthropods — comparison of approaches
It seems unlikely that technical approaches for the same purpose, that is, the discovery of viruses, should differ between fields. However, the huge new interest in virus discovery has made researchers use opportunities within reach. In mammalian and avian viruses, the incentive coming from the medical field has triggered huge investments in technology. Paramount virus discoveries in human medicine, starting from hepatitis B virus in the 1960s through human immunodeficiency virus, hepatitis C virus, and human herpes virus 8 in the early 1990s, have reassured us in our belief that undiscovered viruses would be agents difficult to culture or entirely unculturable. On the contrary, cell culture has been a key technology in the arthropod virus field and it has been long known that unclassified, unknown viruses are routinely cultured from insects and victims of arbovirus infections alike. As a matter of fact, the historical classification of arbovirues in groups a (alphaviruses), b (mainly flaviviruses) and c (other viruses) includes the presumption that a large part of isolates will not be classifiable using traditional methods such as serology. Out of this background, arbovirus researchers have covered continents in their fieldwork and spent decades on collecting viruses.
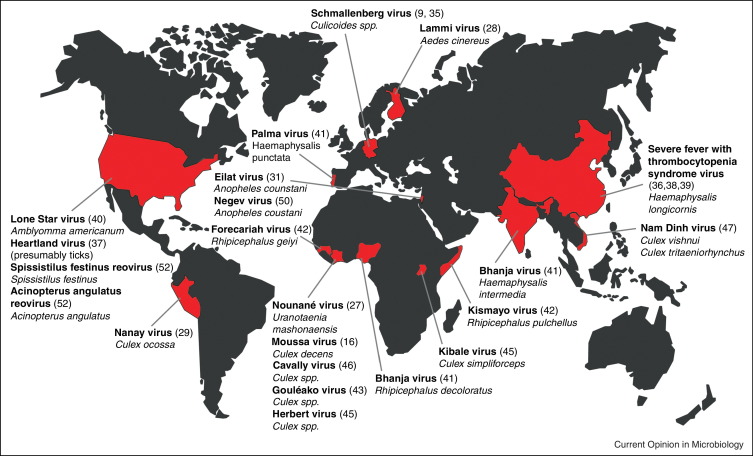
Consequentially, while the investigation of original field samples has come into focus in the mammal and bird virus field, arbovirus researchers have gone back to their collected cultures, particularly those that remained untypable [14, 15]. The collection maintained at the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at University of Texas Medical Branch is probably the largest and surely the most prominent of such collections. Other groups have followed similar technical approaches in that they used cell culture as a first-line tool to gain virus isolates, before applying methods aimed at their further characterization [16, 17, 18, 19]. There have been a number of important recent studies based on RT-PCR, but these were mainly aimed at the description of novel regions of distribution of known viruses, or close relatives thereof [20, 21, 22, 23]. On the contrary, and in spite of demonstrated technical feasibility, there is today not a single exemplary study applying undirected, hypothesis-free deep sequencing on original field samples of arthropods. All studies based on this promising methodology used cell culture isolates as their primary entry point (Figure 1 ).
Figure 1.
Geographic distribution of recent discoveries of novel arthropod-associated viruses. Countries where prototype viruses have been identified are marked in red. Virus names are shown in bold and host species in italic. Numbers in parenthesis are references.
Recent results in arthropod virus discovery — viral diversity in arthropods
Flaviviridae (genus Flavivirus)
Viruses that seem to replicate only in mosquito but not in vertebrate cells were first discovered within the genus Flavivirus, family Flaviviridae. Well-known members of this so-called insect-specific group include cell fusing agent virus, Kamiti River virus, and Culex flavivirus (reviewed in [24]). These insect-specific viruses are thought to be maintained in insects only, and to represent ancestral or primordial forms of the vector-borne flaviviruses. Insect-specific flaviviruses have been divided into Aedes-associated and Culex-associated viruses, respectively. Recent findings of related flaviviruses in mosquito species of other genera [25, 26] suggest a much higher diversity of yet-to-be discovered insect-specific flaviviruses. Interestingly, a clade of potentially insect-specific viruses that replicate in mosquito cells but not in vertebrate cells or newborn mice was discovered within the diversity of mosquito-borne viruses. This clade currently has now three members including Nounané virus isolated from Uranotaenia mashonaensis mosquitoes originating from the Taï National Park in Côte d’Ivoire [27], Lammi virus isolated from Aedes cinereus mosquitoes collected in Finland [28], and Nanay virus, a virus recently isolated from Culex ocossa collected in the Amazonian rainforest near Iquitos, Peru [29]. The diversity of mosquito-associated flaviviruses was further extended by the detection of flavivirus-like sequences integrated within the genomes of Aedes mosquitoes [30].
Togaviridae (genus Alphavirus)
In parallel to the findings within flaviviruses, the first insect-specific alphavirus, named Eilat virus, was discovered in Anopheles coustani mosquitoes collected in the Negev desert, Israel [31••]. Phylogenetic analyses placed Eilat virus on a long branch between the Western equine encephalitis serocomplex and Trocara virus, a virus isolated from Aedes serratus mosquitoes from the Amazon Basin, the prototype of a new antigenic complex [32]. Alphaviruses are divided in two groups, one large group infecting terrestrial mammals and birds via mosquito bites and a second distant related group with two members only infecting fish [33]. The finding of further insect-specific alphaviruses in the future, potentially placed between the aquatic and terrestrial viruses, is very likely.
Bunyaviridae (putative novel genera)
Although the family Bunyaviridae is already one of the largest virus families with about 350 members divided into five genera of plant (genus Tospovirus) or vertebrate (genus Orthobunya-, Phlebo-, Nairo-, and Hantavirus) pathogenic viruses [34], still many new viruses are being discovered, contributing further to the immense bunyavirus diversity. The case of Schmallenberg virus is only one example why we should continue studying the diversity of arthropod-associated bunyaviruses [9, 35]. More examples are provided by severe fever with thrombocytopenia syndrome virus (SFTSV), a novel tick-borne phlebovirus from China isolated from Haemaphysalis longicornis ticks, as well as its cousin in North America, the Heartland virus [36, 37, 38, 39]. Interestingly, also for the SFTSV and Heartland virus clade, closely related sister taxa named Bhanja virus, Palma virus and Lone Star virus, have now been described in Haemaphysalis intermadia, H. puncatata, Rhipicephalus decoloratus, R. geiyn, R. pulchellus, and Amblyomma americanum, respectively [40, 41, 42].
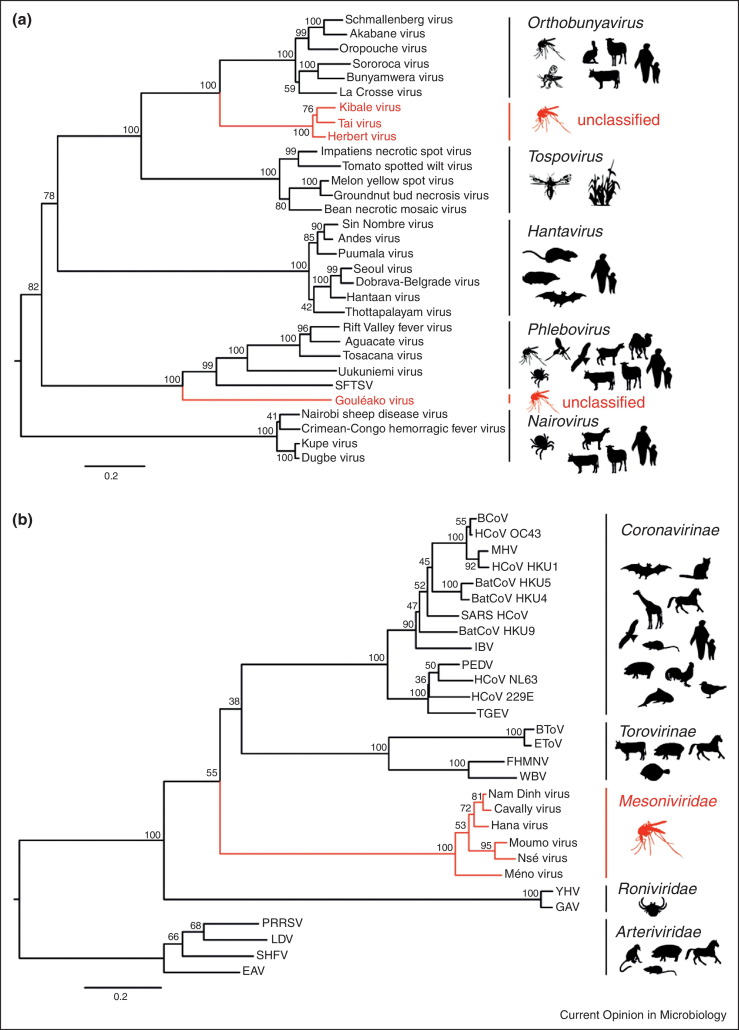
Moreover, a mosquito-associated phylogenetic outlier virus, termed Gouléako virus, was isolated from different mosquito species (mostly Culex nebulosus) collected in the Taï National Park in Côte d’Ivoire [43•]. Gouléako virus did not grow in various primate, bat, bird, rodent, and amphibian cell lines but grew to high titres in mosquito cells suggesting this agent to constitute the first putative insect-specific bunyavirus. Furthermore, a novel clade in phylogenetic sister relationship to orthobunyaviruses and almost equidistant to the established bunyavirus genera has been discovered in mosquitoes originating from Côte d’Ivoire and Uganda [44, 45•]. These viruses termed Herbert virus, Tai virus and Kibale virus did also not infect vertebrate cells but replicated well in mosquito cells. Gouléako virus, Herbert virus, Tai virus and Kibale virus may represent clades of insect-specific viruses in co-ancestral relationship to the vertebrate-infecting phlebo- and orthobunyaviruses, respectively. The diversity of the newly described bunyaviruses suggests the existence of further taxa in these clades, as well as the existence of novel clades in basal phylogenetic relationship to other bunyavirus genera (Figure 2 ).
Figure 2.
Phylogenetic relationship of prototype arthropod-associated viruses and avian and mammalian viruses. Maximum likelihood analyses of the family Bunyaviridae (a) and the order Nidovirales (b). Arthropod-viruses are shown in red. Associated hosts are indicated by silhouettes to the right.
Nidovirales (novel family Mesoniviridae)
Despite the detection of viruses with distant relationship to established clades and genera, even a novel RNA virus family, named Mesoniviridae, was discovered in mosquitoes collected in Côte d’Ivoire and in Vietnam [46••, 47••, 48]. The monogeneric family now contains at least four virus species that seem only to be able to infect insect cells, but not vertebrate cells [49]. The order Nidovirales heretofore comprised three families of fish-, bird-, and mammal-infecting viruses, namely the Coronaviridae (subfamilies Coronavirinae and Torovirinae) and the Arteriviridae. A third family included crustacean-infecting viruses, the Roniviridae. The now expanded nidovirus phylogeny with roniviruses and mesoniviruses branching from deepest tree nodes suggests an origin in arthropods for the whole order Nidoviales [46••, 49] (Figure 2).
Others (new families, taxa)
Novel insect-specific viruses, collectively termed Negevirus, have been detected in mosquitoes and sandflies captured in Brazil, Peru, the United States, Ivory Coast, Israel, and Indonesia [50•]. These are distantly related to viruses of the genus Cilevirus and may represent a novel genus within the same family as cileviruses [50•]. The recently characterized and classified Citrus leprosis virus, a plant virus transmitted by Brevipalpus mites, is the type species of the genus Cilevirus of an as yet unclassified novel virus family [51]. The wide geographic distribution and the detection in mosquitoes of different genera and sandflies suggest negeviruses to be widely distributed and to potentially infect nonbiting dipteran species widely.
The family Reoviridae is the only family of vertebrate-infecting viruses known to occur in a number of non-blood-feeding arthropod species. Two novel viruses were discovered in the three cornered alfalfa hopper (Spissistilus festinus) and in the angulate leafhopper (Acinopterus angulatus) collected in CA, USA, respectively [52•]. Both viruses were proposed to belong to a new genus of the subfamily Spinareovirinae within the family Reoviridae. Other distantly related reoviruses have been isolated from a grass carp (Ctenopharyngodon idella) in China [53], from a mud crab (Scylla serrata) in China [54], as well as from the winter moth Operophtera brumata [55].
Conclusions
Recent efforts to discover arthropod viruses have yielded widely divergent taxa that sometimes even define novel families. While much more effort is currently invested in mammal and bird virus detection, the results there seem to have been restricted to the discovery of variants of virus species, sister species to known viruses, and rarely genera.
Reasons for a seemingly wider genetic range of novel arthropod viruses may be technical. Virus culture as employed in most studies should amplify diverse novel viruses — in contrast to molecular biology techniques with their inherent sequence bias. However, studies combining cell culture and next generation sequencing on mammalian and bird viruses still yield a rather limited diversity of novel viruses. Moreover, while a large range of vertebrate cell lines is available, most arthropod-based virus isolation studies relied on only one cell line, namely C6/36 cells from Aedes albopictus. These cells have recently been shown to be deficient in elemental RNA interference (RNAi) components suggesting the cells to be much more susceptible for virus infections [56]. But again, also for mammal and bird viruses there are several cell lines available with defects in their interferon system facilitating virus infection and replication. It is therefore unlikely that differences in our ability to culture viruses could suffice as an explanation for the higher genetic diversity in novel virus discoveries in insects, as opposed to vertebrates. Notably, while undirected studies based on next generation sequencing are not available for arthropods, they have indeed been conducted intensely for mammals and birds. Even if some of those studies mentioned sequence reads that could neither be ascribed to the host genome or transcriptome, nor to known viral families, the absence of discoveries of highly divergent novel taxa including novel families in mammals and birds is striking. The obvious and most likely explanation is sampling bias. Our current coverage of host diversity is highly limited. In mammalian and bird virus discovery, huge efforts have been spent historically on human hosts, as well as companion and livestock animals [12]. Triggered by findings of important zoonotic agents such as SARS-CoV, Hanta-, and influenza viruses, the array of mammal and bird species of interest has been extended to wild rodents, bats, and migratory birds [1, 2]. Many of the more important recent findings of novel vertebrate viruses have been made there, confirming conjectures that taxa with huge social group sizes, population turnover, and migratory behaviour favour the emergence and maintenance of viruses.
On the contrary, no such considerations have been made so far in the design of sampling for arthropod virus discovery. Sampling has been determined (and limited) by our interest in blood-feeding arthropods, such as mosquitoes, sandflies, and ticks. These are not closely related, with an evolutionary distance of ca. 706 million years [57], in contrast to birds and mammals that separated ca. 160 mya [58]. If we accept virus and host diversity to be roughly correlated, we have to admit that we are probably overlooking the largest part of existing arthropod virus diversity. Arthropods are the largest animal phylum including the class of insects which comprises more than half of all described species [59].
Critically, it has to be considered that viruses may not only be acquired from insects via the blood-borne route but also by the ingestion of infectious diet. Arthropods are a major dietary component for a large range of small vertebrates, and we would be well advised to conduct virus discovery studies on those taxa that are an attractive prey for small mammals and birds — such as moths, butterflies, caterpillars, bugs, or spiders.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Funding was received from European Union DG Research through the programs EMPERIE (grant agreement no. 223498), ANTIGONE (grant agreement no. 278976), and by the Deutsche Forschungsgemeinschaft (grant agreement no. JU2857/3-1).
Contributor Information
Sandra Junglen, Email: junglen@virology-bonn.de.
Christian Drosten, Email: drosten@virology-bonn.de.
References
- 1.Deeks S., Drosten C., Picker L., Subbarao K., Suzich J. Roadblocks to translational challenges on viral pathogenesis. Nat Med. 2013;19:30–34. doi: 10.1038/nm.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smits S.L., Osterhaus A.D. Virus discovery: one step beyond. Curr Opin Virol. 2013 doi: 10.1016/j.coviro.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delwart E. Animal virus discovery: improving animal health, understanding zoonoses, and opportunities for vaccine development. Curr Opin Virol. 2012;2:344–352. doi: 10.1016/j.coviro.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A., Osterhaus A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briese T., Paweska J.T., McMullan L.K., Hutchison S.K., Street C., Palacios G., Khristova M.L., Weyer J., Swanepoel R., Egholm M. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann B., Scheuch M., Hoper D., Jungblut R., Holsteg M., Schirrmeier H., Eschbaumer M., Goller K.V., Wernike K., Fischer M. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 11.Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A., Chen L.M., Recuenco S., Ellison J.A., Davis C.T., York I.A. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drexler J.F., Corman V.M., Muller M.A., Maganga G.D., Vallo P., Binger T., Gloza-Rausch F., Rasche A., Yordanov S., Seebens A. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Vijayendran D., Bonning B.C. Next generation sequencing technologies for insect virus discovery. Viruses. 2011;3:1849–1869. doi: 10.3390/v3101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasilakis N., Widen S., Travassos da Rosa A.P., Wood T.G., Walker P.J., Holmes E.C., Tesh R.B. Malpais spring virus is a new species in the genus vesiculovirus. Virol J. 2013;10:69. doi: 10.1186/1743-422X-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhary R., Street C., Travassos da Rosa A., Nunes M.R., Tee K.K., Hutchison S.K., Vasconcelos P.F., Tesh R.B., Lipkin W.I., Briese T. Genetic characterization of the Wyeomyia group of orthobunyaviruses and their phylogenetic relationships. J Gen Virol. 2012;93:1023–1034. doi: 10.1099/vir.0.039479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan P.L., Junglen S., Tashmukhamedova A., Conlan S., Hutchison S.K., Kurth A., Ellerbrok H., Egholm M., Briese T., Leendertz F.H. Moussa virus: a new member of the Rhabdoviridae family isolated from Culex decens mosquitoes in Cote d’Ivoire. Virus Res. 2010;147:17–24. doi: 10.1016/j.virusres.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhioua E., Moureau G., Chelbi I., Ninove L., Bichaud L., Derbali M., Champs M., Cherni S., Salez N., Cook S. Punique virus, a novel phlebovirus, related to sandfly fever Naples virus, isolated from sandflies collected in Tunisia. J Gen Virol. 2010;91:1275–1283. doi: 10.1099/vir.0.019240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv X., Mohd Jaafar F., Sun X., Belhouchet M., Fu S., Zhang S., Tong S.X., Lv Z., Mertens P.P., Liang G. Isolates of Liao ning virus from wild-caught mosquitoes in the Xinjiang province of China in 2005. PLoS ONE. 2012;7:e37732. doi: 10.1371/journal.pone.0037732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belaganahalli M.N., Maan S., Maan N.S., Nomikou K., Pritchard I., Lunt R., Kirkland P.D., Attoui H., Brownlie J., Mertens P.P. Full genome sequencing and genetic characterization of Eubenangee viruses identify Pata virus as a distinct species within the genus Orbivirus. PLoS ONE. 2012;7:e31911. doi: 10.1371/journal.pone.0031911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker N., Jost H., Ziegler U., Eiden M., Hoper D., Emmerich P., Fichet-Calvet E., Ehichioya D.U., Czajka C., Gabriel M. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PLoS ONE. 2012;7:e32604. doi: 10.1371/journal.pone.0032604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jost H., Bialonski A., Maus D., Sambri V., Eiden M., Groschup M.H., Gunther S., Becker N., Schmidt-Chanasit J. Isolation of usutu virus in Germany. Am J Trop Med Hyg. 2011;85:551–553. doi: 10.4269/ajtmh.2011.11-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jost H., Bialonski A., Schmetz C., Gunther S., Becker N., Schmidt-Chanasit J. Isolation and phylogenetic analysis of Batai virus, Germany. Am J Trop Med Hyg. 2011;84:241–243. doi: 10.4269/ajtmh.2011.10-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moureau G., Bichaud L., Salez N., Ninove L., Hamrioui B., Belazzoug S., de Lamballerie X., Izri A., Charrel R.N. Molecular and serological evidence for the presence of novel phleboviruses in sandflies from northern Algeria. Open Virol J. 2010;4:15–21. doi: 10.2174/1874357901004010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook S., Moureau G., Kitchen A., Gould E.A., de Lamballerie X., Holmes E.C., Harbach R.E. Molecular evolution of the insect-specific flaviviruses. J Gen Virol. 2012;93:223–234. doi: 10.1099/vir.0.036525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calzolari M., Ze-Ze L., Ruzek D., Vazquez A., Jeffries C., Defilippo F., Osorio H.C., Kilian P., Ruiz S., Fooks A.R. Detection of mosquito-only flaviviruses in Europe. J Gen Virol. 2012;93:1215–1225. doi: 10.1099/vir.0.040485-0. [DOI] [PubMed] [Google Scholar]
- 26.Hobson-Peters J., Yam A.W., Lu J.W., Setoh Y.X., May F.J., Kurucz N., Walsh S., Prow N.A., Davis S.S., Weir R. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE. 2013;8:e56534. doi: 10.1371/journal.pone.0056534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junglen S., Kopp A., Kurth A., Pauli G., Ellerbrok H., Leendertz F.H. A new flavivirus and a new vector: characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J Virol. 2009;83:4462–4468. doi: 10.1128/JVI.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huhtamo E., Putkuri N., Kurkela S., Manni T., Vaheri A., Vapalahti O., Uzcategui N.Y. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J Virol. 2009;83:9532–9540. doi: 10.1128/JVI.00529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evangelista J., Cruz C., Guevara C., Astete H., Carey C., Kochel T.J., Morrison A.C., Williams M., Halsey E.S., Forshey B.M. Characterization of a novel flavivirus isolated from Culex (Melanoconion) ocossa mosquitoes from Iquitos, Peru. J Gen Virol. 2013 doi: 10.1099/vir.0.050575-0. [DOI] [PubMed] [Google Scholar]
- 30.Crochu S., Cook S., Attoui H., Charrel R.N., De Chesse R., Belhouchet M., Lemasson J.J., de Micco P., de Lamballerie X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85:1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- 31••.Nasar F., Palacios G., Gorchakov R.V., Guzman H., Da Rosa A.P., Savji N., Popov V.L., Sherman M.B., Lipkin W.I., Tesh R.B. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci U S A. 2012;109:14622–14627. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery and characterization of the first insect-restricted alphavirus.
- 32.Travassos da Rosa A.P., Turell M.J., Watts D.M., Powers A.M., Vasconcelos P.F., Jones J.W., Klein T.A., Dohm D.J., Shope R.E., Degallier N. Trocara virus: a newly recognized Alphavirus (Togaviridae) isolated from mosquitoes in the Amazon Basin. Am J Trop Med Hyg. 2001;64:93–97. doi: 10.4269/ajtmh.2001.64.93. [DOI] [PubMed] [Google Scholar]
- 33.Forrester N.L., Palacios G., Tesh R.B., Savji N., Guzman H., Sherman M., Weaver S.C., Lipkin W.I. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J Virol. 2012;86:2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmaljohn C.S., Nichol S.T. Bunyaviridae. In: Knipe D.M., Howley P.M., editors. vol 5. Lippincott Williams & Wilkins; 2007. pp. 1741–1790. (Fields Virology). [Google Scholar]
- 35.Rasmussen L.D., Kristensen B., Kirkeby C., Rasmussen T.B., Belsham G.J., Bodker R., Botner A. Culicoids as vectors of Schmallenberg virus. Emerg Infect Dis. 2012;18:1204–1206. doi: 10.3201/eid1807.120385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X.J., Liang M.F., Zhang S.Y., Liu Y., Li J.D., Sun Y.L., Zhang L., Zhang Q.F., Popov V.L., Li C. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMullan L.K., Folk S.M., Kelly A.J., MacNeil A., Goldsmith C.S., Metcalfe M.G., Batten B.C., Albarino C.G., Zaki S.R., Rollin P.E. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med. 2012;367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- 38.Xu B., Liu L., Huang X., Ma H., Zhang Y., Du Y., Wang P., Tang X., Wang H., Kang K. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog. 2011;7:e1002369. doi: 10.1371/journal.ppat.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y.Z., He Y.W., Dai Y.A., Xiong Y., Zheng H., Zhou D.J., Li J., Sun Q., Luo X.L., Cheng Y.L. Hemorrhagic fever caused by a novel Bunyavirus in China: pathogenesis and correlates of fatal outcome. Clin Infect Dis. 2012;54:527–533. doi: 10.1093/cid/cir804. [DOI] [PubMed] [Google Scholar]
- 40.Swei A., Russell B.J., Naccache S.N., Kabre B., Veeraraghavan N., Pilgard M.A., Johnson B.J., Chiu C.Y. The genome sequence of lone star virus, a highly divergent bunyavirus found in the Amblyomma americanum tick. PLoS ONE. 2013;8:e62083. doi: 10.1371/journal.pone.0062083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dilcher M., Alves M.J., Finkeisen D., Hufert F., Weidmann M. Genetic characterization of Bhanja virus and Palma virus, two tick-borne phleboviruses. Virus Genes. 2012;45:311–315. doi: 10.1007/s11262-012-0785-y. [DOI] [PubMed] [Google Scholar]
- 42.Matsuno K., Weisend C., Travassos da Rosa A.P., Anzick S.L., Dahlstrom E., Porcella S.F., Dorward D.W., Yu X.J., Tesh R.B., Ebihara H. Characterization of the Bhanja serogroup viruses (Bunyaviridae): a novel species of the genus Phlebovirus and its relationship with other emerging tick-borne phleboviruses. J Virol. 2013;87:3719–3728. doi: 10.1128/JVI.02845-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Marklewitz M., Handrick S., Grasse W., Kurth A., Lukashev A., Drosten C., Ellerbrok H., Leendertz F.H., Pauli G., Junglen S. Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J Virol. 2011;85:9227–9234. doi: 10.1128/JVI.00230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery of a mosquito-associated bunyavirus in almost equidistant relationship to established genera.
- 44.Junglen S., Kurth A., Kuehl H., Quan P.L., Ellerbrok H., Pauli G., Nitsche A., Nunn C., Rich S.M., Lipkin W.I. Examining landscape factors influencing relative distribution of mosquito genera and frequency of virus infection. Ecohealth. 2009;6:239–249. doi: 10.1007/s10393-009-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Marklewitz M, Zirkel F, Heidemann H, Rwego IB, Kurth A, Kallies R, Briese T, Drosten C, Gillespie TR, Junglen S: Discovery of a novel cluster of mosquito-associated bunyaviruses in sister relationship to the genus Orthobunyavirus. Unpublished data; Discovery of a novel cluster of mosquito-associated bunyavirus in almost equidistant relationship to established genera.
- 46••.Zirkel F., Kurth A., Quan P.L., Briese T., Ellerbrok H., Pauli G., Leendertz F.H., Lipkin W.I., Ziebuhr J., Drosten C. An insect nidovirus emerging from a primary tropical rainforest. MBio. 2011;2 doi: 10.1128/mBio.00077-11. e00077–e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Same as for reference [47••].
- 47••.Nga P.T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two articles (Zirkel, Nga) independently describe a novel RNA virus family in mosquitoes.
- 48.Lauber C., Ziebuhr J., Junglen S., Drosten C., Zirkel F., Nga P.T., Morita K., Snijder E.J., Gorbalenya A.E. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol. 2012;157:1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zirkel F., Roth H., Kurth A., Drosten C., Ziebuhr J., Junglen S. Identification and characterization of genetically divergent members of the newly established family mesoniviridae. J Virol. 2013;87:6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Vasilakis N., Forrester N.L., Palacios G., Nasar F., Savji N., Rossi S.L., Guzman H., Wood T.G., Popov V., Gorchakov R. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J Virol. 2013;87:2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detection of insect-restricted viruses that potentially establish a novel RNA virus family, together with an earlier-described plant-infecting virus.
- 51.Locali-Fabris E.C., Freitas-Astua J., Souza A.A., Takita M.A., Astua-Monge G., Antonioli-Luizon R., Rodrigues V., Targon M.L., Machado M.A. Complete nucleotide sequence, genomic organization and phylogenetic analysis of Citrus leprosis virus cytoplasmic type. J Gen Virol. 2006;87:2721–2729. doi: 10.1099/vir.0.82038-0. [DOI] [PubMed] [Google Scholar]
- 52•.Spear A., Sisterson M.S., Stenger D.C. Reovirus genomes from plant-feeding insects represent a newly discovered lineage within the family Reoviridae. Virus Res. 2012;163:503–511. doi: 10.1016/j.virusres.2011.11.015. [DOI] [PubMed] [Google Scholar]; Characterization of viruses from hemipterans that may belong to a proposed new reovirus genus.
- 53.Ye X., Tian Y.Y., Deng G.C., Chi Y.Y., Jiang X.Y. Complete genomic sequence of a reovirus isolated from grass carp in China. Virus Res. 2012;163:275–283. doi: 10.1016/j.virusres.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Deng X.X., Lu L., Ou Y.J., Su H.J., Li G., Guo Z.X., Zhang R., Zheng P.R., Chen Y.G., He J.G. Sequence analysis of 12 genome segments of mud crab reovirus (MCRV) Virology. 2012;422:185–194. doi: 10.1016/j.virol.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 55.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brackney D.E., Scott J.C., Sagawa F., Woodward J.E., Miller N.A., Schilkey F.D., Mudge J., Wilusz J., Olson K.E., Blair C.D. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wheat C.W., Wahlberg N. Phylogenomic insights into the cambrian explosion, the colonization of land and the evolution of flight in arthropoda. Syst Biol. 2013;62:93–109. doi: 10.1093/sysbio/sys074. [DOI] [PubMed] [Google Scholar]
- 58.Luo Z.X., Yuan C.X., Meng Q.J., Ji Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 59.Grimaldi D.A., Engel M.S. Cambridge University Press; Cambridge: 2005. Evolution of the Insects. [Google Scholar]