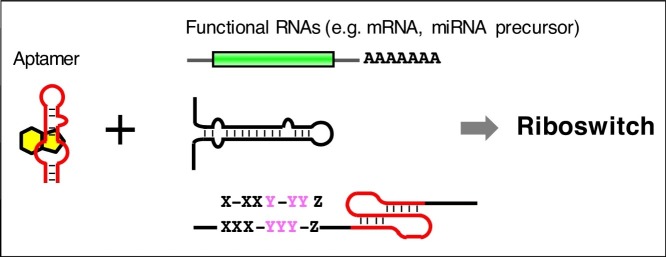
Graphical abstract
Abstract
Molecular recognition by RNA aptamers has been exploited to control gene expression in response to small molecules in mammalian cells. These mammalian synthetic riboswitches offer attractive features such as small genetic size and lower risk of immunological complications compared to protein-based transcriptional gene switches. The diversity of gene regulatory mechanisms that involve RNA has also inspired the development of mammalian riboswitches that harness various regulatory mechanisms. In this report, recent advances in synthetic riboswitches that function in mammalian cells are reviewed focusing on the regulatory mechanisms they exploit such as mRNA degradation, microRNA processing, and programmed ribosomal frameshifting.
Current Opinion in Chemical Biology 2019, 52:72–78
This review comes from a themed issue on Synthetic biology
Edited by Hirohide Saito and Yohei Yokobayashi
For a complete overview see the Issue and the Editorial
Available online 22nd June 2019
https://doi.org/10.1016/j.cbpa.2019.05.018
1367-5931/© 2019 The Author. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
Envisioned practical applications of mammalian synthetic biology frequently require gene switches that recognize endogenous or exogenous chemical signals and turn on or turn off expression of proteins, which in turn regulate synthetic genetic circuits inside the cell. These chemical gene switches need to be flexible enough to be tailored to diverse chemical species, engineered to function as ON-switch or OFF-switch, fine-tuned to adjust the sensitivity, and have a minimal genetic and metabolic footprint. Protein-based engineered transcription factors (TFs), such as Tet-ON and Tet-OFF systems derived from a bacterial TF, are among the most widely used tools to control mammalian gene expression in response to small molecule triggers [1]. However, there are a number of drawbacks of TF-based gene switches as generally applicable switches for mammalian and biomedical applications [2, 3, 4]: (i) adapting an engineered TF to respond to a new compound is challenging, (ii) expression of engineered TFs can trigger immunogenicity, (iii) genetic size and expression of an engineered TF can burden the host cell or the vector, and (iv) switch performance can be influenced by the expression level of the TF; therefore, optimization may be required. RNA aptamers for desired ligands can be obtained relatively easily by in vitro selection (SELEX) [5,6]; therefore, riboswitches can potentially be engineered to respond to a variety of compounds more readily than engineered TFs. Moreover, aptamers are typically small (∼20 to 100 nt). Even with the additional nucleotides necessary to regulate gene expression, genetic footprints of riboswitches (few hundred nt) are small enough to satisfy vectors that have limited capacity (e.g. adeno associated virus). Furthermore, the lack of any translated proteins is expected to result in lower immunogenicity and metabolic burden on the host cell.
In 1998, Werstuck and Green [7] demonstrated that insertion of an RNA aptamer selected to bind the Hoechst dye H33342 in the 5′ untranslated region of mRNA allows repression of gene expression in response to the ligand in CHO cells, presumably by blocking ribosome binding or scanning. It should be noted that this work predated the discovery of natural bacterial riboswitches [8,9], demonstrating that small molecules can directly and specifically affect gene expression in the absence of mediator proteins. Importantly, RNA aptamer-based regulation of gene expression allows bioengineers to harness the rich diversity of gene regulatory mechanisms that involve RNA, for example, translation initiation, RNA interference and microRNAs, and RNA splicing. For each RNA mediated gene regulatory mechanism, there are multiple ways to couple its outcome with aptamer–ligand interaction, further enriching the potential diversity of synthetic riboregulators. An excellent review on RNA-based gene switches in mammalian cells was recently published by Ausländer and Fussenegger [10]. Therefore, this article reviews more recent developments in synthetic mammalian riboswitches with a focus on the diversity of the regulatory mechanisms harnessed by the riboswitches.
Controlling mRNA cleavage by aptazymes
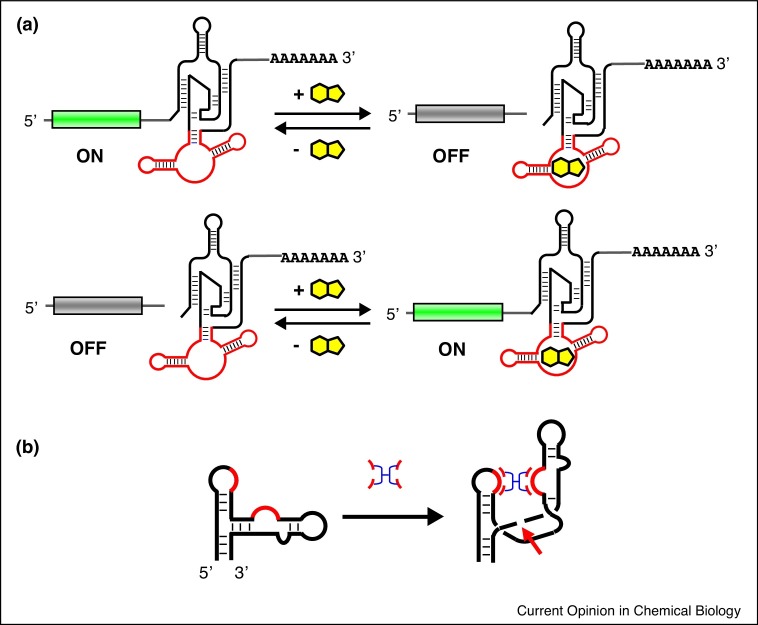
In 2004, Yen et al. demonstrated chemical regulation of gene expression in mammalian cells by embedding a hammerhead ribozyme in the untranslated regions (UTRs) of an mRNA [11]. As the 5′ and 3′ UTRs are indispensable for translation of eukaryotic mRNAs, self-cleavage within the mRNA was expected to suppress gene expression. Addition of toyocamycin to the cell culture medium resulted in nonspecific incorporation of the antiviral nucleotide analog into the mRNA and statistical inactivation of the ribozyme activity [12]. While the ribozyme was not specifically regulated by a small molecule via an aptamer, this work paved the way for the subsequent riboswitches that employ allosterically regulated ribozymes (aptazymes) embedded in the 5′ and/or 3′ UTR to chemically regulate gene expression in mammalian cells (Figure 1 a) [13, 14, 15, 16]. This strategy continues to be popular, and the recent advances highlight new approaches to design and optimize aptazymes.
Figure 1.
Regulatory mechanisms of synthetic riboswitches. (a) Aptazymes inserted in untranslated regions of mRNAs regulate the integrity of mRNA via chemically controlled self-cleavage. (b) Schematic illustration of the ‘molecular glue’ that activates a hammerhead ribozyme by restoring loop–loop tertiary interaction.
Earlier aptazyme-based riboswitches in mammalian cells were designed via trial-and-error, or based on medium-throughput or high-throughput screening in bacteria or yeast systems. However, it has been reported that ribozyme activity in living cells are not highly correlated among different cell types (bacteria, yeast, or mammalian cells) which is understandable considering the differences in translational mechanism, intracellular environment (RNA binding proteins, ribonucleases, etc.), and mode of gene regulation by ribozymes in different cell types [17•]. Because high-throughput screening of riboswitches directly in mammalian cells is technically challenging, alternative design strategies focusing on aptazymes that function in mammalian cells are desirable.
The Farzan group demonstrated an intriguing strategy that combines empirical experimental data with quantitative modeling [18]. They first synthesized a panel of 32 aptazymes and measured their riboswitch performance in cultured mammalian cells, and attempted to correlate the experimental results with various design parameters such as calculated annealing energy and the number of hydrogen bonds in the communication module. The researchers concluded that the proximity of base pairing (or lack thereof) within the communication module to the ribozyme affects the ribozyme activity, and devised ‘Weighted Hydrogen Bond Score (WHBS)’ as a calculable parameter that correlates with the ribozyme performance. They used WHBS as a guide to design aptazyme-based riboswitches using three aptamers with good switching characteristics [18]. It remains to be seen if the strategy can be extrapolated to different aptamers, ribozymes, and aptazyme architectures.
Dohno et al. addressed the aptazyme design problem with a unique approach [19••]. Instead of starting from an existing aptamer, they started with a rationally designed molecule targeted to mismatched DNA/RNA sequences. The naphthyridine carbamate tetramer with Z-stilbene linker (Z-NCTS) designed by the group is known to bind XGG/XGG mismatches in DNA and RNA through Watson-Crick type hydrogen bonding between the naphthyridine moieties with the unpaired guanines [20]. They used this ligand as a ‘molecular glue’ to induce a tertiary contact between loops I and II of a hammerhead ribozyme that is critical for ribozyme activity (Figure 1b). The tertiary contact between the two loops was disrupted by introducing an AGG/UGG mismatch that is recognized by Z-NCTS. The engineered ribozyme was inserted into the 3′ UTR of an mRNA encoding firefly luciferase whose expression was reduced by ∼4-fold upon addition of Z-NCTS (0.6 μM) in HeLa cells. While it is notable that the switching was observed with a low ligand concentration, Z-NCTS exhibited cellular toxicity (LC50 = 0.8 μM) probably due to nonspecific interactions with cellular RNAs.
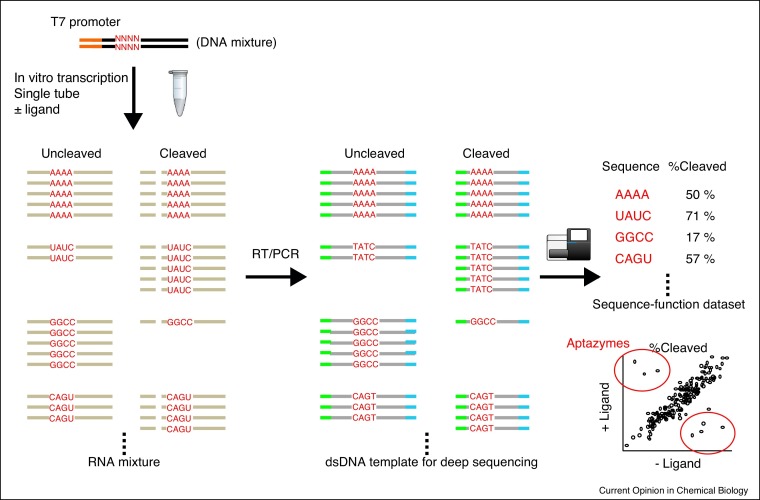
In contrast to these rational or semi-rational design efforts, high-throughput screening of aptazymes has mostly been executed in Escherichia coli or Saccharomyces cerevisiae. However, screening is generally labor and cost intensive while yielding sequence information for a handful of ‘hits’. Screening also yields limited sequence–function information that can be exploited for further optimization or library design. To address the lack of comprehensive sequence–function relationship data for ribozymes, we developed an in vitro high-throughput ribozyme assay method using deep sequencing (Figure 2 ) [21••,22,23]. A library of ribozyme or aptazyme mutants are transcribed in vitro in a single tube as a mixture, and the resulting ribozymes (cleaved or uncleaved) are converted to DNA templates for deep sequencing. The number of cleaved and uncleaved reads for each ribozyme variant are then counted to calculate cleavage efficiency under the reaction condition. This strategy yields a complete sequence–function relationship for all members of the ribozyme library up to 104 variants or more [24], depending on library preparation and sequencing output. This method can unambiguously identify functional aptazymes if they exist, and provide a broader view of the sequence–function relationship that can be used to build a mechanistic model and/or to further optimize the aptazyme design. We used deep sequencing to identify guanine-activated ribozymes based on an HDV-like ribozyme which were subsequently used to control gene expression in HEK 293 cells [21••]. Use of a low Mg2+ concentration during in vitro transcription may have contributed to the positive correlation of the ribozyme activity in vitro and in mammalian cells. Similar methods were also used to fine tune mammalian gene expression levels [25] and to screen for active ribozymes (but not aptazymes) directly in mammalian cells [26].
Figure 2.
Schematic illustration of in vitro aptazyme assay strategy based on deep sequencing. An aptazyme library is prepared as a DNA mixture which is transcribed into RNA in vitro by T7 RNA polymerase. The aptazyme pool is then converted to DNA sequencing templates by reverse transcription and PCR. Deep sequencing yields read counts of the cleaved and uncleaved fragments for every mutant in the library from which cleavage efficiency (% cleaved) is calculated. Plotting cleavage efficiency of the mutants in the presence and absence of the ligand reveals aptazyme candidates which are subsequently evaluated as riboswitches.
Controlling ribosomal frameshifting by aptamers
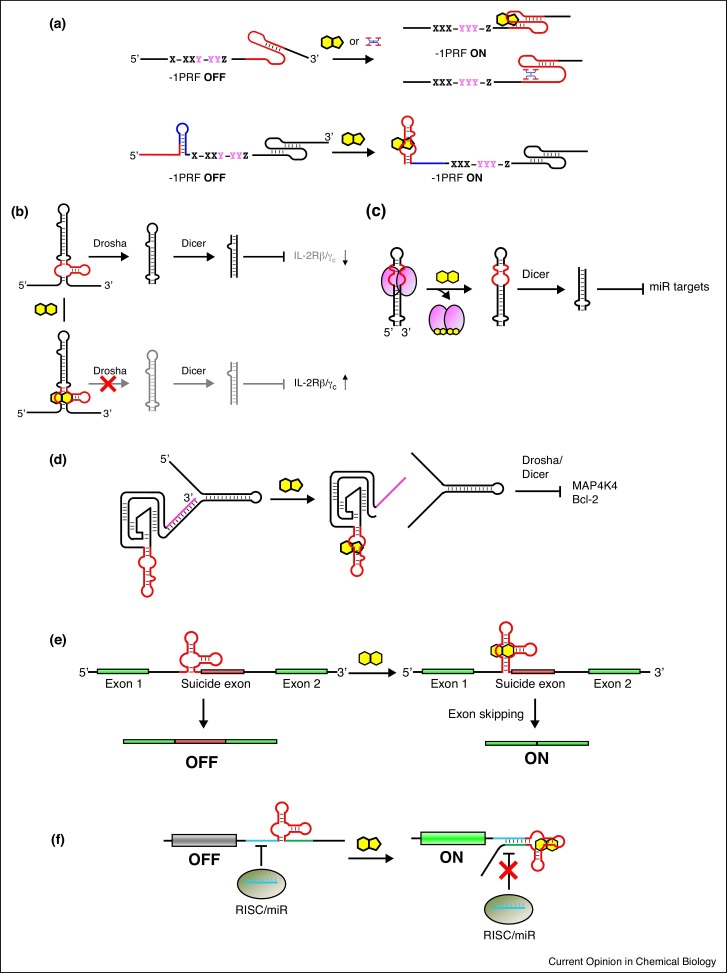
Programmed -1 ribosomal frameshifting (-1PRF) [27] can result in translation of an alternative polypeptide from an mRNA by shifting the translation reading frame by -1 nucleotide. A canonical -1PRF element consists of a 7-nt slippery sequence X XXY YYZ (trinucleotides XXY and YYZ representing the original frame and XXX YYY representing the -1 frameshift) followed by a stable secondary structure such as a pseudoknot or a stem-loop. The Chang group first controlled -1PRF in mammalian cells (Figure 3 a) by incorporating theophylline and S-adenosyl-l-homocysteine (SAH) aptamers [28]. An ON/OFF ratio of ∼6 was reported but the frameshifting efficiency (FE) remained low. The same group used another -1PRF stimulating pseudoknot from SARS coronavirus to engineer theophylline inducible -1PRF switches with up to fivefold activation of gene expression [29]. More recently, Matsumoto et al. used the analogs of naphthyridine carbamate tetramers described above to control the formation of a pseudoknot, thereby chemically inducing -1PRF (Figure 3a, top) [30]. Although up to ∼9-fold activation was observed in HeLa cells, the maximum FE was low (3.2%) in the presence of the pseudoknot inducing ligand. It is possible that the toxicity of the ligand prevented observation of higher FEs in HeLa cells, as an FE as high as 24% was observed in rabbit reticulocyte lysate.
Figure 3.
Regulatory mechanisms of synthetic riboswitches. (a) Regulation of -1PRF by a small molecule via stabilizing the downstream pseudoknot element or interfering with formation of the upstream stem loop that negatively affect -1PRF. (b) Interfering with the cleavage of pri-miRNA by Drosha by an aptamer–ligand complex near the cleavage site. (c) TetR protein bound to the aptamer embedded in pre-miRNA prevents Dicer cleavage. Addition of doxycycline removes TetR and upregulates miRNA maturation. (d) The 3′ single-stranded region of a pri-miRNA that is necessary for Drosha processing is unmasked by an aptazyme. (e) An aptamer incorporated near the 3′ splice site of an intron followed by a suicide exon containing a stop codon can induce exon skipping in the presence of the aptamer ligand. (f) A miRNA target site inserted in the 3′ UTR of an mRNA is occluded by aptamer–ligand interaction, resulting in increased protein expression.
Controlling microRNA maturation pathway by aptamers
Besides controlling translation via an aptamer embedded in the targeted mRNA, another strategy aims to control the microRNA (miRNA) processing pathway by an RNA aptamer. The miRNA product subsequently targets one or more mRNAs in trans. An et al. first demonstrated that an aptamer incorporated in a miRNA precursor allows chemical regulation of RNA interference (RNAi), in this case, by modulating the Dicer mediated cleavage of a short-hairpin RNA (shRNA) [31]. The Smolke group has developed an alternative strategy that controls the Drosha mediated processing of primary miRNA substrates by embedding an RNA aptamer in the vicinity of the Drosha cleavage site [32]. Binding of the ligand to the aptamer inhibits Drosha cleavage and upregulates the expression of the gene targeted by the miRNA. Recently, the group adapted an aptamer that they selected for (6R)-folinic acid (FA) to control a synthetic miRNA precursor targeting IL-2 receptors (Figure 3b) [33]. Although the modulation of the targeted gene expression by the ligand was rather modest (up to 35% activation), it was sufficient to observe a robust regulation of cell proliferation.
The Suess group reported a new strategy to control the Dicer processing by inserting an aptamer that binds to TetR protein [34] near the cleavage site blocking the reaction in the presence of TetR [35•]. They showed that addition of the TetR ligand doxycycline dissociates TetR from the miRNA precursor and increases the formation of the mature miRNA (Figure 3c). Doxycycline was able to tightly control the mature miRNA level (1%–40% relative to unmodified miRNA precursor), although the ON/OFF ratio of the targeted reporter gene was more moderate (∼3). Although this system requires an exogenous protein factor, it allows the use of doxycycline which has been used extensively in mammalian cells and animals.
The Pei group adapted another Drosha modulation strategy originally reported by our group [36,37] to control endogenous gene expression in cancer cell lines to induce apoptosis. A theophylline activated aptazyme was inserted between an inhibitory strand that blocks the 3′ single-stranded region of the pri-miRNA, thereby preventing the RNA from being processed by Drosha (Figure 3d). The researchers targeted MAP4K4 in HepG2 cells [38] and Bcl-2 in MCF-7 cells [39], and observed fourfold to fivefold increase in apoptosis.
Controlling other gene regulatory processes by aptamers
Vogel et al. showed that exon skipping can be controlled by a tetracycline RNA aptamer inserted near the 3′ splice site along with a suicide exon (Figure 3e) [40]. Tetracycline induces exon skipping resulting in ∼5-fold activation of gene expression. In combination with an aptazyme device, the dynamic range increased to ∼7-fold in HEK 293 cells.
An mRNA can be targeted by an endogenous miRNA by inserting one or more miRNA target sequences in the 3′ UTR, a strategy sometimes used to achieve cell type-dependent gene expression. Mou et al. controlled the accessibility of the miRNA target sequence by an aptamer embedded near the target site in the mRNA [41••]. Addition of tetracycline occluded the miRNA target site resulting in upregulation of gene expression (Figure 3f). The performance of the switch, as expected, was dependent on the miRNAs and cell types used, but they observed tight regulation of gene expression with up to 19-fold activation in HeLa cells with a miR-21 target site.
Conclusions
The diversity of gene regulatory mechanisms that involve RNA has and will continue to inspire synthetic RNA-based gene switches in mammalian cells. It can be anticipated that riboswitches based on different regulatory mechanisms have advantages for different applications. For example, miRNA-based riboswitches are more convenient for controlling endogenous gene expression because they operate in trans as opposed to the UTR embedded aptazymes that function in cis. However, the existing mammalian riboswitches still exhibit poor and variable dynamic range and ligand sensitivity compared to the conventional TF-based gene switches. With further optimization and increasing availability of new aptamers and ligands for cellular applications, synthetic riboswitches should emerge as useful tools in synthetic biology of mammalian cells.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgement
Work in the author’s laboratory mentioned in the article was financially supported by Okinawa Institute of Science and Technology Graduate University.
References
- 1.Clackson T. Controlling mammalian gene expression with small molecules. Curr Opin Chem Biol. 1997;1:210–218. doi: 10.1016/s1367-5931(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 2.Welman A., Barraclough J., Dive C. Generation of cells expressing improved doxycycline-regulated reverse transcriptional transactivator rtTA2S-M2. Nat Protoc. 2006;1:803–811. doi: 10.1038/nprot.2006.117. [DOI] [PubMed] [Google Scholar]
- 3.Das A.T., Zhou X., Metz S.W., Vink M.A., Berkhout B. Selecting the optimal Tet-On system for doxycycline-inducible gene expression in transiently transfected and stably transduced mammalian cells. Biotechnol J. 2016;11:71–79. doi: 10.1002/biot.201500236. [DOI] [PubMed] [Google Scholar]
- 4.Das A.T., Tenenbaum L., Berkhout B. Tet-On systems for doxycycline-inducible gene expression. Curr Gene Ther. 2016;16:156–167. doi: 10.2174/1566523216666160524144041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 6.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Werstuck G., Green M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 8.Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 9.Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 10.Ausländer S., Fussenegger M. Synthetic RNA-based switches for mammalian gene expression control. Curr Opin Biotechnol. 2017;48:54–60. doi: 10.1016/j.copbio.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Yen L., Svendsen J., Lee J.S., Gray J.T., Magnier M., Baba T., D’Amato R.J., Mulligan R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 12.Yen L.S., Magnier M., Weissleder R., Stockwell B.R., Mulligan R.C. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high-throughput screening of chemical libraries. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura Y., Zhou L., Miu A., Yokobayashi Y. Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth Biol. 2013;2:684–689. doi: 10.1021/sb400037a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.Y., Jensen M.C., Smolke C.D. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beilstein K., Wittmann A., Grez M., Suess B. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth Biol. 2015;4:526–534. doi: 10.1021/sb500270h. [DOI] [PubMed] [Google Scholar]
- 16.Ausländer S., Ketzer P., Hartig J.S. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst. 2010;6:807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 17•.Wurmthaler L.A., Klauser B., Hartig J.S. Highly motif- and organism-dependent effects of naturally occurring hammerhead ribozyme sequences on gene expression. RNA Biol. 2018;15:231–241. doi: 10.1080/15476286.2017.1397870. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work surveyed a panel of ribozymes in various cellular and genetic contexts and showed that ribozyme activity is highly context dependent. The results imply that care must be taken when translating screening results obtained in nonmammalian systems to mammalian cells.
- 18.Zhong G., Wang H., Bailey C.C., Gao G., Farzan M. Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. eLife. 2016;5 doi: 10.7554/eLife.18858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Dohno C., Kimura M., Nakatani K. Restoration of ribozyme tertiary contact and function by using a molecular glue for RNA. Angew Chem Int Ed Engl. 2018;57:506–510. doi: 10.1002/anie.201709041. [DOI] [PubMed] [Google Scholar]; The authors used a ‘molecular glue’ to restore a tertiary contact in a hammerhead ribozyme, presenting an alternative to canonical aptamers for controlling gene expression in mammalian cells.
- 20.Dohno C., Kohyama I., Hong C., Nakatani K. Naphthyridine tetramer with a pre-organized structure for 1:1 binding to a CGG/CGG sequence. Nucleic Acids Res. 2012;40:2771–2781. doi: 10.1093/nar/gkr1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Kobori S., Nomura Y., Miu A., Yokobayashi Y. High-throughput assay and engineering of self-cleaving ribozymes by sequencing. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv265. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deep sequencing was used to exhaustively assay 512 aptazyme mutants without screening or selection in vitro. The aptazymes were then shown to function as riboswitches in mammalian cells.
- 22.Kobori S., Yokobayashi Y. High-throughput mutational analysis of a twister ribozyme. Angew Chem Int Ed. 2016;55:10354–10357. doi: 10.1002/anie.201605470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokobayashi Y. Applications of high-throughput sequencing to analyze and engineer ribozymes. Methods. 2019;161:41–45. doi: 10.1016/j.ymeth.2019.1002.1001. [DOI] [PubMed] [Google Scholar]
- 24.Kobori S., Takahashi K., Yokobayashi Y. Deep sequencing analysis of aptazyme variants based on a pistol ribozyme. ACS Synth Biol. 2017;6:1283–1288. doi: 10.1021/acssynbio.7b00057. [DOI] [PubMed] [Google Scholar]
- 25.Kobori S., Yokobayashi Y. Analyzing and tuning ribozyme activity by deep sequencing to modulate gene expression level in mammalian cells. ACS Synth Biol. 2018;7:371–376. doi: 10.1021/acssynbio.7b00367. [DOI] [PubMed] [Google Scholar]
- 26.Nomura Y., Chien H.-C., Yokobayashi Y. Direct screening for ribozyme activity in mammalian cells. Chem Commun. 2017;53:12540–12543. doi: 10.1039/c7cc07815c. [DOI] [PubMed] [Google Scholar]
- 27.Caliskan N., Peske F., Rodnina M.V. Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem Sci. 2015;40:265–274. doi: 10.1016/j.tibs.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu H.-T., Lin Y.-H., Chang K.-Y. Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells. Nucleic Acids Res. 2014;42:14070–14082. doi: 10.1093/nar/gku1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y.H., Chang K.Y. Rational design of a synthetic mammalian riboswitch as a ligand-responsive -1 ribosomal frame-shifting stimulator. Nucleic Acids Res. 2016;44:9005–9015. doi: 10.1093/nar/gkw718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto S., Caliskan N., Rodnina M.V., Murata A., Nakatani K. Small synthetic molecule-stabilized RNA pseudoknot as an activator for -1 ribosomal frameshifting. Nucleic Acids Res. 2018;46:8079–8089. doi: 10.1093/nar/gky689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An C.I., Trinh V.B., Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beisel C.L., Chen Y.Y., Culler S.J., Hoff K.G., Smolke C.D. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2011;39:2981–2994. doi: 10.1093/nar/gkq954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong R.S., Chen Y.Y., Smolke C.D. Regulation of T cell proliferation with drug-responsive microRNA switches. Nucleic Acids Res. 2018;46:1541–1552. doi: 10.1093/nar/gkx1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunsicker A., Steber M., Mayer G., Meitert J., Klotzsche M., Blind M., Hillen W., Berens C., Suess B. An RNA aptamer that induces transcription. Chem Biol. 2009;16:173–180. doi: 10.1016/j.chembiol.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 35•.Atanasov J., Groher F., Weigand J.E., Suess B. Design and implementation of a synthetic pre-miR switch for controlling miRNA biogenesis in mammals. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx858. [DOI] [PMC free article] [PubMed] [Google Scholar]; Although this system includes a protein component (TetR), tight control of natural microRNA maturation was demonstrated using a widely used small molecule (doxycycline) in mammalian cells.
- 36.Tuleuova N., An C.I., Ramanculov E., Revzin A., Yokobayashi Y. Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochem Biophys Res Commun. 2008;376:169–173. doi: 10.1016/j.bbrc.2008.08.112. [DOI] [PubMed] [Google Scholar]
- 37.Kumar D., An C.I., Yokobayashi Y. Conditional RNA interference mediated by allosteric ribozyme. J Am Chem Soc. 2009;131:13906–13907. doi: 10.1021/ja905596t. [DOI] [PubMed] [Google Scholar]
- 38.Cheng H., Zhang Y., Wang H., Sun N., Liu M., Chen H., Pei R. Regulation of MAP4K4 gene expression by RNA interference through an engineered theophylline-dependent hepatitis delta virus ribozyme switch. Mol Biosyst. 2016;12:3370–3376. doi: 10.1039/c6mb00540c. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Wang J., Cheng H., Sun N., Liu M., Wu Z., Pei R. Inducible Bcl-2 gene RNA interference mediated by aptamer-integrated HDV ribozyme switch. Integr Biol. 2017;9:619–626. doi: 10.1039/c7ib00029d. [DOI] [PubMed] [Google Scholar]
- 40.Vogel M., Weigand J.E., Kluge B., Grez M., Suess B. A small, portable RNA device for the control of exon skipping in mammalian cells. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Mou H.H., Zhong G.C., Gardner M.R., Wang H.M., Wang Y.W., Cheng D.C., Farzan M. Conditional regulation of gene expression by ligand-induced occlusion of a MicroRNA target sequence. Mol Ther. 2018;26:1277–1286. doi: 10.1016/j.ymthe.2018.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; A new mode of engineered RNA-based gene regulation in mammalian cells was demonstrated by controlling the accessibility of a miRNA target site by aptamer–ligand interaction.